Abstract
Nonclassical MHC class Ib (class Ib) genes are heterogeneous genes encoding molecules that are structurally similar to classical MHC class Ia molecules but with limited tissue distribution and polymorphism. Mammalian class Ib genes have diverse and often uncharacterized functions, and because of their rapid rate of evolution, class Ib phylogeny is difficult to establish. We have conducted an extensive genomic, molecular, and phylogenetic characterization of class Ib genes in two Xenopodinae amphibian species of different genera that diverged from a common ancestor as long ago as primates and rodents (~65 million years). In contrast with the unsteadiness of mammalian class Ib genes, our results reveal an unusual degree of conservation of most Xenopodinae class Ib gene lineages, including a novel monogenic lineage represented by the divergent Xenopus laevis XNC10 gene and its unequivocal Silurana (Xenopus) tropicalis orthologue, SNC10. The preferential expression of this gene lineage by thymocytes themselves from the onset of thymic organogenesis is consistent with a specialized role of class Ib in early T cell development and suggests such a function is conserved in all tetrapods.
Major histocompatibility complex class I and class II genes encode proteins that are involved in immune surveillance by presenting antigenic peptides to CD8 and CD4 T lymphocytes, respectively. MHC class I genes are further subdivided into classical MHC class Ia (class Ia) and nonclassical MHC class Ib (class Ib) based on structural and functional differences. Class Ia genes are central to the function of the immune system because they are involved in classical peptide Ag presentation. They are minimally polygenic, highly polymorphic, and codominantly expressed on cells in almost all tissues. Class Ib genes, in contrast, are structurally similar to class Ia genes and often function in immune responses by acting as indicators of intracellular stress and malignancy (1). This is partly due to the ability of different class Ib molecules to bind a diverse array of ligands including stimulatory or inhibitory receptors expressed on T, NK, and/or NKT lymphocytes (2). Although class Ib genes are defined as heterogeneous genes encoding molecules with a limited tissue distribution and low polymorphism, there are many exceptions to this definition. For example, some class Ib proteins, such as MIC-A and MIC-B in humans, are polymorphic (3), and yet others, such as HLA-E, are ubiquitously expressed (4).
The distinction between class Ia and class Ib genes becomes even more blurred when their evolutionary relationships are analyzed. Genomic and phylogenetic analysis of class Ia and class Ib genes reveals that in general, different orders or families of mammals have different numbers of genes or genetic loci. For example, the human class Ia loci A, B, and C are shared only by hominoid species (e.g., human, gorilla, and orangutan), but the New World monkeys (e.g., tamarin) and nonprimate mammals do not have these human orthologues (5). In other words, different families or orders of mammals usually do not have orthologous genes. This pattern of evolution of MHC class I genes has been explained by the birth and death model of evolution (6) in which new genes are created by gene duplication, and some duplicated genes are maintained in the genome for a long time, whereas others are deleted or become nonfunctional through harmful mutations. Because of this rapid rate of evolution, it becomes difficult to translate directly to humans what is learned from studies of class Ib structure and function in nonhuman animal model systems. MHC class I genes (both class Ia and class Ib) have been isolated from all studied representatives of jawed vertebrates (7). Comparative analyses of MHC class I genes across distantly related nonmammalian species thus provides the opportunity to relate the evolution of structure with function and promotes understanding of the impact of convergent evolution on MHC class I molecules.
Species from the Anuran amphibian subfamily Xenopodinae exemplify one of the few documented cases in vertebrates of speciation by genome duplication via allopolyploidization (8); this taxon includes various species ranging in ploidy from diploid to dodecaploid. Although all of these species are grouped into one taxon, multiple lines of evidence indicate that they belong to two divergent genera named Xenopus and Silurana (9, 10). Xenopus laevis is a representative species of the Xenopus genus, whereas Xenopus tropicalis is a representative species of the Silurana genus and as such is now named Silurana tropicalis. These two species, X. laevis and S. tropicalis, whose divergence from a common ancestor is estimated to have occurred more than 65 million years ago (MYA), are serving as valuable nonmammalian animal models in the study of early vertebrate development and comparative immunology (9). To date, the tetraploid frog X. laevis has served as the most widely used nonmammalian comparative animal model for the study of immunity in general, including class Ib genes (11). The MHC of this organism has been extensively studied at the functional, biochemical, and molecular levels. And although it is not known precisely how many X. laevis non-classical MHC class Ib (XNC) genes there are, XNC genes are expressed and can be categorized into subfamilies based on sequence similarity; sequences within a subfamily are greater than 90% identical in amino acid sequence in their α1 domains (12). Eleven XNC subfamilies have been identified (12, 13), although the precise number of genes in each subfamily is not known because a genome sequence is not currently available for X. laevis.
