Abstract
Signal transducer and activator of transcription-3 (STAT-3) is one of six members of a family of transcription factors. It was discovered almost 15 years ago as an acute-phase response factor. This factor has now been associated with inflammation, cellular transformation, survival, proliferation, invasion, angiogenesis, and metastasis of cancer. Various types of carcinogens, radiation, viruses, growth factors, oncogenes, and inflammatory cytokines have been found to activate STAT-3. STAT-3 is constitutively active in most tumor cells but not in normal cells. Phosphorylation of STAT-3 at tyrosine 705 leads to its dimerization, nuclear translocation, DNA binding, and gene transcription. The phosphorylation of STAT-3 at serine 727 may regulate its activity negatively or positively. STAT-3 regulates the expression of genes that mediate survival (survivin, bcl-xl, mcl-1, cellular FLICE-like inhibitory protein), proliferation (c-fos, c-myc, cyclin D1), invasion (matrix metalloproteinase-2), and angiogenesis (vascular endothelial growth factor). STAT-3 activation has also been associated with both chemoresistance and radioresistance. STAT-3 mediates these effects through its collaboration with various other transcription factors, including nuclear factor-κB, hypoxia-inducible factor-1, and peroxisome proliferator activated receptor-γ. Because of its critical role in tumorigenesis, inhibitors of this factor’s activation are being sought for both prevention and therapy of cancer. This has led to identification of small peptides, oligonucleotides, and small molecules as potential STAT-3 inhibitors. Several of these small molecules are chemo-preventive agents derived from plants. This review discusses the intimate relationship between STAT-3, inflammation, and cancer in more detail.
Keywords: STAT-3, inflammation, cancer, chemoresistance
Introduction
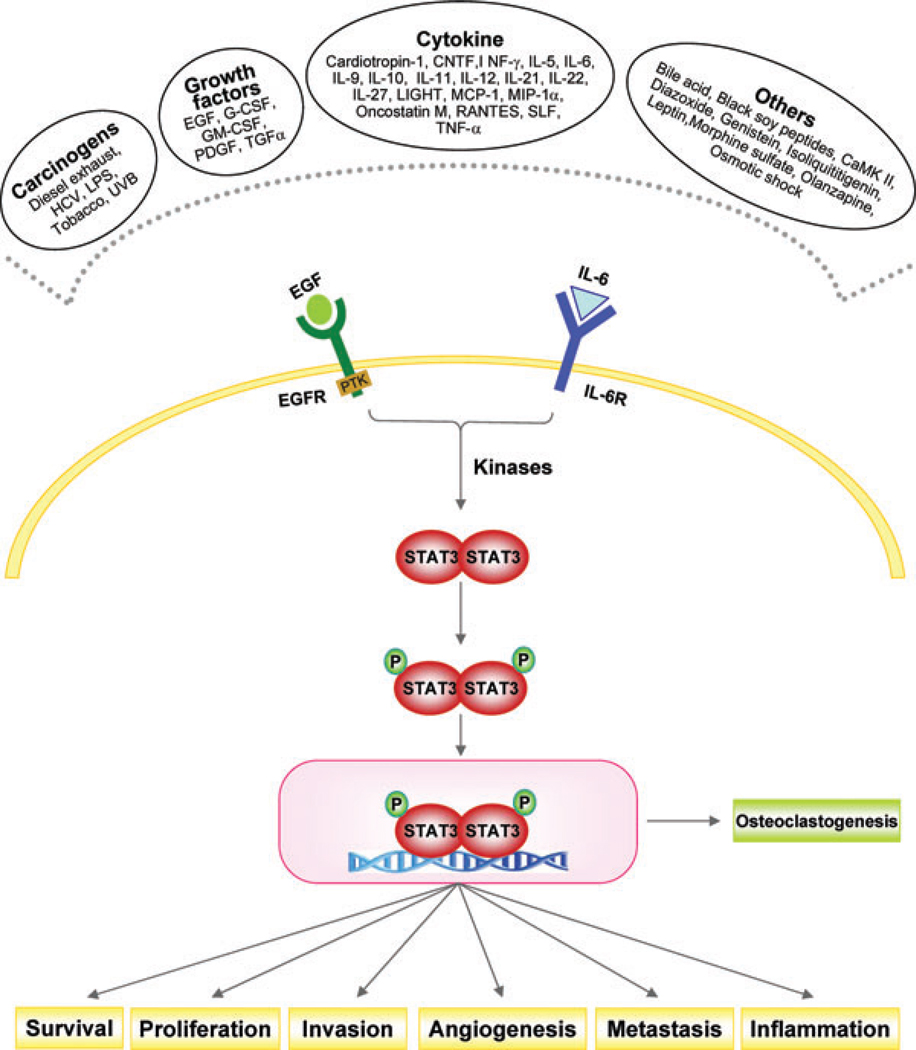
Signal transducer and activator of transcription (STAT)-3 is one of the members of a family of transcription factors. It was first identified in 1994 as a DNA-binding factor that selectively binds to the IL-6-responsive element in the promoter of acute-phase genes from IL-6-stimulated hepatocytes.1 STAT-3 was also independently identified as a DNA-binding protein in response to epidermal growth factor.2 The gene that encodes STAT-3 is located on chromosome 17q21. The 92-kDa protein is 770 amino acids long with sequential N-terminal coiled-coil domain, DNA-binding domain, a linker, SH2 domain, and C-terminal transactivation domain. The latter contains a tyrosine residue at position 705 and a serine residue at position 727, which undergoes phosphorylation when activated (Fig. 1).
Figure 1.
Signaling pathway leading to signal transducer and activator of transcription (STAT)-3 activation (see text for definitions of abbreviations).
STAT-3 is activated by many cytokines and growth factors, including epidermal growth factor,3 platelet-derived growth factor,4 and IL-61 as well as by oncogenic proteins, such as Src5 and Ras6 (Table 1). In addition numerous carcinogens, such as cigarette smoke7 and tumor promoters, have been identified that can activate STAT-3.8,9
TABLE 1.
Activators and Inhibitors of Signal Transducer and Activator of Transcription (STAT)-3
| Activators | Others | • Sodium salicylate64 |
| Cytokines | • Bile acids34 | • Statin65 |
| • Cardiotrophin-110 | • Black soy peptides35 | • T4021466 |
| • CNTF11 | • CaMKIIg36 | • UCN-0167 |
| • IFN-γ12 | • Diazoxide37 | • WP-103468 |
| • IL-513 | • Genistein38 | Natural |
| • IL-62 | • Isoliquiritigenin39 | • Caffeic acid49 |
| • IL-914 | • Leptin40 | • Capsaicin69 |
| • IL-1015 | • Morphine sulfate41 | • CDDO-Me70 |
| • IL-1116 | • Olanzapine42 | • Chalcone71 |
| • IL-1217 | • Osmotic shock43 | • Cucurbitacin72 |
| • IL-2118 | Inhibitors | • Curcumin73 |
| • IL-2219 | Synthetic | • Deoxytetrangiomycin74 |
| • IL-2720 | • AG 49044 | • EGCG75 |
| • LIGHT21 | • Atiprimod45 | • Emodin76 |
| • MCP-122 | • Auranofin46 | • Flavopiridol77 |
| • MIP-1α23 | • Aurothiomalate47 | • Galiellalactone78 |
| • Oncostatin M24 | • BMS-35482548 | • Genistein79 |
| • RANTES23 | • CADPE49 | • Guggulsterone80 |
| • SLF25 | • Stattic50 | • Indirubin81 |
| • TNF-α26 | • Dobesilate51 | • Magnolol82 |
| Growth Factors | • Ethanol52 | • Parthenolide83 |
| • EGF2 | • NCX-401653 | • Piceatannol84 |
| • G-CSF27 | • Nelfinavir54 | • Resveratrol85 |
| • GM-CSF13 | • PDP55 | • Silibinin86 |
| • PDGF4 | • Platinum compounds56 | • Ursolic acid87 |
| • TGF-α28 | • PS-34157 | Others |
| Carcinogens | • Y(p)LPQTV58 | • EKB56988 |
| • Diesel exhaust particles29 | • R11577759 | • GQ-ODN66 |
| • HCV30 | • S31-M200160 | • Retinoic acid89 |
| • LPS31 | • S-3I-20161 | • Rituximab90 |
| • Tobacco32 | • SCH6633662 | • STA-2191 |
| • UVB33 | • SD-102963 | • TKS 05079 |
CaMKII, calmodulin-dependent protein kinase II; CAPDE, caffeic acid phenyl ethyl ester; CDDO-Me, methyl-2-cyano-3,12-dioxooleana-1,9-dien-28-oate; CNTF, ciliary neurotrophic factor; EGCG, (−)-epigallocatechin-3-gallate; EGF, epidermal growth factor; G-CSF, granulocyte colony-stimulating factor; GM-CSF, granulocyte-macrophage colony-stimulating factor; GQ-ODN, G-quartet oligodeoxynucleotide; HCV, hepatitis C virus; IFN-γ, interferon gamma; IL, interleukin; LIGHT, lymphotoxin homologue, inducible and competes with HSV glycoprotein D for HveA and is expressed on T lymphocytes; LPS, lipopolysaccharide; MCP-1, monocyte chemoattractant protein 1; MIP-1α, macrophage inflammatory protein-1-α; PDGF, platelet-derived growth factor; PDP, phosphododecapeptides; PS-341, bortezomib; RANTES, regulated on activation normal T cell expressed and presumably secreted; SLF, steel factor; TGF-α, transforming growth factor α; TKS 050, N-{4-[(3,4-dichloro-6-fluoro-phenyl)amino]-quinazoline-6-yl}-2-chloroacetamide; TNF-α, tumor necrosis factor α; UVB, ultraviolet B radiation.
The activation of STAT-3 is regulated by phosphorylation of tyrosine 705 by receptor and nonreceptor protein tyrosine kinases (Table 2). These include epidermal growth factor receptor (EGFR) kinase,92 Src,5 Janus-activated kinases (JAK),93–95 and extracellular signal-regulated kinase (ERK).96 The phosphorylation of STAT-3 in the cytoplasm leads to its dimerization, translocation into the nucleus, and DNA binding; as a result genes that regulate cell proliferation, differentiation, and apoptosis are expressed. In addition, numerous serine kinases have been implicated in the phosphorylation of STAT-3 at serine 727. These include protein kinase C (PKC),97 mitogen-activated protein kinases, and CDK5.98 PKC-ε has been shown to interact with STAT-3 directly and phosphorylate serine 727,99 which maximizes its transcriptional activity.100,101
TABLE 2.