The amphibian S. tropicalis, in contrast, has been selected as a model organism for a whole genome sequencing project because of its important phylogenetic position and because it is the only diploid species of the Xenopodinae subfamily (http://genome.jgi-psf.org/Xentr4/Xentr4.home.html). Analysis of the S. tropicalis genome reveals an extensive degree of conserved gene synteny with human and chicken genomes and also a high degree of conservation of genes associated with human disease (14). Scaffolds containing the MHC locus have been annotated and characterized in detail (15), and these studies indicate that compared with the mammalian MHCs, the amphibian MHC is evolutionarily highly conserved. These studies also demonstrate that, compared with S. tropicalis MHC, the vertebrate MHC experienced a vigorous rearrangement in the bony fish and with some modification in bird lineages and a translocation and expansion of the MHC class I genes in the mammalian lineage. For this reason, the amphibian is considered to be a valuable model to study the evolution of the MHC. We have therefore identified and characterized a large family of S. tropicalis nonclassical MHC class Ib (SNC) genes and compared them with XNC subfamilies to evaluate the degree of evolutionary conservation of class Ib genes within the Xenopodinae subfamily.
Materials and Methods
Animals
Outbred X. laevis and S. tropicalis adults and larvae were obtained from our breeding colony (http://www.urmc.rochester.edu/smd/mbi/xenopus/index.htm). Animals were sacrificed by immersion in tricaine methane sulfonate (5 g/l for adults and 1 g/l for tadpoles). Sublethal gamma-irradiation (10 Gy) was delivered to premetamorphic larvae stages 56–58 with a cobalt source. All animals were handled under strict laboratory and university committee on animal resources regulations (approval no. 100577/2003-151 and 2004-199), minimizing discomfort at all times.
RNA extraction, rapid amplification of cDNA ends-PCR, and RT-PCR
Total RNA was isolated from tissues of pooled adults (10 to 20 individuals) and larvae (30 individuals). All RNA extractions were carried out using 1 ml TRIzol reagent. For rapid amplification of cDNA ends (RACE), 5′ and 3′ rapid amplification of cDNA ends-PCR (RACE-PCR)–ready cDNAs were prepared using the SuperSMART PCR cDNA synthesis kit from Clontech (Mountain View, CA). RACE-PCR was carried out using the Advantage 2 PCR Enzyme System from Clontech. RACE-PCR primers used were IbConsTrop-5′ RACE-1 (5′-CCC TCC TCT GGT GTT ACC TCC AC-3′) and IbConsTrop-5′ RACE-2 (5′-GCC ACT CTC TGA CTC TGA GCT GG-3′). cDNA was synthesized using the iScript cDNA synthesis kit from Bio-Rad (Hercules, CA) and diluted 2×. RT-PCR primers specific for the SNC genes 2.2, 4, 6.1, 6.2, 7.1, 12, and 13.3 as well as for EF-1α were designed, and the annealing temperatures were determined using gradient PCR. All RT-PCRs included water and reverse transcriptase (RT)-minus controls (omission of RT during cDNA synthesis).
Southern blotting
Genomic DNA from Xenopus erythrocytes was isolated as described (16) and digested to completion with restriction endonuclease. The digested DNA was separated on a 1% agarose gel and transferred onto nylon membranes by the capillary blotting technique in 10× SSC. Increasing amounts of DNA were loaded in higher-ploidy animals according to the ploidy level.
Bioinformatics tools
Nucleotide and amino acid sequences were analyzed using utilities at the National Center for Biotechnology Information Web site (http://www.ncbi.nlm.nih.gov). Nucleotide and amino acid sequences were aligned using ClustalX (http://www.clustal.org/), and alignments were edited and shaded with the GeneDoc program (http://www.nrbsc.org/gfx/genedoc/). The nucleotide and amino acid sequences of known genes were retrieved from GenBank using ENTREZ at http://www.ncbi.nlm.nih.gov. Genomic sequences were retrieved from the S. tropicalis JGI Web site (http://genome.jgi-psf.org/Xentr4/Xentr4.home.html). Homology searches were performed using TBLASTN and BLASTN programs. Phylogenetic analysis was performed using Molecular Evolutionary Genetics Analysis (MEGA, version 4.0; http://www.megasoftware.net/). Phylogenetic trees were generated by the neighbor-joining method of Saitou and Nei (17). Genetic distances were calculated by estimating the number of amino acid substitutions using the p-distance method with pairwise deletion of gaps. Numbers on nodes represent percentages of 1000 bootstrap replicates supporting each partition.
Results
Genomic characterization of SNC genes
XNC gene transcripts can be grouped into at least 11 subfamilies, numbered XNC1 through XNC11, based on amino acid sequence similarity of their α1 domains (12, 13). Although X. laevis is estimated to have roughly 20 XNC genes per haplotype by Southern blot analysis (12), in absence of genomic sequences their exact total number and the respective number of XNC genes within each subfamily is unknown. In contrast, the genome of the only diploid frog of the Xenopodinae subfamily, S. tropicalis, has recently been fully sequenced and annotated. Because class Ia and class Ib genes of mammalian species studied to date are generally not conserved between animals as divergent as X. laevis and S. tropicalis, we sought to identify and characterize SNC genes in S. tropicalis and compare them with XNC gene subfamilies of X. laevis. We used in silico analysis and searched the S. tropicalis genomic database version 4.1 (http://genome.jgi-psf.org/Xentr4/Xentr4.home.html) using the TBLASTN algorithm with deduced amino acid sequences of XNC transcripts. The BLASTN algorithm was also used to search the genome with nucleotide sequences of XNC α1, α2, and α3 domain exons.
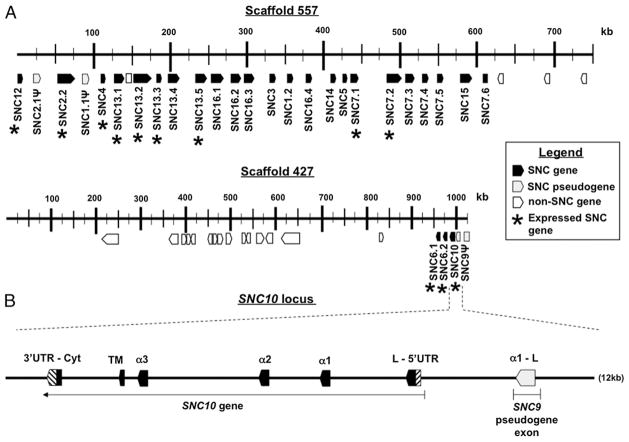
Two scaffolds, 557 and 427, encompassing 1.76 Mbp, contain a total of 29 automatically predicted SNC genes, including pseudogenes and gene fragments (Fig. 1, Table I). Some of these genes were missing exons containing either Leader or Transmembrane and Cytoplasmic domains. Manual searches were performed around these SNC genes, and if the exons were present, they were manually annotated. Several class I-like sequences were also found in scaffolds 2166, 145, and 3581. However, their sequences were mainly incomplete and not supported by expressed sequence tag (EST) databases. The characterization of these class I-like sequences was therefore not considered further in this study. In summary, a total of 29 SNC genes, including some pseudogenes and gene fragments, was retained for further analysis.
FIGURE 1.
Genomic organization of the predicted SNC gene models. A, Organization of the 29 SNC genes, pseudogenes, and gene fragments over two scaffolds encompassing a total of 1.76 Mbp (drawn to scale). *, expressed gene (supported by EST, RACE-PCR, or RT-PCR). Note difference in scale between the two scaffolds. B, Manual annotation (drawn to scale) and correction of the SNC10 gene model that is located near an XNC9-like α1 domain gene fragment.
Table I.