Intracellular Modulators of STAT-3 Activity
| Protein kinases |
| • JAK1 and JAK2 phosphorylate STAT-3.2,93,94 |
| • Src kinase family of kinases (Src, Hck, Lyn, Fyn, and Fgr) binds STAT-3 and induces tyrosine phosphorylation.5,102 |
| • Bcr-Abl induces tyrosine phosphorylation and DNA-binding activity of STAT-3.103 |
| • JAK3 binds CD40 and phopshorylates STAT-3.104 |
| • ERK binds STAT-3 and phosphorylates at ser 727, which negatively regulates Tyr 702 phosphorylation.105 |
| • Fes binds and induces tyrosine phosphorylation of STAT-3.106,107 |
| • PKCδ binds STAT-3, induces Ser727 phosphorylation, and inhibits its activity.97,108,109 |
| • p94 (fer) binds and causes the tyrosine phosphorylation of STAT-3.110 |
| • mTOR or p70 S6 kinase activated by PI3K/AKT mediates the serine 727 phosphorylation of STAT-3 by CNTF.101 |
| • IRAK1 binds and causes the Ser 727 phosphorylation of STAT-3.111 |
| • CDK9 binds STAT-3 and leads to human γ-fibrinogen gene expression.77,112 |
| • ZIP kinase binds STAT-3 in the nucleus and enhances its transcriptional activity via phosphorylation of Ser727.113 |
| • TGF-β-activated kinase 1 (TAK1) binds STAT-3 and increases ser 727 phosphorylation.114 |
| • NIK binds STAT-3 in response to LIGHT.21 |
| • Protein kinase C-ε binds and phosphorylates STAT-3 at Ser727.99 |
| • Bruton’s tyrosine kinase binds STAT-3 and prevents its activation.115 |
| • Peptidyl-prolyl cis/trans isomerase 1 (Pin1) binds STAT-3, induces ser 727 phosphorylation, and enhances its activity.116 |
| Protein phosphatases |
| • SHP-1 and SHP-2 prevents the phoshporylation of STAT-3 by negatively regulating JAK activity.117 |
| • LMW-PTPase is negative regulator of STAT-3 phosphorylation.118 |
| • Protein phosphatase 2 A translocates to nucleus and dephosphorylates STAT-3 at serine 727.119,120 |
| • Protein-tyrosine phosphatase D1 activates STAT-3 through interaction with Etk.121 |
| • Cytosolic isoform of PTPε inhibits STAT-3 activation by inactivating JAKs.122 |
| • CD45 directly dephosphorylates and binds to JAKs.123 |
| • PTEN is a negative regulator of STAT-3 activation through inhibition of PI3K/AKT pathway.124,125 |
| • PTP1 B is a negative regulator of JAK2.126 |
| • T-cell PTP inhibits IL-6-induced tyrosine phosphorylation and activation of STAT-3.127 |
| • LMW-DSP2 regulates IL-6/LIF-mediated signaling through dephosphorylation of Jaks and STAT-3.128 |
| • Receptor protein tyrosine phosphatase T dephosphorylates STAT-3.129 |
| Viral proteins |
| • EZI, a novel nuclear zinc finger protein, binds nuclear STAT-3 and augments its activity.130 |
| • Kaposi sarcoma-associated viral cyclin K binds nuclear STAT-3 and inhibits its activity.131 |
| • Herpes virus saimiri subgroup A strain 11 (STP-A11) binds STAT-3 and increases its transcriptional activity.132 |
| • Kaposi’s sarcoma-associated herpes virus (KSHV)-encoded latency-associated nuclear antigen (LANA) binds STAT-3 and enhances its transcriptional activity.133 |
| Others |
| • c-Jun binds STAT-3 β and enhances promoter activity.134 |
| • IFNAR-1 chain binds to STAT-3 directly and enhances its activity.135 |
| • SOCS family of proteins binds JAK and negatively regulates JAK-STAT pathway.136 |
| • Glucocorticoid receptor binds to STAT-3 and forms a transactivating/signaling complex.137 |
| • Protein inhibitor of activated STAT (PIAS)-3, an E3 ligase, binds STAT-3 and blocks its DNA-binding and gene expression.105 |
| • SSI-1 [(for STAT-induced STAT inhibitor/SOCS)-1] binds Jak2 and Tyk2, and negatively regulates STAT-3 activation.138 |
| • STAT-3 binds NF-κB p65 and inactivates its transcriptional activity.139–141 |
| • CREB-binding protein (CBP)/P300 binds STAT-3, induces acetylation at Lys 685, and induces dimerization.142–145 |
| • STAT-3-interacting protein, StIP1, binds STAT-3 and prevents nuclear translocation.146 |
| • EGFR binds STAT-3 and stimulates its activity.92,147 |
| • IL-2 receptor β chain binds to STAT-3.148 |
| • Cyclin-dependent kinase inhibitor p21 binds to STAT-3 and inhibits its activity.149 |
| • Cyclin D1 binds to nuclear STAT-3 and inhibits its activity.150 |
| • Co-activator NcoA/SRC1a binds to STAT-3 through 752–761 region, phosphorylates ser 727, and enhances its activity.151,152 |
| • Grb2 binds STAT-3 and inhibits its interaction with EGFR.55,147,153 |
| • Rac1 GTPase binds and stimulates STAT-3 phosphorylation at tyrosine and serine residues.154 |
| • MyoD binds STAT-3 and inhibits its activity.155 |
| • Promyelocytic leukemia protein (PML) binds STAT-3 and inhibits cell proliferation.156 |
| • GRIM-19 binds STAT-3 and negatively regulates its activity.157 |
| • Prothymosine-α binds STAT-3 and enhances its activity.158 |
| • PPARγ binds STAT-3 and inactivates its transcriptional activity.159 |
| • Osteospecific transcription factor Runx2 binds nuclear STAT-3 and inhibits its activity.160 |
| • Proline-, glutamic acid-, and leucine-rich protein-1 (PELP1) is a novel estrogen receptor co-activator that binds to STAT-3 in the nucleus and increases its activity.161 |
| • PAX3-FKHR binds STAT-3 and its transcriptional activity.162 |
| • A Ras homologue member I (ARHI) binds STAT-3 and inhibits its activity.163 |
| • Histone deacetylase (HDAC)-1 binds STAT-3 and induces deacetylation.142 |
| • SP1 binds STAT-3 and increases its transcriptional activity.164 |
| • HIF-1α and p300 binds to STAT-3 and leads to VEGF expression.165 |
| • Importin α5 and α7 bind to STAT-3 and enhance its activity.166 |
| • G-CSFR phosphotyrosine peptide ligands pY704VLQ and pY744LRC bind to STAT-3.167 |
| • Duplin, a negative regulator of Wnt signaling, binds STAT-3 and inhibits its DNA-binding activity.168 |
| • Daxx binds STAT-3 in the nucleus and downregulates its transcriptional activation.133 |
| • Unphosphorylated STAT-3 accumulates in response to IL-6 and activates transcription by binding to NF-κB.169 |
| • Nescient helix-loop-helix 2 interacts with STAT-3 to regulate transcription of prohormone convertase 1/3.170 |
| • Kruppel-associated box zinc-finger protein (KAP) 1 binds STAT-3 and regulates its transcriptional activity.171 |
| • Binder of ADP-ribosylation factor-like two (BART) augments STAT-3 activity by keeping it in the nucleus.172 |
CDK9, cyclin-dependent kinase 9; CNTF, ciliary neurotrophic factor; DSP, dual specificity phosphatase; EGFR, epidermal growth factor receptor; G-CSFR, granulocyte colony-stimulating factor receptor; HIF-1α, hypoxia-inducible factor 1 subunit α; IFNAR-1, interferon (α, β, and ω) receptor 1; IRAK1, interleukin-1 receptor-associated kinase 1; JAK, Janus kinase; LMW, low molecular weight; PKC, protein kinase C; mTOR, mammalian target of rapamycin; PI3K, phosphatidylinositol 3-kinase; NIK, NF-κB-inducing kinase; PPARγ, peroxisome proliferator-activated receptor γ; PTEN, phosphatase and tensin homologue; PTP, protein tyrosine phosphatase; SOCS, suppressors of cytokine signaling; STAT, signal transducers and activators of transcription; EZI, endothelial cell-derived zinc finger protein; ZIP, leucine zipper kinase; GRIM-19, gene associated with retinoid-IFN-induced mortality-19; MyoD, myogenic differentiation; PAX3-FKHR, paired box 3-FKHR-Forkhead (Drosophila) homolog 1 (rhabdomyosarcoma).
Besides phosphorylation on tyrosine and serine sites within the carboxyl-terminal region, STAT-3 is also acetylated on a single lysine residue 685 by histone acetyltransferase p300142 (Table 2). STAT-3 acetylation is reversible by type I histone deacetylase (HDAC). The acetylation of STAT-3 was found to be critical for it to form stable dimers, which are required for cytokine-stimulated DNA binding and transcriptional regulation.
STAT-3 activation is negatively regulated through numerous mechanisms (Table 2). These involve the suppressors of cytokine signaling (SOCS),136 protein inhibitor of activated STAT (PIAS),105 protein phosphatases,173 and ubiquitination-dependent proteosomal degradation174 (Table 2). The SOCS proteins were shown to bind to the JAK activation loop as pseudosubstrate inhibitors through their SH2 domain, thereby blocking subsequent signaling that requires phosphorylation and activation of STAT-3.175 Eight SOCS proteins with similar structures have been identified so far.176 SOCS-3 negatively regulates the gp130-STAT-3 pathway in mouse skin wound healing, suggesting that STAT-3 is required for wound healing.177 Different SOCS family members, however, have distinct mechanisms of inhibition of JAK/STAT signaling. Recently, the involvement of SOCS-1 in carcinogenesis has been reported.178 Frequent hypermethylation in CpG islands of the functional SOCS-3 promoter correlates with its transcription silencing in cell lines (lung cancer, breast cancer, and mesothelioma) and primary lung cancer tissue samples.179–181 Restoration of SOCS-3 in lung cancer cells where SOCS-3 was silenced by methylation resulted in the downregulation of active STAT-3, induction of apoptosis, and growth suppression.181 Methylation silencing of SOCS-3 is an important mechanism of constitutive activation of the STAT-3 pathway in cancer pathogenesis.178,179
In contrast to SOCS, the PIAS-3 are nuclear factors that are able to interact with phos-phorylated STAT-3 and block transcription.105 Smad4 has been shown to suppress the tyrosine phosphorylation of STAT-3 in pancreatic cancer cells.182
STAT-3 activation is also negatively regulated by various protein tyrosine phosphatases, including CD45,123 PTEN,124 SHP-1,183 SHP-2184 (Table 2).
The ubiquitin-proteasome pathway is responsible for selective degradation of shortlived cellular proteins and is critical for the regulation of many cellular processes. STAT-3 has been shown to undergo degradation through this pathway.174,185,186 In IL-6-dependent KT-3 cells, the transcription factor was found to be conjugated by exogenous biotinylated Ub and degraded in a proteasome-dependent manner.174 Additionally, caspases have been found to directly cleave STAT-3.187 STAT-3 cleavage was accompanied by reductions in STAT-3– DNA binding, STAT-3-driven reporter protein (luciferase) activity, and the expression of selected STAT-3-dependent genes and correlated with increased sensitivity to apoptotic stimuli.
The ablation of STAT-3 leads to embryonic lethality,188 and tissue-specific ablation of the transcription factor yields important defects in hepatocytes,189 macrophages,190 keratinocytes,191 and thymic or mammary epithelial cells.192
STAT-3 is an oncogenic protein that is constitutively activated in many human cancers. For instance, in 30–60% of primary breast cancers, STAT-3 is constitutively active.193 Constitutive activation of STAT-3 has also been reported in several other primary cancers, in tumor cell lines, and in many oncogene-transformed cells. Inactivation of STAT-3 in most of these cell lines leads to inhibition of cell proliferation. The critical role of this factor in cancer is indicated by the fact that β4 integrin actively contributes to the initiation, growth, and invasion of ErbB2-induced mammary tumors in transgenic mice by promoting the activation of STAT-3.194 The evidence below shows that STAT-3 activation is intimately connected with all aspects of tumorigenesis.
STAT-3 Activation Mediates Inflammation
Several lines of evidence suggest that STAT-3 is a mediator of inflammation.195 First, STAT-3 was initially discovered as an acute-phase response protein, thus suggesting its link to inflammation. Second, most proinflammatory agents have been shown to activate this factor. IL-6 is a major mediator of inflammation and mediates its effects through the activation of the STAT-3 pathway.2 Similarly, tumor promoters, lipopolysaccharides, and cigarette smoke can activate the STAT-3 pathway.7,196 Third, the DNA binding for STAT-3 in the promoter of acute-phase proteins was found to compete with that of NF-κB, another pro-inflammatory transcription factor.139 Fourth, STAT-3 has been shown to regulate NF-κB recruitment to the IL-12p40 promoter in dendritic cells.197 Fifth, recently it was shown that IL-11 and its glycoprotein 130 (gp130) receptor in inflammation-associated gastric epithelial cell oncogenic transformation is mediated by and dependent on increased activation of STAT-3.198 Sixth, in some cell types IL-6-induced STAT-3 activation has been shown to be dependent on cyclooxygenase 2, a pro-inflammatory enzyme.199 All this evidence supports the role of the STAT-3 pathway in inflammation.