Twenty-nine SNC genes, pseudogenes, and gene fragments among two scaffolds
| Scaffold No. (genome assembly 4.1) | Protein ID | Position | Gene Name |
|---|---|---|---|
| 427a | 178628 | 963091–973347 | SNC6.1 |
| 178629 | 977506–984314 | SNC6.2 | |
| 374185 | 990632–998282 | SNC10 | |
| 992046–1004069 | SNC9Ψ | ||
| 557b | 386267 | 253–6190 | SNC12 |
| 386272 | 16483–37957 | SNC2.1Ψ | |
| 386185 | 57583–76814 | SNC2.2 | |
| 386177 | 85023–95237 | SNC1.1Ψ | |
| 386268 | 110253–116573 | SNC4 | |
| 458818 | 126280–134843 | SNC13.1 | |
| 182055 | 153164–176381 | SNC13.2 | |
| 386203 | 180370–187959 | SNC13.3 | |
| 386310 | 196609–203924 | SNC13.4 | |
| 182058 | 232324–249472 | SNC13.5 | |
| 182059 | 256738–273249 | SNC16.1 | |
| 386304 | 279463–291543 | SNC16.2 | |
| 386297 | 297124–306904 | SNC16.3 | |
| 386192 | 331557–334917 | SNC3 | |
| 386184 | 354429–364805 | SNC1.2 | |
| 386302 | 377723–386183 | SNC16.4 | |
| 386241 | 410224–415542 | SNC14 | |
| 386246 | 425413–427874 | SNC5 | |
| 386189 | 433522–448178 | SNC7.1 | |
| 386240 | 487368–502788 | SNC7.2 | |
| 386261 | 511135–521252 | SNC7.3 | |
| 386244 | 529093–535288 | SNC7.4 | |
| 386294 | 548537–557354 | SNC7.5 | |
| 386214 | 583078–595673 | SNC15 | |
| 386300 | 610756–614712 | SNC7.6 |
Transmembrane/cytoplasmic domain manually annotated for SNCs 4, 5, 1.2, 2.1, 13.5.
SNC10 manually annotated and corrected.
pseudogene.
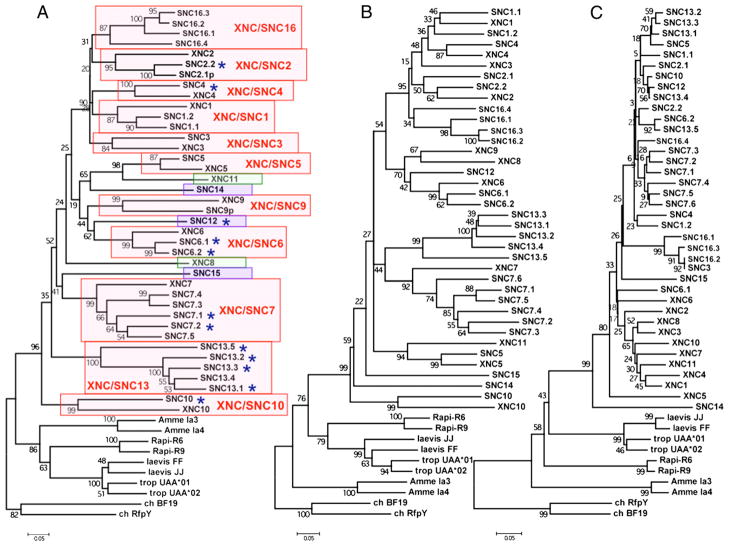
Because our objective was to assess orthologous relationships between XNC gene subfamilies in X. laevis and SNC genes in S. tropicalis, we chose a nomenclature and the methodology used in X. laevis that is based on the most divergent α1 domain to discriminate subfamilies (12). To assign SNC genes to particular subfamilies that correspond with XNC subfamilies, we first performed a phylogenetic analysis based on an amino acid alignment of α1 domains among SNC genes. This resulted in 14 separate subfamilies defined by separate clades of SNC genes with bootstrap support <70. To determine orthologies to XNC subfamilies, an amino acid alignment of SNC and XNC α1 domains was made. This alignment was used to generate a neighbor-joining phylogenetic tree. Orthologous XNC and SNC gene subfamilies were defined as any clade having a bootstrap value of 75 or higher. As such, the amino acid sequence similarity of individual α1 domain sequences within each clade is greater than 60%. Each S. tropicalis gene model was named with a three-letter code, SNC, followed by a distinctive number. We matched the numbers between XNC subfamilies and SNC orthologues [or co-orthologues, defined as two or more S. tropicalis SNC genes that are collectively orthologous to one X. laevis XNC subfamily, likely due to recent gene duplication (18)] based on sequence similarity determined by nucleotide (data not shown) and amino acid alignments of the α1 (Supplemental Fig. 1) and α2 (Supplemental Fig. 2) domains, as well as by phylogenetic analysis of the α1 domains (Fig. 2A). SNC genes without an orthologous XNC subfamily were given a number higher than 11 (i.e., because of the 11 XNC subfamilies in X. laevis characterized to date). Pseudogenes were labeled with the character “Ψ” In a few cases, particular genes could have been grouped into one of two subfamilies. For example, SNC genes SNC2/3.3, SNC2/3.4, and SNC2/3.5 were similar to both XNC2 and XNC3. We decided to name the genes with an “SNC2/3” prefix simply because this group of genes was not different enough from either XNC2 (α1 domain) or XNC3 (α3 domain) to be grouped into its own unique subfamily and because regardless of whether these particular SNC genes would have been grouped into the XNC2 or XNC3 subfamily, SNC lineages 1, 2, 3, 4, and 16 all evolved from a recent common ancestor. Our analysis indicates that S. tropicalis SNC genes group into 14 subfamilies based on sequence similarity in their α1 domains (Fig. 2A). Most SNC subfamilies are monogenic, and a few others have experienced lineage-specific expansions. Surprisingly, most SNC genes are orthologous (or co-orthologous) to XNC subfamilies. Of the 14 SNC subfamilies, 9 are conserved with X. laevis. Although most SNC genes are conserved with XNC subfamilies, there are some SNC genes that do not have a corresponding orthologue in X. laevis, and vice versa, and therefore represent species-specific class Ib genes.