STAT-3 Activation Can Transform Cells
The transformation of cells by various oncogenes, protein tyrosine kinases, and viruses accompanies the activation of STAT-3.200 Yu et al. showed that transformation of cells by src protein kinase is mediated through the activation of STAT-3.5,201 Similarly the transformation of T cells by human T-cell lymphotropic virus I was also mediated through the activation of STAT-3.95 Hepatitis C virus core protein has also been shown to transform the cells through activation of STAT-3.30 The STAT-3 activation is induced by v-Fps; by polyoma virus middle T antigen, which activates Src family kinases; and by v-Sis, which acts as a ligand for the platelet-derived growth factor receptor.92 STAT-3 signaling is also required for hepatocyte growth factor/scatter factor-Met-mediated tumorigenes.202 Moreover, a constitutively activated form of STAT-3 induces cell transformation, growth in soft agar, and tumors in nude mice, further confirming the importance of the activated form detected in tumors. Thus, STAT-3 is considered an oncogene.203
STAT-3 Activation Can Suppress Apoptosis
Evidence indicates that oncogenic transformation of the cells leads to activation of STAT-3, which then provides the survival signal. Conditional inactivation of STAT-3 shows that it has proapoptotic functions during mammary gland involution.192 In most cells, STAT-3 activation can suppress apoptosis. These effects are mediated through the expression of various cell survival gene products that are regulated by STAT-3. These include bcl-xl,204,205 bcl-2,206 survivin,207 Mcl-1,208 and cIAP2.209 Additionally, most tumor cells that exhibit constitutive activation of STAT-3 also express these cell survival gene products.210,211 Thus, suppression of STAT-3 activation can suppress the expression of all these cell survival gene products and potentiate apoptosis.212 The downregulation of STAT-3 also leads to expression of fas protein, which can promote apoptosis.213
STAT-3 Activation Can Lead to Cellular Proliferation
STAT-3 activation has also been linked with proliferation of tumor cells. This effect of STAT-3 is mediated through its ability to induce the expression of cyclin D1.214 STAT-3 has also been shown to upregulate the expression of several growth-promoting genes, such as myc215 and pim-1.216 The proapoptotic factors, such as Fas, are downmodulated by STAT-3 activation.213 There are other reports, however, which suggest that this transcription factor can activate the expression of the cell cycle inhibitor p21(waf1),217 suggesting that STAT-3 can also block cell cycle progression and prevent abnormal cell proliferation. During cellular transformation, however, phosphatidylinositol 3-kinase/Akt pathway was found to inhibit the transcriptional activation of the p21(waf1) gene by STAT-3 proteins without altering the regulation of the myc promoter.218
STAT-3 Activation Can Mediate Cellular Invasion
Numerous reports indicate STAT-3 activation plays a major role in tumor cell invasion, and inhibition of STAT-3 reduces invasion.182,219–221 STAT-3 activation regulates the expression of matrix metalloproteinase (MMP)-2 and MMP-1, which then mediate tumor invasion and metastasis.222,223 STAT-3 upregulates the transcription of MMP-2 through direct interaction with the MMP-2 promoter. Furthermore, blockade of activated STAT-3 in highly metastatic cells significantly suppresses the invasiveness of the tumor cells, inhibits tumor growth, and prevents metastasis in nude mice. Also, overexpression of phosphorylated STAT-3 correlates with the invasion and metastasis of cutaneous squamous cell carcinoma.224 STAT-3, however, is also known to upregulate tissue inhibitors of metalloproteinase (TIMP)-1, a cytokine known to block metalloproteinases and decrease invasiveness in certain cancer cell types.225 STAT-3 also controls the expression of the MUC1 gene, which can mediate tumor invasion.226 Thus, STAT-3 mediates tumor invasion through numerous mechanisms.
STAT-3 Activation Can Mediate Angiogenesis and Metastasis
One of the first pieces of evidence to suggest that STAT-3 is linked with angiogenesis was from granulocyte-macrophage colony-stimulating factor-induced angiogenetic activity in chick chorioallantoic membrane.227 It was shown that constitutive STAT-3 activity upregulates vascular endothelial growth factor (VEGF) expression and tumor angiogenesis.228 Most tumor cells that exhibit constitutively active STAT-3 also express VEGF.229,230 Thus, downmodulation of STAT-3 activation can suppress the expression of VEGF and inhibit angiogenesis. Indeed, Li et al. found an inhibition of growth and metastasis of human hepatocellular carcinoma by antisense oligonucleotide targeting of STAT-3.231 The metastasis of human melanoma to brain was also linked to STAT-3 activation.232 Besides VEGF, it has been shown that TWIST, another mediator of tumor metastasis, is regulated by STAT-3.233
Role of STAT-3 in Carcinogenesis
STAT-3 can mediate both the tumor initiation and the tumor promotion phases of carcinogenesis. While deletion of STAT-3 suppressed skin carcinogenesis,9 forced expression enhanced malignant progression.234,235 STAT-3-deficient mice were completely resistant to skin tumor development when 9,10-dimethylbenz-[a-]anthracene was used as the initiator and 12-O-tetradecanolyphorbol-13-acetate as the promoter.9 Activation of STAT-3 has also been shown to be an early event in tobacco-chewing-mediated oral carcinogenesis in human samples.32 The activation of STAT-3 has also been linked with hepatocarcino-genesis, as suggested by SOCS-3 deficiency in mice.236
Role of STAT-3 in Chemoresistance and Radioresistance
Activation of STAT-3 has been linked with resistance of tumor cells to chemotherapeutic agents.80,237 Work from our laboratory and others have shown constitutive activation of STAT-3 in multiple myeloma can mediate chemoresistance.238 This is mediated through the upregulation of antiapoptotic gene products regulated by STAT-3, as shown in metastatic breast cancer cells.239 Thus, downmodulation of STAT-3 can overcome chemoresistance.85
The resistance of tumor cells to γ radiation has also been associated with STAT-3 activation. STAT-3-deleted B cells are highly susceptible to irradiation.240 In vivo experiments with gene-targeted mice showed that IL-6 and, to a lesser extent, IL-10 are the relevant stimuli that combine with B-cell receptor (BCR) ligands to promote B-1 cell radioresistance. STAT-3 promotes cell survival in response to selected growth factors and is activated by combined BCR cross-linking and IL-6 (IL-10). Importantly, STAT-3−/− B-1 cells become susceptible to irradiation, indicating that STAT-3 activation by BCR accounts for the inherent radioresistance of peritoneal B-1 B cells. Kim et al. showed that DN-STAT-3 and DN-survivin together result in the greatest radiosensitization of MDA-MB-231 (breast cancer cell line), decreasing angiogenesis, and cell survival.241
Chemopreventive Agents Inhibit STAT-3 Activation
Several natural agents known to be chemo-preventive are quite effective in suppressing STAT-3 activation (Table 1). These include curcumin,73,242 resveratrol,85 ursolic acid,87 guggulsterone,80 capsaicin,69 cucurbitacin,72 indirubin,81 flavopiridol,77 epigallocatechin gallate,75 CDDO-Me (methyl-2-cyano-3,12-dioxooleana-1,9-dien-28-oate),70 emodin,76 silibinin,86 and chalcone.71 How these phytochemicals suppress STAT-3 activation has been investigated. For instance, guggulsterone, ursolic acid, and capsaicin have been shown to transcriptionally upregulate the expression of SHP2, which leads to inactivation of STAT-3.69,80,87 Other mechanisms have also been described. For instance, luteolin has been shown to promote the degradation in STAT-3 in human hepatoma cells.243 Indirubin was found to inhibit STAT-3 activation through inhibition of Src kinase activity.81
Conclusions
This description, overall, shows that STAT-3 activation plays a very intimate role in tumorigenesis. Inhibitors of the STAT-3 pathway thus have enormous potential in the treatment of cancer. Whether STAT-3 can be exploited as a prognostic factor in human cancers remains to be examined.
Acknowledgments
This work was supported by a grant from the Clayton Foundation for Research (to B.B.A.). A core grant from the National Institutes of Health (CA-16 672), and a program project grant from National Institute of Health (NIH CA-124787-01A2).
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Akira S, et al. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 2.Zhong Z, Wen Z, Darnell JE., Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 3.Cao X, et al. Activation and association of Stat3 with Src in v-Src-transformed cell lines. Mol. Cell Biol. 1996;16:1595–1603. doi: 10.1128/mcb.16.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vignais ML, et al. Platelet-derived growth factor induces phosphorylation of multiple JAK family kinases and STAT proteins. Mol. Cell Biol. 1996;16:1759–1769. doi: 10.1128/mcb.16.4.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu CL, et al. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science. 1995;269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 6.Giordano V, et al. Shc mediates IL-6 signaling by interacting with gp130 and Jak2 kinase. J. Immunol. 1997;158:4097–4103. [PubMed] [Google Scholar]
- 7.Arredondo J, et al. Receptor-mediated tobacco toxicity: cooperation of the Ras/Raf-1/MEK1/ERK and JAK-2/STAT-3 pathways downstream of alpha7 nicotinic receptor in oral ker-atinocytes. FASEB J. 2006;20:2093–2101. doi: 10.1096/fj.06-6191com. [DOI] [PubMed] [Google Scholar]
- 8.Tharappel JC, et al. Regulation of cell proliferation, apoptosis, and transcription factor activities during the promotion of liver carcinogenesis by polychlorinated biphenyls. Toxicol. Appl. Pharmacol. 2002;179:172–184. doi: 10.1006/taap.2001.9360. [DOI] [PubMed] [Google Scholar]
- 9.Chan KS, et al. Disruption of Stat3 reveals a critical role in both the initiation and the promotion stages of epithelial carcinogenesis. J. Clin. Invest. 2004;114:720–728. doi: 10.1172/JCI21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurdi M, Booz GW. Can the protective actions of JAK-STAT in the heart be exploited therapeutically? Parsing the regulation of interleukin-6-type cytokine signaling. J. Cardiovasc. Pharmacol. 2007;50:126–141. doi: 10.1097/FJC.0b013e318068dd49. [DOI] [PubMed] [Google Scholar]
- 11.Ji JZ, et al. CNTF promotes survival of retinal ganglion cells after induction of ocular hypertension in rats: the possible involvement of STAT3 pathway. Eur. J. Neurosci. 2004;19:265–272. doi: 10.1111/j.0953-816x.2003.03107.x. [DOI] [PubMed] [Google Scholar]
- 12.Kordula T, et al. Activation of signal transducer and activator of transcription-3 (Stat3) expression by interferon-gamma and interleukin-6 in hepatoma cells. Biochem. Biophys. Res. Commun. 1995;216:999–1005. doi: 10.1006/bbrc.1995.2719. [DOI] [PubMed] [Google Scholar]
- 13.Stout BA, et al. IL-5 and granulocyte-macrophage colony-stimulating factor activate STAT3 and STAT5 and promote Pim-1 and cyclin D3 protein expression in human eosinophils. J. Immunol. 2004;173:6409–6417. doi: 10.4049/jimmunol.173.10.6409. [DOI] [PubMed] [Google Scholar]
- 14.Demoulin JB, et al. A single tyrosine of the interleukin-9 (IL-9) receptor is required for STAT activation, antiapoptotic activity, and growth regulation by IL-9. Mol. Cell Biol. 1996;16:4710–4716. doi: 10.1128/mcb.16.9.4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams L, et al. Signal transducer and activator of transcription 3 is the dominant mediator of the anti-inflammatory effects of IL-10 in human macrophages. J. Immunol. 2004;172:567–576. doi: 10.4049/jimmunol.172.1.567. [DOI] [PubMed] [Google Scholar]
- 16.Yanagisawa M, et al. Astrocyte differentiation of fetal neuroepithelial cells by interleukin-11 via activation of a common cytokine signal transducer, gp130, and a transcription factor, STAT3. J. Neurochem. 2000;74:1498–1504. doi: 10.1046/j.1471-4159.2000.0741498.x. [DOI] [PubMed] [Google Scholar]
- 17.Jacobson NG, et al. Interleukin 12 signaling in T helper type 1 (Th1) cells involves tyrosine phosphorylation of signal transducer and activator of transcription (Stat)3 and Stat4. J. Exp. Med. 1995;181:1755–1762. doi: 10.1084/jem.181.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei L, et al. IL-21 is produced by Th17 cells and drives IL-17 production in a STAT3-dependent manner. J. Biol. Chem. 2007;282:34605–34610. doi: 10.1074/jbc.M705100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radaeva S, et al. Interleukin 22 (IL-22) plays a protective role in T cell-mediated murine hepatitis: IL-22 is a survival factor for hepatocytes via STAT3 activation. Hepatology. 2004;39:1332–1342. doi: 10.1002/hep.20184. [DOI] [PubMed] [Google Scholar]
- 20.Lang R. Tuning of macrophage responses by Stat3-inducing cytokines: molecular mechanisms and consequences in infection. Immunobiology. 2005;210:63–76. doi: 10.1016/j.imbio.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 21.Nadiminty N, et al. LIGHT, a member of the TNF superfamily, activates Stat3 mediated by NIK pathway. Biochem. Biophys. Res. Commun. 2007;359:379–384. doi: 10.1016/j.bbrc.2007.05.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellado M, et al. The chemokine monocyte chemotactic protein 1 triggers Janus kinase 2 activation and tyrosine phosphorylation of the CCR2B receptor. J. Immunol. 1998;161:805–813. [PubMed] [Google Scholar]
- 23.Wong M, Fish EN. RANTES and MIP-1alpha activate stats in T cells. J. Biol. Chem. 1998;273:309–314. doi: 10.1074/jbc.273.1.309. [DOI] [PubMed] [Google Scholar]
- 24.Hintzen C, et al. Box 2 region of the oncostatin m receptor determines specificity for recruitment of Janus kinases and STAT5 activation. J. Biol . Chem. 2008;283:19465–19477. doi: 10.1074/jbc.M710157200. [DOI] [PubMed] [Google Scholar]
- 25.Gotoh A, et al. Steel factor induces serine phosphorylation of Stat3 in human growth factor-dependent myeloid cell lines. Blood. 1996;88:138–145. [PubMed] [Google Scholar]
- 26.Miscia S, et al. Tumor necrosis factor alpha (TNF-alpha) activates Jak1/Stat3-Stat5B signaling through TNFR-1 in human B cells. Cell Growth Differ. 2002;13:13–18. [PubMed] [Google Scholar]
- 27.Nishiki S, et al. Selective activation of STAT3 in human monocytes stimulated by G-CSF: implication in inhibition of LPS-induced TNF-alpha production. Am. J. Physiol. Cell Physiol. 2004;286:C1302–C1311. doi: 10.1152/ajpcell.00387.2003. [DOI] [PubMed] [Google Scholar]
- 28.Grandis JR, et al. Requirement of Stat3 but not Stat1 activation for epidermal growth factor receptor- mediated cell growth in vitro. J. Clin. Invest. 1998;102:1385–1392. doi: 10.1172/JCI3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao D, et al. Diesel exhaust particulate-induced activation of Stat3 requires activities of EGFR and Src in airway epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2007;292:L422–L429. doi: 10.1152/ajplung.00204.2006. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida T, et al. Activation of STAT3 by the hepatitis C virus core protein leads to cellular transformation. J. Exp. Med. 2002;196:641–653. doi: 10.1084/jem.20012127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carl VS, et al. Role of endogenous IL-10 in LPS-induced STAT3 activation and IL-1 receptor antagonist gene expression. J. Leukoc. Biol. 2004;76:735–742. doi: 10.1189/jlb.1003526. [DOI] [PubMed] [Google Scholar]
- 32.Nagpal JK, Mishra R, Das BR. Activation of Stat-3 as one of the early events in tobacco chewing-mediated oral carcinogenesis. Cancer. 2002;94:2393–2400. doi: 10.1002/cncr.10499. [DOI] [PubMed] [Google Scholar]
- 33.Ahsan H, Aziz MH, Ahmad N. Ultraviolet B exposure activates Stat3 signaling via phosphorylation at tyrosine705 in skin of SKH1 hairless mouse: a target for the management of skin cancer? Biochem. Biophys. Res. Commun. 2005;333:241–246. doi: 10.1016/j.bbrc.2005.05.106. [DOI] [PubMed] [Google Scholar]
- 34.Dvorak K, et al. Activation of the interleukin-6/STAT3 antiapoptotic pathway in esophageal cells by bile acids and low pH: relevance to barrett’s esophagus. Clin. Cancer Res. 2007;13:5305–5313. doi: 10.1158/1078-0432.CCR-07-0483. [DOI] [PubMed] [Google Scholar]
- 35.Jang EH, et al. Novel black soy peptides with antiobesity effects: activation of leptin-like signaling and AMP-activated protein kinase. Int. J. Obes. (Lond.) 2008;32:1161–1170. doi: 10.1038/ijo.2008.60. [DOI] [PubMed] [Google Scholar]
- 36.Si J, Collins SJ. Activated Ca2+/ calmodulin-dependent protein kinase IIgamma is a critical regulator of myeloid leukemia cell proliferation. Cancer Res. 2008;68:3733–3742. doi: 10.1158/0008-5472.CAN-07-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsieh YJ, et al. Cardioplegia and diazoxide modulate STAT3 activation and DNA binding. Ann. Thorac. Surg. 2007;84:1272–1278. doi: 10.1016/j.athoracsur.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chau MN, et al. Physiologically achievable concentrations of genistein enhance telomerase activity in prostate cancer cells via the activation of STAT3. Carcinogenesis. 2007;28:2282–2290. doi: 10.1093/carcin/bgm148. [DOI] [PubMed] [Google Scholar]
- 39.An W, Yang J, Ao Y. Metallothionein mediates cardioprotection of isoliquiritigenin against ischemia-reperfusion through JAK2/STAT3 activation. Acta Pharmacol. Sin. 2006;27:1431–1437. doi: 10.1111/j.1745-7254.2006.00419.x. [DOI] [PubMed] [Google Scholar]
- 40.Vaisse C, et al. Leptin activation of Stat3 in the hypothalamus of wild-type and ob/ob mice but not db/db mice. Nat. Genet. 1996;14:95–97. doi: 10.1038/ng0996-95. [DOI] [PubMed] [Google Scholar]
- 41.Weber ML, et al. Morphine induces mesangial cell proliferation and glomerulopathy via kappa-opioid receptors. Am. J. Physiol. Renal. Physiol. 2008;294:F1388–F1397. doi: 10.1152/ajprenal.00389.2007. [DOI] [PubMed] [Google Scholar]
- 42.Muma NA, et al. Chronic olanzapine activates the Stat3 signal transduction pathway and alters expression of components of the 5-HT2A receptor signaling system in rat frontal cortex. Neuropharmacology. 2007;53:552–562. doi: 10.1016/j.neuropharm.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gatsios P, et al. Activation of the Janus kinase/signal transducer and activator of transcription pathway by osmotic shock. J. Biol. Chem. 1998;273:22962–22968. doi: 10.1074/jbc.273.36.22962. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen M, et al. Constitutive activation of a slowly migrating isoform of Stat3 in mycosis fungoides: tyrphostin AG490 inhibits Stat3 activation and growth of mycosis fungoides tumor cell lines. Proc. Natl. Acad. Sci. USA. 1997;94:6764–6769. doi: 10.1073/pnas.94.13.6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amit-Vazina M, et al. Atiprimod blocks STAT3 phosphorylation and induces apoptosis in multiple myeloma cells. Br. J. Cancer. 2005;93:70–80. doi: 10.1038/sj.bjc.6602637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim NH, et al. Auranofin blocks interleukin-6 signalling by inhibiting phosphorylation of JAK1 and STAT3. Immunology. 2007;122:607–614. doi: 10.1111/j.1365-2567.2007.02679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stuhlmeier KM. The anti-rheumatic gold salt aurothiomalate suppresses interleukin-1beta-induced hyaluronan accumulation by blocking HAS1 transcription and by acting as a COX-2 transcriptional repressor. J. Biol. Chem. 2007;282:2250–2258. doi: 10.1074/jbc.M605011200. [DOI] [PubMed] [Google Scholar]
- 48.Chen Z, et al. Potent inhibition of platelet-derived growth factor-induced responses in vascular smooth muscle cells by BMS-354825 (dasatinib) Mol. Pharmacol. 2006;69:1527–1533. doi: 10.1124/mol.105.020172. [DOI] [PubMed] [Google Scholar]
- 49.Jung JE, et al. Caffeic acid and its synthetic derivative CADPE suppress tumor angiogenesis by blocking STAT3-mediated VEGF expression in human renal carcinoma cells. Carcinogenesis. 2007;28:1780–1787. doi: 10.1093/carcin/bgm130. [DOI] [PubMed] [Google Scholar]
- 50.Schust J, et al. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem. Biol. 2006;13:1235–1242. doi: 10.1016/j.chembiol.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 51.Cuevas P, et al. Dobesilate inhibits the activation of signal transducer and activator of transcription 3, and the expression of cyclin D1 and bcl-XL in glioma cells. Neurol. Res. 2006;28:127–130. doi: 10.1179/016164106X97982. [DOI] [PubMed] [Google Scholar]
- 52.Chen J, Kunos G, Gao B. Ethanol rapidly inhibits IL-6-activated STAT3 and C/EBP mRNA expression in freshly isolated rat hepatocytes. FEBS Lett. 1999;457:162–168. doi: 10.1016/s0014-5793(99)01031-5. [DOI] [PubMed] [Google Scholar]
- 53.Selvendiran K, et al. NCX-4016, a nitro-derivative of aspirin, inhibits EGFR and STAT3 signaling and modulates Bcl-2 proteins in cisplatin-resistant human ovarian cancer cells and xenografts. Cell Cycle. 2008;7:81–88. doi: 10.4161/cc.7.1.5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Y, et al. HIV-1 protease inhibitor induces growth arrest and apoptosis of human prostate cancer LNCaP cells in vitro and in vivo in conjunction with blockade of androgen receptor STAT3 and AKT signaling. Cancer Sci. 2005;96:425–433. doi: 10.1111/j.1349-7006.2005.00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shao H, et al. Identification and characterization of signal transducer and activator of transcription 3 recruitment sites within the epidermal growth factor receptor. Cancer Res. 2003;63:3923–3930. [PubMed] [Google Scholar]