FIGURE 2.
Conservation of S. tropicalis SNC genes and X. laevis XNC subfamilies. Bootstrap consensus neighbor-joining trees were constructed from amino acid alignments of either α1 (A), α2 (B), or α3 (C) domain amino acid sequences from XNC, SNC, and class Ia genes from X. laevis, S. tropicalis, as well as the northern leopard frog (AN: AF185587 and AF185588) and the axolotl (AN: U83137 and U83138) class Ia genes. Trees were rooted with chicken class Ia and class Ib genes (AN: M84766 and AF218783, respectively). Pairwise deletion of gaps and p-distance were used to estimate the number of amino acid substitutions. Support for each node was assessed using 1000 bootstrap replicates. Trees generated using nucleotide sequences gave similar results (data not shown). Red box, Orthologous (or co-orthologous) SNC genes and XNC subfamilies. Green box, Xenopus-specific subfamily. Blue box, Silurana-specific gene. *, expressed SNC gene. Note that SNC7.6 does not have an α1 domain and therefore is not present in A. AN, accession number.
Conservation of Xenopodinae class Ib gene lineages
Analysis of class Ib genes of S. tropicalis and X. laevis reveal an unprecedented degree of conservation of multiple class Ib gene lineages between two divergent vertebrate species. These results suggest that compared with mammalian taxon, class Ib genes in Xenopodinae underwent different selective pressures, favoring maintenance of individual class Ib lineages. To address this hypothesis further, we evaluated the degree of similarity of the different S. tropicalis SNC genes with X. laevis XNC subfamilies and class Ia through nucleotide and amino acid alignments and phylogenetic analyses. To strengthen this analysis, we included MHC class I sequences of two other amphibian species available in GenBank; the northern leopard frog, Rana pipiens, and the axolotl, Ambystoma mexicanum, organisms whose most recent common ancestors with species from the Xenopodinae subfamily date back 220 and 270 MYA, respectively, as determined by both molecular and paleontological evidence (19, 20). Neighbor-joining consensus trees of amino acid alignments (i.e., nucleotide data gave similar results; data not shown) of individual α1, α2, and α3 extracellular domains were constructed with 1000 bootstrap replicates (Fig. 2A–C). Rather than forming species-specific clusters, the majority of class Ib loci of S. tropicalis and X. laevis cluster in a locus-specific manner (Fig. 2A, 2B). These gene lineages have thus maintained their genetic identities at least since the S. tropicalis and X. laevis speciation event, more than 65 MYA (9, 10). Notably, both the α1 and α2 domain trees (Fig. 2A, 2B) demonstrate that Xenopodinae class Ia sequences are more similar to class Ia sequences from the Anuran amphibian R. pipiens than they are to class Ib gene sequences from their own species. Assuming similar rates of evolution, this result suggests that Anuran class Ia and class Ib genes diverged more than 200 MYA. This conclusion is, however, subjected to the limitation that the rates of evolution of these two gene families have probably not been constant and have varied independently. Almost certainly, particular regions in the α1 and α2 domains have been selected for divergence (e.g., interaction with peptides), whereas other regions have been selected to stay the same (e.g., interactions with β2-microglobulin [β2m], TCRs, NK cell receptors, etc.). Moreover, the class Ib lineage should have been subjected to fast evolutionary changes when it specialized into new functions, but afterward changes may have been selected against. Importantly, and irrespective of the variation in the rate of evolution, this phylogeny also implies that the SNC10 gene along with its X. laevis XNC10 orthologue were the first class Ib genes to have diverged from the XNC/SNC common ancestor.