- 56.Littlefield SL, et al. Synthesis, characterization and Stat3 inhibitory properties of the prototypical platinum(IV) anticancer drug, [PtCl3(NO2)(NH3)2] (CPA-7) Inorg. Chem. 2008;47:2798–2804. doi: 10.1021/ic702057q. [DOI] [PubMed] [Google Scholar]
- 57.Hideshima T, et al. Proteasome inhibitor PS-341 abrogates IL-6 triggered signaling cascades via caspase-dependent downregulation of gp130 in multiple myeloma. Oncogene. 2003;22:8386–8393. doi: 10.1038/sj.onc.1207170. [DOI] [PubMed] [Google Scholar]
- 58.Ren Z, et al. Identification of a high-affinity phosphopeptide inhibitor of Stat3. Bioorg. Med. Chem. Lett. 2003;13:633–636. doi: 10.1016/s0960-894x(02)01050-8. [DOI] [PubMed] [Google Scholar]
- 59.Venkatasubbarao K, Choudary A, Freeman JW. Farnesyl transferase inhibitor (R115777)-induced inhibition of STAT3(Tyr705) phosphorylation in human pancreatic cancer cell lines require extracellular signal-regulated kinases. Cancer Res. 2005;65:2861–2871. doi: 10.1158/0008-5472.CAN-04-2396. [DOI] [PubMed] [Google Scholar]
- 60.Siddiquee KA, et al. An oxazole-based small-molecule Stat3 inhibitor modulates Stat3 stability and processing and induces antitumor cell effects. ACS Chem. Biol. 2007;2:787–798. doi: 10.1021/cb7001973. [DOI] [PubMed] [Google Scholar]
- 61.Siddiquee K, et al. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc. Natl. Acad. Sci. USA. 2007;104:7391–7396. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dowlati A, et al. SCH66336, inhibitor of protein farnesylation, blocks signal transducer and activators of transcription 3 signaling in lung cancer and interacts with a small molecule inhibitor of epidermal growth factor receptor/human epidermal growth factor receptor 2. Anticancer Drugs. 2008;19:9–16. doi: 10.1097/CAD.0b013e3282f1a908. [DOI] [PubMed] [Google Scholar]
- 63.Duan Z, et al. SD-1029 inhibits signal transducer and activator of transcription 3 nuclear translocation. Clin. Cancer Res. 2006;12:6844–6852. doi: 10.1158/1078-0432.CCR-06-1330. [DOI] [PubMed] [Google Scholar]
- 64.Wang Z, Jiang B, Brecher P. Selective inhibition of STAT3 phosphorylation by sodium salicylate in cardiac fibroblasts. Biochem. Pharmacol. 2002;63:1197–1207. doi: 10.1016/s0006-2952(02)00853-5. [DOI] [PubMed] [Google Scholar]
- 65.Arnaud C, et al. Statins reduce interleukin-6-induced C-reactive protein in human hepatocytes: new evidence for direct antiinflammatory effects of statins. Arterioscler Thromb. Vasc. Biol. 2005;25:1231–1236. doi: 10.1161/01.ATV.0000163840.63685.0c. [DOI] [PubMed] [Google Scholar]
- 66.Jing N, et al. Targeting Stat3 with G-quartet oligodeoxynucleotides in human cancer cells. DNA Cell Biol. 2003;22:685–696. doi: 10.1089/104454903770946665. [DOI] [PubMed] [Google Scholar]
- 67.Bhonde MR, et al. The broad-range cyclin-dependent kinase inhibitor UCN-01 induces apoptosis in colon carcinoma cells through transcriptional suppression of the Bcl-x(L) protein. Oncogene. 2005;24:148–156. doi: 10.1038/sj.onc.1207842. [DOI] [PubMed] [Google Scholar]
- 68.Faderl S, et al. WP-1034, a novel JAK-STAT inhibitor, with proapoptotic and antileukemic activity in acute myeloid leukemia (AML) Anticancer Res. 2005;25:1841–1850. [PubMed] [Google Scholar]
- 69.Bhutani M, et al. Capsaicin is a novel blocker of constitutive and interleukin-6-inducible STAT3 activation. Clin. Cancer Res. 2007;13:3024–3032. doi: 10.1158/1078-0432.CCR-06-2575. [DOI] [PubMed] [Google Scholar]
- 70.Ling X, et al. The novel triterpenoid C-28 methyl ester of 2-cyano-3, 12-dioxoolen-1, 9-dien-28-oic acid inhibits metastatic murine breast tumor growth through inactivation of STAT3 signaling. Cancer Res. 2007;67:4210–4218. doi: 10.1158/0008-5472.CAN-06-3629. [DOI] [PubMed] [Google Scholar]
- 71.Liu YC, et al. Chalcone inhibits the activation of NF-kappaB and STAT3 in endothelial cells via endogenous electrophile. Life Sci. 2007;80:1420–1430. doi: 10.1016/j.lfs.2006.12.040. [DOI] [PubMed] [Google Scholar]
- 72.van Kester MS, et al. Cucurbitacin I inhibits Stat3 and induces apoptosis in Sezary cells. J. Invest . Dermatol. 2008;128:1691–1695. doi: 10.1038/sj.jid.5701246. [DOI] [PubMed] [Google Scholar]
- 73.Bharti AC, Donato N, Aggarwal BB. Curcumin (diferuloylmethane) inhibits constitutive and IL-6-inducible STAT3 phosphorylation in human multiple myeloma cells. J. Immunol. 2003;171:3863–3871. doi: 10.4049/jimmunol.171.7.3863. [DOI] [PubMed] [Google Scholar]
- 74.Deng J, Grande F, Neamati N. Small molecule inhibitors of Stat3 signaling pathway. Curr. Cancer Drug Targets. 2007;7:91–107. doi: 10.2174/156800907780006922. [DOI] [PubMed] [Google Scholar]
- 75.Shimizu M, et al. EGCG inhibits activation of the insulin-like growth factor (IGF)/IGF-1 receptor axis in human hepatocellular carcinoma cells. Cancer Lett. 2007;206:10–18. doi: 10.1016/j.canlet.2007.11.026. [DOI] [PubMed] [Google Scholar]
- 76.Muto A, et al. Emodin has a cytotoxic activity against human multiple myeloma as a Janus-activated kinase 2 inhibitor. Mol. Cancer Ther. 2007;6:987–994. doi: 10.1158/1535-7163.MCT-06-0605. [DOI] [PubMed] [Google Scholar]
- 77.Hou T, Ray S, Brasier AR. The functional role of an interleukin 6-inducible CDK9.STAT3 complex in human gamma-fibrinogen gene expression. J. Biol. Chem. 2007;282:37091–37102. doi: 10.1074/jbc.M706458200. [DOI] [PubMed] [Google Scholar]
- 78.Weidler M, et al. Inhibition of interleukin-6 signaling by galiellalactone. FEBS Lett. 2000;484:1–6. doi: 10.1016/s0014-5793(00)02115-3. [DOI] [PubMed] [Google Scholar]
- 79.Shushan A, et al. Inhibition of leiomyoma cell proliferation in vitro by genistein and the protein tyrosine kinase inhibitor TKS050. Fertil. Steril. 2007;87:127–135. doi: 10.1016/j.fertnstert.2006.05.056. [DOI] [PubMed] [Google Scholar]
- 80.Ahn KS, et al. Guggulsterone, a farnesoid X receptor antagonist, inhibits constitutive and inducible STAT3 activation through induction of a protein tyrosine phosphatase SHP-1. Cancer Res. 2008;68:4406–4415. doi: 10.1158/0008-5472.CAN-07-6696. [DOI] [PubMed] [Google Scholar]
- 81.Nam S, et al. Indirubin derivatives inhibit Stat3 signaling and induce apoptosis in human cancer cells. Proc. Natl. Acad. Sci. USA. 2005;102:5998–6003. doi: 10.1073/pnas.0409467102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen SC, et al. Herbal remedy magnolol suppresses IL-6-induced STAT3 activation and gene expression in endothelial cells. Br. J. Pharmacol. 2006;148:226–232. doi: 10.1038/sj.bjp.0706647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sobota R, et al. Parthenolide inhibits activation of signal transducers and activators of transcription (STATs) induced by cytokines of the IL-6 family. Biochem. Biophys. Res. Commun. 2000;267:329–333. doi: 10.1006/bbrc.1999.1948. [DOI] [PubMed] [Google Scholar]
- 84.Su L, David M. Distinct mechanisms of STAT phosphorylation via the interferon-alpha/beta receptor. Selective inhibition of STAT3 and STAT5 by piceatannol. J. Biol. Chem. 2000;275:12661–12666. doi: 10.1074/jbc.275.17.12661. [DOI] [PubMed] [Google Scholar]
- 85.Bhardwaj A, et al. Resveratrol inhibits proliferation, induces apoptosis, and overcomes chemoresistance through down-regulation of STAT3 and nuclear factor-kappaB-regulated antiapoptotic and cell survival gene products in human multiple myeloma cells. Blood. 2007;109:2293–2302. doi: 10.1182/blood-2006-02-003988. [DOI] [PubMed] [Google Scholar]
- 86.Agarwal C, et al. Silibinin inhibits constitutive activation of Stat3, and causes caspase activation and apoptotic death of human prostate carcinoma DU145 cells. Carcinogenesis. 2007;28:1463–1470. doi: 10.1093/carcin/bgm042. [DOI] [PubMed] [Google Scholar]
- 87.Pathak AK, et al. Ursolic acid inhibits STAT3 activation pathway leading to suppression of proliferation and chemosensitization of human multiple myeloma cells. Mol Cancer Res. 2007;5:943–955. doi: 10.1158/1541-7786.MCR-06-0348. [DOI] [PubMed] [Google Scholar]
- 88.Nunes M, Shi C, Greenberger LM. Phosphorylation of extracellular signal-regulated kinase 1 and 2, protein kinase B, and signal transducer and activator of transcription 3 are differently inhibited by an epidermal growth factor receptor inhibitor, EKB-569, in tumor cells and normal human keratinocytes. Mol. Cancer Ther. 2004;3:21–27. [PubMed] [Google Scholar]
- 89.Tighe AP, Gudas LJ. Retinoic acid inhibits leukemia inhibitory factor signaling pathways in mouse embryonic stem cells. J. Cell Physiol. 2004;198:223–229. doi: 10.1002/jcp.10424. [DOI] [PubMed] [Google Scholar]
- 90.Alas S, Bonavida B. Rituximab inactivates signal transducer and activation of transcription 3 (STAT3) activity in B-non-Hodgkin’s lymphoma through inhibition of the interleukin 10 autocrine/paracrine loop and results in down-regulation of Bcl-2 and sensitization to cytotoxic drugs. Cancer Res. 2001;61:5137–5144. [PubMed] [Google Scholar]
- 91.Song H, et al. A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. Proc. Natl. Acad. Sci. USA. 2005;102:4700–4705. doi: 10.1073/pnas.0409894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Garcia R, et al. Constitutive activation of Stat3 in fibroblasts transformed by diverse onco-proteins and in breast carcinoma cells. Cell Growth Differ. 1997;8:1267–1276. [PubMed] [Google Scholar]
- 93.Lutticken C, et al. Association of transcription factor APRF and protein kinase Jak1 with the interleukin-6 signal transducer gp130. Science. 1994;263:89–92. doi: 10.1126/science.8272872. [DOI] [PubMed] [Google Scholar]
- 94.Tian SS, et al. Multiple signaling pathways induced by granulocyte colony-stimulating factor involving activation of JAKs, STAT5, and/or STAT3 are required for regulation of three distinct classes of immediate early genes. Blood. 1996;88:4435–4444. [PubMed] [Google Scholar]
- 95.Migone TS, et al. Constitutively activated Jak-STAT pathway in T cells transformed with HTLV-I. Science. 1995;269:79–81. doi: 10.1126/science.7604283. [DOI] [PubMed] [Google Scholar]
- 96.Megeney LA, et al. bFGF and LIF signaling activates STAT3 in proliferating myoblasts. Dev. Genet. 1996;19:139–145. doi: 10.1002/(SICI)1520-6408(1996)19:2<139::AID-DVG5>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 97.Jain N, et al. Protein kinase C delta associates with and phosphorylates Stat3 in an interleukin-6-dependent manner. J. Biol. Chem. 1999;274:24392–24400. doi: 10.1074/jbc.274.34.24392. [DOI] [PubMed] [Google Scholar]
- 98.Fu AK, et al. Cyclin-dependent kinase 5 phosphorylates signal transducer and activator of transcription 3 and regulates its transcriptional activity. Proc. Natl. Acad. Sci. USA. 2004;101:6728–6733. doi: 10.1073/pnas.0307606100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aziz MH, et al. Protein kinase Cepsilon interacts with signal transducers and activators of transcription 3 (Stat3), phosphorylates Stat3Ser727, and regulates its constitutive activation in prostate cancer. Cancer Res. 2007;67:8828–8838. doi: 10.1158/0008-5472.CAN-07-1604. [DOI] [PubMed] [Google Scholar]
- 100.Wen Z, Darnell JE., Jr Mapping of Stat3 serine phosphorylation to a single residue (727) and evidence that serine phosphorylation has no influence on DNA binding of Stat1 and Stat3. Nucleic Acids Res. 1997;25:2062–2067. doi: 10.1093/nar/25.11.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yokogami K, et al. Serine phosphorylation and maximal activation of STAT3 during CNTF signaling is mediated by the rapamycin target mTOR. Curr. Biol. 2000;10:47–50. doi: 10.1016/s0960-9822(99)00268-7. [DOI] [PubMed] [Google Scholar]
- 102.Schreiner SJ, Schiavone AP, Smithgall TE. Activation of STAT3 by the Src family kinase Hck requires a functional SH3 domain. J. Biol. Chem. 2002;277:45680–45687. doi: 10.1074/jbc.M204255200. [DOI] [PubMed] [Google Scholar]
- 103.Ilaria RL, Jr, Van Etten RA. P210 and P190(BCR/ABL) induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J. Biol. Chem. 1996;271:31704–31710. doi: 10.1074/jbc.271.49.31704. [DOI] [PubMed] [Google Scholar]
- 104.Hanissian SH, Geha RS. Jak3 is associated with CD40 and is critical for CD40 induction of gene expression in B cells. Immunity. 1997;6:379–387. doi: 10.1016/s1074-7613(00)80281-2. [DOI] [PubMed] [Google Scholar]
- 105.Chung CD, et al. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 1997;278:1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- 106.Nelson KL, et al. Activation of STAT3 by the c-Fes protein-tyrosine kinase. J. Biol. Chem. 1998;273:7072–7077. doi: 10.1074/jbc.273.12.7072. [DOI] [PubMed] [Google Scholar]
- 107.Park WY, et al. c-Fes tyrosine kinase binds to and activates STAT3 after granulocyte-macrophage colony-stimulating factor stimulation. Cancer Lett. 1998;129:29–37. doi: 10.1016/s0304-3835(98)00077-9. [DOI] [PubMed] [Google Scholar]
- 108.Bhattacharjee A, et al. Monocyte 15-lipoxygenase expression is regulated by a novel cytosolic signaling complex with protein kinase C delta and tyrosine-phosphorylated Stat3. J. Immunol. 2006;177:3771–3781. doi: 10.4049/jimmunol.177.6.3771. [DOI] [PubMed] [Google Scholar]
- 109.Novotny-Diermayr V, et al. Protein kinase C delta associates with the interleukin-6 receptor sub-unit glycoprotein (gp) 130 via Stat3 and enhances Stat3-gp130 interaction. J. Biol. Chem. 2002;277:49134–49142. doi: 10.1074/jbc.M206727200. [DOI] [PubMed] [Google Scholar]
- 110.Priel-Halachmi S, et al. FER kinase activation of Stat3 is determined by the N-terminal sequence. J. Biol. Chem. 2000;275:28902–28910. doi: 10.1074/jbc.M003402200. [DOI] [PubMed] [Google Scholar]
- 111.Huang Y, et al. IRAK1 serves as a novel regulator essential for lipopolysaccharide-induced interleukin-10 gene expression. J. Biol. Chem. 2004;279:51697–51703. doi: 10.1074/jbc.M410369200. [DOI] [PubMed] [Google Scholar]
- 112.Giraud S, et al. Implication of BRG1 and cdk9 in the STAT3-mediated activation of the p21waf1 gene. Oncogene. 2004;23:7391–7398. doi: 10.1038/sj.onc.1207972. [DOI] [PubMed] [Google Scholar]
- 113.Sato N, et al. Physical and functional interactions between STAT3 and ZIP kinase. Int. Immunol. 2005;17:1543–1552. doi: 10.1093/intimm/dxh331. [DOI] [PubMed] [Google Scholar]
- 114.Kojima H, et al. STAT3 regulates Nemolike kinase by mediating its interaction with IL-6-stimulated TGFbeta-activated kinase 1 for STAT3 Ser-727 phosphorylation. Proc. Natl. Acad. Sci. USA. 2005;102:4524–4529. doi: 10.1073/pnas.0500679102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Uckun F, Ozer Z, Vassilev A. Bruton’s tyrosine kinase prevents activation of the anti-apoptotic transcription factor STAT3 and promotes apoptosis in neoplastic B-cells and B-cell precursors exposed to oxidative stress. Br. J. Haematol. 2007;136:574–589. doi: 10.1111/j.1365-2141.2006.06468.x. [DOI] [PubMed] [Google Scholar]
- 116.Lufei C, et al. Pin1 is required for the Ser727 phosphorylation-dependent Stat3 activity. Oncogene. 2007;26:7656–7664. doi: 10.1038/sj.onc.1210567. [DOI] [PubMed] [Google Scholar]
- 117.Kim H, et al. Protein tyrosine phosphatase 2 (SHP-2) moderates signaling by gp130 but is not required for the induction of acute-phase plasma protein genes in hepatic cells. Mol. Cell Biol. 1998;18:1525–1533. doi: 10.1128/mcb.18.3.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chiarugi P, et al. The Src and signal transducers and activators of transcription pathways as specific targets for low molecular weight phosphotyrosine-protein phosphatase in platelet-derived growth factor signaling. J. Biol. Chem. 1998;273:6776–6785. doi: 10.1074/jbc.273.12.6776. [DOI] [PubMed] [Google Scholar]
- 119.Liang H, et al. Regulation of angiotensin II-induced phosphorylation of STAT3 in vascular smooth muscle cells. J. Biol. Chem. 1999;274:19846–19851. doi: 10.1074/jbc.274.28.19846. [DOI] [PubMed] [Google Scholar]
- 120.Woetmann A, et al. Inhibition of protein phosphatase 2A induces serine/threonine phosphorylation, subcellular redistribution, and functional inhibition of STAT3. Proc. Natl. Acad. Sci. USA. 1999;96:10620–10625. doi: 10.1073/pnas.96.19.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jui HY, et al. Protein-tyrosine phosphatase D1, a potential regulator and effector for Tec family kinases. J. Biol. Chem. 2000;275:41124–41132. doi: 10.1074/jbc.M007772200. [DOI] [PubMed] [Google Scholar]
- 122.Tanuma N, et al. Protein tyrosine phosphatase epsilonC selectively inhibits interleukin-6-and interleukin- 10-induced JAK-STAT signaling. Blood. 2001;98:3030–3034. doi: 10.1182/blood.v98.10.3030. [DOI] [PubMed] [Google Scholar]
- 123.Irie-Sasaki J, et al. CD45 is a JAK phosphatase and negatively regulates cytokine receptor signalling. Nature. 2001;409:349–354. doi: 10.1038/35053086. [DOI] [PubMed] [Google Scholar]
- 124.Sun S, Steinberg BM. PTEN is a negative regulator of STAT3 activation in human papillomavirus-infected cells. J. Gen . Virol. 2002;83:1651–1658. doi: 10.1099/0022-1317-83-7-1651. [DOI] [PubMed] [Google Scholar]
- 125.Zhou J, et al. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc. Natl. Acad. Sci. USA. 2007;104:16158–16163. doi: 10.1073/pnas.0702596104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cheng A, et al. Attenuation of leptin action and regulation of obesity by protein tyrosine phosphatase 1B. Dev. Cell. 2002;2:497–503. doi: 10.1016/s1534-5807(02)00149-1. [DOI] [PubMed] [Google Scholar]
- 127.Yamamoto T, et al. The nuclear isoform of protein-tyrosine phosphatase TC-PTP regulates interleukin-6-mediated signaling pathway through STAT3 dephosphorylation. Biochem. Biophys. Res. Commun. 2002;297:811–817. doi: 10.1016/s0006-291x(02)02291-x. [DOI] [PubMed] [Google Scholar]
- 128.Sekine Y, et al. Modulation of TLR4 signaling by a novel adaptor protein signal-transducing adaptor protein-2 in macrophages. J. Immunol. 2006;176:380–389. doi: 10.4049/jimmunol.176.1.380. [DOI] [PubMed] [Google Scholar]
- 129.Zhang X, et al. Identification of STAT3 as a substrate of receptor protein tyrosine phosphatase T. Proc. Natl. Acad. Sci. USA. 2007;104:4060–4064. doi: 10.1073/pnas.0611665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nakayama K, Kim KW, Miyajima A. A novel nuclear zinc finger protein EZI enhances nuclear retention and transactivation of STAT3. EMBO J. 2002;21:6174–6184. doi: 10.1093/emboj/cdf596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lundquist A, et al. Kaposi sarcoma-associated viral cyclin K overrides cell growth inhibition mediated by oncostatin M through STAT3 inhibition. Blood. 2003;101:4070–4077. doi: 10.1182/blood-2002-07-1994. [DOI] [PubMed] [Google Scholar]
- 132.Chung YH, et al. Activation of Stat3 transcription factor by Herpesvirus saimiri STP-A on-coprotein. J. Virol. 2004;78:6489–6497. doi: 10.1128/JVI.78.12.6489-6497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Muromoto R, et al. Physical and functional interactions between Daxx and STAT3. Oncogene. 2006;25:2131–21316. doi: 10.1038/sj.onc.1209235. [DOI] [PubMed] [Google Scholar]
- 134.Schaefer TS, Sanders LK, Nathans D. Cooperative transcriptional activity of Jun and Stat3 beta, a short form of Stat3. Proc. Natl. Acad. Sci. USA. 1995;92:9097–9101. doi: 10.1073/pnas.92.20.9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yang CH, et al. Direct association of STAT3 with the IFNAR-1 chain of the human type I interferon receptor. J. Biol. Chem. 1996;271:8057–8061. doi: 10.1074/jbc.271.14.8057. [DOI] [PubMed] [Google Scholar]
- 136.Starr R, et al. A family of cytokine-inducible inhibitors of signalling. Nature. 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 137.Zhang Z, et al. STAT3 acts as a co-activator of glucocorticoid receptor signaling. J. Biol. Chem. 1997;272:30607–30610. doi: 10.1074/jbc.272.49.30607. [DOI] [PubMed] [Google Scholar]
- 138.Naka T, et al. Structure and function of a new STAT-induced STAT inhibitor. Nature. 1997;387:924–929. doi: 10.1038/43219. [DOI] [PubMed] [Google Scholar]
- 139.Zhang Z, Fuller GM. The competitive binding of STAT3 and NF-kappaB on an overlapping DNA binding site. Biochem. Biophys. Res. Commun. 1997;237:90–94. doi: 10.1006/bbrc.1997.7082. [DOI] [PubMed] [Google Scholar]
- 140.Yu Z, Zhang W, Kone BC. Signal transducers and activators of transcription 3 (STAT3) inhibits transcription of the inducible nitric oxide synthase gene by interacting with nuclear factor kappaB. Biochem. J. 2002;367:97–105. doi: 10.1042/BJ20020588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yoshida Y, et al. Interleukin 1 activates STAT3/nuclear factor-kappaB cross-talk via a unique TRAF6- and p65-dependent mechanism. J. Biol. Chem. 2004;279:1768–1776. doi: 10.1074/jbc.M311498200. [DOI] [PubMed] [Google Scholar]
- 142.Yuan ZL, et al. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 143.Nakashima K, et al. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- 144.Paulson M, et al. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J. Biol. Chem. 1999;274:25343–25349. doi: 10.1074/jbc.274.36.25343. [DOI] [PubMed] [Google Scholar]
- 145.Wang R, Cherukuri P, Luo J. Activation of Stat3 sequence-specific DNA binding and transcription by p300/CREB-binding protein-mediated acetylation. J. Biol. Chem. 2005;280:11528–11534. doi: 10.1074/jbc.M413930200. [DOI] [PubMed] [Google Scholar]
- 146.Collum RG, et al. A Stat3-interacting protein (StIP1) regulates cytokine signal transduction. Proc. Natl. Acad. Sci. USA. 2000;97:10120–10125. doi: 10.1073/pnas.170192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lo HW, et al. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell. 2005;7:575–589. doi: 10.1016/j.ccr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 148.Delespine-Carmagnat M, Bouvier G, Bertoglio J. Association of STAT1, STAT3 and STAT5 proteins with the IL-2 receptor involves different subdomains of the IL-2 receptor beta chain. Eur. J. Immunol. 2000;30:59–68. doi: 10.1002/1521-4141(200001)30:1<59::AID-IMMU59>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 149.Coqueret O, Gascan H. Functional interaction of STAT3 transcription factor with the cell cycle inhibitor p21WAF1/CIP1/SDI1. J. Biol . Chem. 2000;275:18794–18800. doi: 10.1074/jbc.M001601200. [DOI] [PubMed] [Google Scholar]