In contrast, a different evolutionary scenario emerges from the analysis of α3 domain sequences. Fig. 2C reveals an overall sequence similarity shared by all XNC and SNC α3 domains suggesting intraspecies homogenization and presumably subsequent selection to maintain important functions of α3 domain sequences of SNC and XNC subfamilies. This is similar to what is seen for class Ia α3 domains (21). Homogenization of α3 domains of MHC class I molecules might occur to maintain important functions, such as association with β2m. It is possible that the difference between α3 domains of class Ia and class Ib may come from their association with different accessory proteins (β2m, CD8). For example, the putative Xenopodinae β2m- and CD8-binding residues of the α3 domain show some distinctive features between class Ia and class Ib. For both β2m- and CD8-binding residues, there are some positions that are conserved between class Ia and class Ib and other positions that have a clear dichotomy (Supplemental Fig. 3). Therefore, for both SNC and XNC genes, α1 and α2 domain sequences, collectively forming the peptide-binding domain, cluster according to loci, whereas α3 Ig domain sequences cluster according to species.
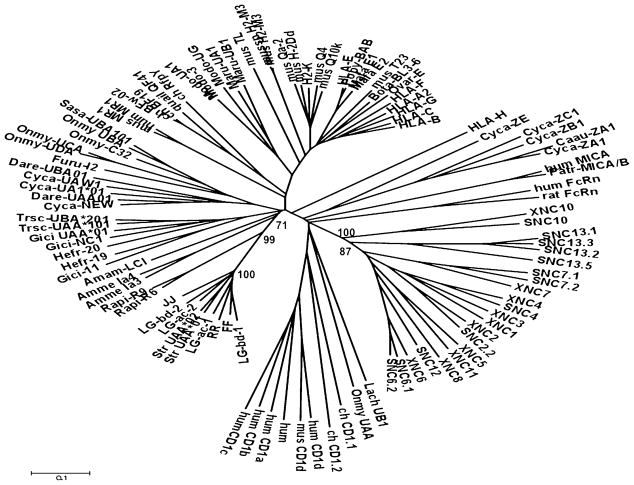
It has been proposed that a few class Ib loci diverged from class Ia loci a long time ago and now have different functions (22). Because our phylogenetic analysis indicates that Xenopodinae class Ia and class Ib genes likely diverged more than 200 MYA, we wanted to compare SNC gene sequences with other jawed vertebrate class Ia and class Ib sequences. A phylogenetic analysis of expressed SNC genes (see the section SNC genes are expressed for more details) and representative jawed vertebrate class Ia and class Ib sequences (Fig. 3) was conducted. We postulated that selective pressures should primarily affect Ag-binding functions of class Ib genes and therefore focused our analysis on α1 and α2 domain sequences. The tree presented in Fig. 3 shows that SNC and XNC genes form a monophyletic clade with respect to other vertebrate class Ia and class Ib genes, including amphibian class Ia. This provides support for a common evolutionary origin of class Ib genes from the Xenopodinae subfamily.
FIGURE 3.
Distinct evolutionary origins of Xenopodinae class Ia and class Ib genes. Phylogenetic analysis was conducted as in Fig. 2 but an unrooted tree was generated using α1–α2 domain amino acid sequences of selected jawed vertebrate class Ia and Ib sequences. Class I sequences used are listed in Supplemental Table I.
In conclusion, phylogenetic analysis indicates that SNC and XNC genes from S. tropicalis and X. laevis evolved from a common ancestor, with the XNC/SNC10 gene lineage being the first to diverge. In addition, although there has been expansion and contraction of individual class Ib genes, there is a remarkable preservation of several class Ib gene lineages over a relatively long period of time in two divergent amphibian species.