- 150.Bienvenu F, Gascan H, Coqueret O. Cyclin D1 represses STAT3 activation through a Cdk4-independent mechanism. J. Biol. Chem. 2001;276:16840–16847. doi: 10.1074/jbc.M100795200. [DOI] [PubMed] [Google Scholar]
- 151.Giraud S, et al. Functional interaction of STAT3 transcription factor with the coactivator NcoA/SRC1a. J. Biol. Chem. 2002;277:8004–8011. doi: 10.1074/jbc.M111486200. [DOI] [PubMed] [Google Scholar]
- 152.Zhao H, et al. Region 752–761 of STAT3 is critical for SRC-1 recruitment and Ser727 phosphorylation. Biochem. Biophys. Res. Commun. 2004;325:541–548. doi: 10.1016/j.bbrc.2004.10.075. [DOI] [PubMed] [Google Scholar]
- 153.Zhang T, Ma J, Cao X. Grb2 regulates Stat3 activation negatively in epidermal growth factor signalling. Biochem. J. 2003;376:457–464. doi: 10.1042/BJ20030668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Simon AR, et al. Regulation of STAT3 by direct binding to the Rac1 GTPase. Science. 2000;290:144–147. doi: 10.1126/science.290.5489.144. [DOI] [PubMed] [Google Scholar]
- 155.Kataoka Y, et al. Reciprocal inhibition between MyoD and STAT3 in theregulationof growth and differentiation of myoblasts. J. Biol. Chem. 2003;278:44178–44187. doi: 10.1074/jbc.M304884200. [DOI] [PubMed] [Google Scholar]
- 156.Kawasaki A, et al. Opposing effects of PML and PML/RAR alpha on STAT3 activity. Blood. 2003;101:3668–3673. doi: 10.1182/blood-2002-08-2474. [DOI] [PubMed] [Google Scholar]
- 157.Lufei C, et al. GRIM-19, a death-regulatory gene product, suppresses Stat3 activity via functional interaction. EMBO J. 2003;22:1325–1335. doi: 10.1093/emboj/cdg135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yang CH, et al. Interferon induces the interaction of prothymosin-alpha with STAT3 and results in the nuclear translocation of the complex. Exp. Cell Res. 2004;298:197–206. doi: 10.1016/j.yexcr.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 159.Wang LH, et al. Transcriptional inactivation of STAT3 by PPARgamma suppresses IL-6-responsive multiple myeloma cells. Immunity. 2004;20:205–218. doi: 10.1016/s1074-7613(04)00030-5. [DOI] [PubMed] [Google Scholar]
- 160.Ziros PG, et al. Growth hormone attenuates the transcriptional activity of Runx2 by facilitating its physical association with Stat3beta. J. Bone Miner. Res. 2004;19:1892–1904. doi: 10.1359/JBMR.040701. [DOI] [PubMed] [Google Scholar]
- 161.Manavathi B, et al. Proline-, glutamic acid-, and leucine-rich protein-1 is essential in growth factor regulation of signal transducers and activators of transcription 3 activation. Cancer Res. 2005;65:5571–5577. doi: 10.1158/0008-5472.CAN-04-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nabarro S, et al. Coordinated oncogenic transformation and inhibition of host immune responses by the PAX3-FKHR fusion oncoprotein. J. Exp. Med. 2005;202:1399–1410. doi: 10.1084/jem.20050730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Nishimoto A, et al. A Ras homologue member I directly inhibits signal transducers and activators of transcription 3 translocation and activity in human breast and ovarian cancer cells. Cancer Res. 2005;65:6701–6710. doi: 10.1158/0008-5472.CAN-05-0130. [DOI] [PubMed] [Google Scholar]
- 164.Loeffler S, et al. Interleukin-6 induces transcriptional activation of vascular endothelial growth factor (VEGF) in astrocytes in vivo and regulates VEGF promoter activity in glioblastoma cells via direct interaction between STAT3 and Sp1. Int. J. Cancer. 2005;115:202–213. doi: 10.1002/ijc.20871. [DOI] [PubMed] [Google Scholar]
- 165.Jung JE, et al. STAT3 is a potential modulator of HIF-1-mediated VEGF expression in human renal carcinoma cells. FASEB J. 2005;19:1296–1298. doi: 10.1096/fj.04-3099fje. [DOI] [PubMed] [Google Scholar]
- 166.Ma J, Cao X. Regulation of Stat3 nuclear import by importin alpha5 and importin alpha7 via two different functional sequence elements. Cell Signal. 2006;18:1117–1126. doi: 10.1016/j.cellsig.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 167.Shao H, et al. Unique structural determinants for Stat3 recruitment and activation by the granulocyte colony-stimulating factor receptor at phosphotyrosine ligands 704 and 744. J. Immunol. 2006;176:2933–2941. doi: 10.4049/jimmunol.176.5.2933. [DOI] [PubMed] [Google Scholar]
- 168.Yamashina K, et al. Suppression of STAT3 activity by Duplin, which is a negative regulator of the Wnt signal. J. Biochem. 2006;139:305–314. doi: 10.1093/jb/mvj033. [DOI] [PubMed] [Google Scholar]
- 169.Yang J, et al. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev. 2007;21:1396–1408. doi: 10.1101/gad.1553707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fox DL, Good DJ. Nescient helix-loop-helix 2 interacts with signal transducer and activator of transcription 3 to regulate transcription of prohormone convertase 1/3. Mol. Endocrinol. 2008;22:1438–1448. doi: 10.1210/me.2008-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tsuruma R, et al. Physical and functional interactions between STAT3 and KAP1. Oncogene. 2008;27:3054–3059. doi: 10.1038/sj.onc.1210952. [DOI] [PubMed] [Google Scholar]
- 172.Muromoto R, et al. BART is essential for nuclear retention of STAT3. Int. Immunol. 2008;20:395–403. doi: 10.1093/intimm/dxm154. [DOI] [PubMed] [Google Scholar]
- 173.Stahl N, et al. Choice of STATs and other substrates specified by modular tyrosine-based motifs in cytokine receptors. Science. 1995;267:1349–1353. doi: 10.1126/science.7871433. [DOI] [PubMed] [Google Scholar]
- 174.Daino H, et al. Induction of apoptosis by extracellular ubiquitin in human hematopoietic cells: possible involvement of STAT3 degradation by proteasome pathway in interleukin 6-dependent hematopoietic cells. Blood. 2000;95:2577–2585. [PubMed] [Google Scholar]
- 175.Zhang JG, et al. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc. Natl. Acad. Sci. USA. 1999;96:2071–2076. doi: 10.1073/pnas.96.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat. Rev. Immunol. 2007;7:454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 177.Zhu BM, et al. SOCS3 negatively regulates the gp130-STAT3 pathway in mouse skin wound healing. J. Invest. Dermatol. 2008;128:1821–1829. doi: 10.1038/sj.jid.5701224. [DOI] [PubMed] [Google Scholar]
- 178.Tischoff I, et al. Methylation of SOCS-3 and SOCS-1 in the carcinogenesis of Barrett’s adenocarcinoma. Gut. 2007;56:1047–1053. doi: 10.1136/gut.2006.111633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Niwa Y, et al. Methylation silencing of SOCS-3 promotes cell growth and migration by enhancing JAK/STAT and FAK signalings in human hepatocellular carcinoma. Oncogene. 2005;24:6406–6417. doi: 10.1038/sj.onc.1208788. [DOI] [PubMed] [Google Scholar]
- 180.Weber A, et al. SOCS-3 is frequently methylated in head and neck squamous cell carcinoma and its precursor lesions and causes growth inhibition. Oncogene. 2005;24:6699–6708. doi: 10.1038/sj.onc.1208818. [DOI] [PubMed] [Google Scholar]
- 181.He B, et al. SOCS-3 is frequently silenced by hypermethylation and suppresses cell growth in human lung cancer. Proc. Natl. Acad. Sci. USA. 2003;100:14133–14138. doi: 10.1073/pnas.2232790100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhao S, et al. Inhibition of STAT3 Tyr705 phosphorylation by Smad4 suppresses transforming growth factor beta-mediated invasion and metastasis in pancreatic cancer cells. Cancer Res. 2008;68:4221–4228. doi: 10.1158/0008-5472.CAN-07-5123. [DOI] [PubMed] [Google Scholar]
- 183.Migone TS, et al. Recruitment of SH2-containing protein tyrosine phosphatase SHP-1 to the interleukin 2 receptor; loss of SHP-1 expression in human T-lymphotropic virus type I-transformed T cells. Proc. Natl. Acad. Sci. USA. 1998;95:3845–3850. doi: 10.1073/pnas.95.7.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Schaper F, et al. Activation of the protein tyro-sine phosphatase SHP2 via the interleukin-6 signal transducing receptor protein gp130 requires tyro-sine kinase Jak1 and limits acute-phase protein expression. Biochem. J. 1998;335(Pt 3):557–565. doi: 10.1042/bj3350557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Perry E, et al. TMF/ARA160 is a BC-box-containing protein that mediates the degradation of Stat3. Oncogene. 2004;23:8908–8919. doi: 10.1038/sj.onc.1208149. [DOI] [PubMed] [Google Scholar]
- 186.Ulane CM, et al. Composition and assembly of STAT-targeting ubiquitin ligase complexes: paramyxovirus V protein carboxyl terminus is an oligomerization domain. J. Virol. 2005;79:10180–10189. doi: 10.1128/JVI.79.16.10180-10189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Darnowski JW, et al. Stat3 cleavage by caspases: impact on full-length Stat3 expression, fragment formation, and transcriptional activity. J. Biol. Chem. 2006;281:17707–17717. doi: 10.1074/jbc.M600088200. [DOI] [PubMed] [Google Scholar]
- 188.Takeda K, et al. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. USA. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Li W, et al. STAT3 contributes to the mitogenic response of hepatocytes during liver regeneration. J. Biol. Chem. 2002;277:28411–28417. doi: 10.1074/jbc.M202807200. [DOI] [PubMed] [Google Scholar]
- 190.Takeda K, et al. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10:39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- 191.Sano S, et al. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. EMBO J. 1999;18:4657–4668. doi: 10.1093/emboj/18.17.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Chapman RS, et al. Suppression of epithelial apoptosis and delayed mammary gland involution in mice with a conditional knockout of Stat3. Genes Dev. 1999;13:2604–2616. doi: 10.1101/gad.13.19.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Desrivieres S, et al. The biological functions of the versatile transcription factors STAT3 and STAT5 and new strategies for their targeted inhibition. J. Mamm. Gland Biol. Neoplasia. 2006;11:75–87. doi: 10.1007/s10911-006-9014-4. [DOI] [PubMed] [Google Scholar]
- 194.Guo W, et al. Beta 4 integrin amplifies ErbB2 signaling to promote mammary tumorigenesis. Cell. 2006;126:489–502. doi: 10.1016/j.cell.2006.05.047. [DOI] [PubMed] [Google Scholar]
- 195.Pfitzner E, et al. The role of STATs in inflammation and inflammatory diseases. Curr. Pharm. Des. 2004;10:2839–2850. doi: 10.2174/1381612043383638. [DOI] [PubMed] [Google Scholar]
- 196.Kobierski LA, Srivastava S, Borsook D. Systemic lipopolysaccharide and interleukin-1beta activate the interleukin 6: STAT intracellular signaling pathway in neurons of mouse trigeminal ganglion. Neurosci. Lett. 2000;281:61–64. doi: 10.1016/s0304-3940(99)00953-2. [DOI] [PubMed] [Google Scholar]
- 197.Hoentjen F, et al. STAT3 regulates NF-kappaB recruitment to the IL-12p40 promoter in dendritic cells. Blood. 2005;105:689–696. doi: 10.1182/blood-2004-04-1309. [DOI] [PubMed] [Google Scholar]
- 198.Ernst M, et al. STAT3 and STAT1 mediate IL-11-dependent and inflammation-associated gastric tumorigenesis in gp130 receptor mutant mice. J. Clin. Invest. 2008;118:1727–1738. doi: 10.1172/JCI34944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Dalwadi H, et al. Cyclooxygenase-2-dependent activation of signal transducer and activator of transcription 3 by interleukin-6 in non-small cell lung cancer. Clin. Cancer Res. 2005;11:7674–7682. doi: 10.1158/1078-0432.CCR-05-1205. [DOI] [PubMed] [Google Scholar]
- 200.Frank DA. STAT3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 2007;251:199–210. doi: 10.1016/j.canlet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 201.Bromberg JF, et al. Stat3 activation is required for cellular transformation by v-src. Mol. Cell Biol. 1998;18:2553–2558. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Zhang YW, et al. Requirement of Stat3 signaling for HGF/SF-Met mediated tumorigenesis. Oncogene. 2002;21:217–226. doi: 10.1038/sj.onc.1205004. [DOI] [PubMed] [Google Scholar]
- 203.Bromberg JF, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 204.Catlett-Falcone R, et al. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10:105–115. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- 205.Karni R, Jove R, Levitzki A. Inhibition of pp60c–Src reduces Bcl-XL expression and reverses the transformed phenotype of cells overexpressing EGF and HER-2 receptors. Oncogene. 1999;18:4654–4662. doi: 10.1038/sj.onc.1202835. [DOI] [PubMed] [Google Scholar]
- 206.Zushi S, et al. STAT3 mediates the survival signal in oncogenic ras-transfected intestinal epithelial cells. Int. J. Cancer. 1998;78:326–330. doi: 10.1002/(SICI)1097-0215(19981029)78:3<326::AID-IJC12>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 207.Mahboubi K, et al. Interleukin-11 up-regulates survivin expression in endothelial cells through a signal transducer and activator of transcription-3 pathway. Lab Invest. 2001;81:327–334. doi: 10.1038/labinvest.3780241. [DOI] [PubMed] [Google Scholar]
- 208.Liu H, et al. Serine phosphorylation of STAT3 is essential for Mcl-1 expression and macrophage survival. Blood. 2003;102:344–352. doi: 10.1182/blood-2002-11-3396. [DOI] [PubMed] [Google Scholar]
- 209.Bhattacharya S, Schindler C. Regulation of Stat3 nuclear export. J. Clin. Invest. 2003;111:553–559. doi: 10.1172/JCI15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Aoki Y, Feldman GM, Tosato G. Inhibition of STAT3 signaling induces apoptosis and decreases survivin expression in primary effusion lymphoma. Blood. 2003;101:1535–1542. doi: 10.1182/blood-2002-07-2130. [DOI] [PubMed] [Google Scholar]
- 211.Kanda N, et al. STAT3 is constitutively activated and supports cell survival in association with survivin expression in gastric cancer cells. Oncogene. 2004;23:4921–4929. doi: 10.1038/sj.onc.1207606. [DOI] [PubMed] [Google Scholar]
- 212.Konnikova L, et al. Knockdown of STAT3 expression by RNAi induces apoptosis in astrocytoma cells. BMC Cancer. 2003;3:23. doi: 10.1186/1471-2407-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ivanov VN, et al. Cooperation between STAT3 and c-jun suppresses Fas transcription. Mol. Cell. 2001;7:517–528. doi: 10.1016/s1097-2765(01)00199-x. [DOI] [PubMed] [Google Scholar]
- 214.Masuda M, et al. Constitutive activation of signal transducers and activators of transcription 3 correlates with cyclin D1 overexpression and may provide a novel prognostic marker in head and neck squamous cell carcinoma. Cancer Res. 2002;62:3351–3355. [PubMed] [Google Scholar]
- 215.Kiuchi N, et al. STAT3 is required for the gp130-mediated full activation of the c-myc gene. J. Exp. Med. 1999;189:63–73. doi: 10.1084/jem.189.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Shirogane T, et al. Synergistic roles for Pim-1 and c-Myc in STAT3-mediated cell cycle progression and antiapoptosis. Immunity. 1999;11:709–719. doi: 10.1016/s1074-7613(00)80145-4. [DOI] [PubMed] [Google Scholar]
- 217.Bellido T, et al. Transcriptional activation of the p21(WAF1,CIP1,SDI1) gene by interleukin-6 type cytokines. A prerequisite for their pro-differentiating and anti-apoptotic effects on human osteoblastic cells. J. Biol. Chem. 1998;273:21137–21144. doi: 10.1074/jbc.273.33.21137. [DOI] [PubMed] [Google Scholar]
- 218.Barre B, Avril S, Coqueret O. Opposite regulation of myc and p21waf1 transcription by STAT3 proteins. J. Biol. Chem. 2003;278:2990–2996. doi: 10.1074/jbc.M210422200. [DOI] [PubMed] [Google Scholar]
- 219.Xiong H, et al. Inhibition of JAK1, 2/STAT3 signaling induces apoptosis, cell cycle arrest, and reduces tumor cell invasion in colorectal cancer cells. Neoplasia. 2008;10:287–297. doi: 10.1593/neo.07971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ma PC, et al. Downstream signalling and specific inhibition of c-MET/HGF pathway in small cell lung cancer: implications for tumour invasion. Br. J. Cancer. 2007;97:368–377. doi: 10.1038/sj.bjc.6603884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Yakata Y, et al. Expression of p-STAT3 in human gastric carcinoma: significant correlation in tumour invasion and prognosis. Int. J. Oncol. 2007;30:437–442. [PubMed] [Google Scholar]
- 222.Xie TX, et al. Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis. Oncogene. 2004;23:3550–3560. doi: 10.1038/sj.onc.1207383. [DOI] [PubMed] [Google Scholar]
- 223.Itoh M, et al. Requirement of STAT3 activation for maximal collagenase-1 (MMP-1) induction by epidermal growth factor and malignant characteristics in T24 bladder cancer cells. Oncogene. 2006;25:1195–1204. doi: 10.1038/sj.onc.1209149. [DOI] [PubMed] [Google Scholar]
- 224.Suiqing C, Min Z, Lirong C. Overexpression of phosphorylated-STAT3 correlated with the invasion and metastasis of cutaneous squamous cell carcinoma. J. Dermatol. 2005;32:354–360. doi: 10.1111/j.1346-8138.2005.tb00906.x. [DOI] [PubMed] [Google Scholar]
- 225.Dien J, et al. Signal transducers and activators of transcription-3 up-regulates tissue inhibitor of metalloproteinase-1 expression and decreases invasiveness of breast cancer. Am. J. Pathol. 2006;169:633–642. doi: 10.2353/ajpath.2006.051109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Gaemers IC, et al. A stat-responsive element in the promoter of the episialin/MUC1 gene is involved in its overexpression in carcinoma cells. J. Biol. Chem. 2001;276:6191–6199. doi: 10.1074/jbc.M009449200. [DOI] [PubMed] [Google Scholar]
- 227.Valdembri D, et al. In vivo activation of JAK2/STAT-3 pathway during angiogenesis induced by GM-CSF. FASEB J. 2002;16:225–227. doi: 10.1096/fj.01-0633fje. [DOI] [PubMed] [Google Scholar]
- 228.Niu G, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21:2000–2008. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 229.Wei D, et al. Stat3 activation regulates the expression of vascular endothelial growth factor and human pancreatic cancer angiogenesis and metastasis. Oncogene. 2003;22:319–329. doi: 10.1038/sj.onc.1206122. [DOI] [PubMed] [Google Scholar]
- 230.Wei LH, et al. Interleukin-6 promotes cervical tumor growth by VEGF-dependent angiogenesis via a STAT3 pathway. Oncogene. 2003;22:1517–1527. doi: 10.1038/sj.onc.1206226. [DOI] [PubMed] [Google Scholar]
- 231.Li WC, et al. Inhibition of growth and metastasis of human hepatocellular carcinoma by antisense oligonucleotide targeting signal transducer and activator of transcription 3. Clin. Cancer Res. 2006;12:7140–7148. doi: 10.1158/1078-0432.CCR-06-0484. [DOI] [PubMed] [Google Scholar]
- 232.Xie TX, et al. Activation of stat3 in human melanoma promotes brain metastasis. Cancer Res. 2006;66:3188–3196. doi: 10.1158/0008-5472.CAN-05-2674. [DOI] [PubMed] [Google Scholar]
- 233.Cheng GZ, et al. Twist is transcriptionally induced by activation of STAT3 and mediates STAT3 oncogenic function. J. Biol. Chem. 2008;283:14665–14673. doi: 10.1074/jbc.M707429200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Chan KS, et al. Forced expression of a constitutively active form of Stat3 in mouse epidermis enhances malignant progression of skin tumors induced by two-stage carcinogenesis. Oncogene. 2008;27:1087–1094. doi: 10.1038/sj.onc.1210726. [DOI] [PubMed] [Google Scholar]
- 235.Kataoka K, et al. Stage-specific disruption of Stat3 demonstrates a direct requirement during both the initiation and promotion stages of mouse skin tumorigenesis. Carcinogenesis. 2008;29:1108–1114. doi: 10.1093/carcin/bgn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Riehle KJ, et al. Regulation of liver regeneration and hepatocarcinogenesis by suppressor of cytokine signaling 3. J. Exp. Med. 2008;205:91–103. doi: 10.1084/jem.20070820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Boehm AL, et al. Combined targeting of epidermal growth factor receptor, signal transducer and activator of transcription-3, and Bcl-X(L) enhances antitumor effects in squamous cell carcinoma of the head and neck. Mol. Pharmacol. 2008;73:1632–1642. doi: 10.1124/mol.107.044636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Bharti AC, et al. Nuclear factor-kappaB and STAT3 are constitutively active in CD138+ cells derived from multiple myeloma patients, and suppression of these transcription factors leads to apoptosis. Blood. 2004;103:3175–3184. doi: 10.1182/blood-2003-06-2151. [DOI] [PubMed] [Google Scholar]
- 239.Real PJ, et al. Resistance to chemotherapy via Stat3-dependent overexpression of Bcl-2 in metastatic breast cancer cells. Oncogene. 2002;21:7611–7618. doi: 10.1038/sj.onc.1206004. [DOI] [PubMed] [Google Scholar]
- 240.Otero DC, et al. Cutting edge: inherent and acquired resistance to radiation-induced apoptosis in B cells: a pivotal role for STAT3. J. Immunol. 2006;177:6593–6597. doi: 10.4049/jimmunol.177.10.6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Kim KW, et al. Inhibition of signal transducer and activator of transcription 3 activity results in down-regulation of Survivin following irradiation. Mol. Cancer Ther. 2006;5:2659–2665. doi: 10.1158/1535-7163.MCT-06-0261. [DOI] [PubMed] [Google Scholar]
- 242.Chakravarti N, Myers JN, Aggarwal BB. Targeting constitutive and interleukin-6-inducible signal transducers and activators of transcription 3 pathway in head and neck squamous cell carcinoma cells by curcumin (diferuloylmethane) Int. J. Cancer. 2006;119:1268–1275. doi: 10.1002/ijc.21967. [DOI] [PubMed] [Google Scholar]
- 243.Selvendiran K, et al. Luteolin promotes degradation in signal transducer and activator of transcription 3 in human hepatoma cells: an implication for the antitumor potential of flavonoids. Cancer Res. 2006;66:4826–4834. doi: 10.1158/0008-5472.CAN-05-4062. [DOI] [PubMed] [Google Scholar]