SNC genes are expressed
Class Ib molecules identified to date tend to have a more restricted tissue expression pattern compared with that of class Ia (4). To determine if the different SNC genes also have a limited tissue-specific distribution, we conducted EST database searches as well as RACE-PCR and RT-PCR analyses. We first searched the National Center for Biotechnology Information GeneBank S. tropicalis EST database to determine which of the 29 SNC genes have EST support. BLAST analyses were performed with the TBLASTN algorithm using SNC consensus α1 and α2 domain amino acid sequences from each of the 14 SNC subfamilies (see alignment in Supplemental Figs. 1 and 2). A total of 47 SNC transcripts were obtained, but only 35 contained sufficient sequence in the α1, α2, or Cytoplasmic domains to assign them to a particular gene. In total, nine genes were represented by ESTs (Table II). In addition, most EST transcripts were from adult thymus and spleen, whereas a few were from adult brain, fat bodies, intestine, liver, lung, skin, skeletal muscle, oviduct, and testes.
Table II.
Summary of adult expression pattern for SNC genes
| SNC Gene | No. EST | No. of Adult RACE-PCR | RT-PCR | SNC Tissue Expression Pattern |
|---|---|---|---|---|
| SNC2.2 | 1 | 8 | Yes | Thymus, spleen, colon, intestine, liver, stomach |
| SNC4 | 0 | 10 | Yes | Thymus, spleen, intestine, skin |
| SNC6.1 | 1 | 7 | Yes | Thymus, spleen |
| SNC6.2 | 0 | 1 | Spleen | |
| SNC7.1 | 12 | 3 | Yes | Ubiquitous |
| SNC7.2 | 1 | 0 | ND | Thymus, spleen |
| SNC10 | 0 | 0 | Yes | Thymus |
| SNC12 | 2 | 5 | Yes | Ubiquitous |
| SNC13.1 | 2 | 0 | ND | Skeletal muscle, heart |
| SNC13.2 | 10 | 0 | ND | Thymus, lung |
| SNC13.3 | 5 | 9 | Yes | Thymus, spleen, heart, liver, skin |
| SNC13.5 | 1 | 0 | ND | Brain |
ND, no data.
To substantiate our EST data, we performed two distinct 5′ RACE-PCR reactions: one with cDNA synthesized from a pool of 20 adult outbred thymuses, and another one with cDNA from a pool of 20 adult spleens. RACE-PCR products were separated by gel electrophoresis, and bands were cloned and sequenced. A total of 43 SNC sequences analyzed from adult thymus and spleen supported 7 SNC genes (Table II), 5 of which were common to those identified by ESTs. Finally, RT-PCR of cDNA from different organs from a pool of 20 adult tissues was performed using SNC gene-specific primers. In addition to the 11 SNC genes already found to be expressed, RT-PCR resulted in detection of a 12th expressed SNC gene; SNC10 transcripts were amplified from adult thymus (Table II).
In summary, evidence for expression for 12, of 29 total, SNC genes was obtained in this study (Fig. 1A, asterisks). As expected, many of these genes, including SNC10, displayed a restricted tissue expression pattern that mostly involved lymphoid organs. In addition, the levels of expression of several SNC genes were higher than those of others, as is the case for X. laevis as well as mammalian class Ib genes (23, 24).
Genes from the divergent Xenopodinae class Ib lineage, SNC10 and XNC10, are orthologous
It has been proposed that class Ib genes that are evolutionarily old might have evolved unique functions that became fixed early during vertebrate evolution (25). The orthologous relationship of the oldest Xenopodinae class Ib genes, SNC10 and XNC10, was investigated with the purpose of gaining functional insight. It is important to note that during our initial inspection of the 29 automatically predicted gene models, we did not identify an obvious XNC10 orthologue. However, it was observed that JGI gene model 374185 had an α1 domain exon similar to XNC9 and an α2 domain exon similar to XNC10. Therefore, this open reading frame was either annotated incorrectly by the gene prediction models or it was a hybrid SNC gene. To address these possibilities, the genomic region surrounding and including this predicted open reading frame was further inspected and manually annotated. Two unannotated exons coding for an XNC10-like Leader and α1 domain were identified in between the automatically predicted XNC9-like Leader-α1 domain-containing exon and the XNC10-like α2 domain exon for gene model 374185 (Fig. 1B). To know whether the S. tropicalis genome contains an XNC10 orthologue, we verified the gene architecture by PCR amplification of the genomic region containing the putative SNC10 gene followed by analytical endonuclease digestion (data not shown). These results confirmed that S. tropicalis contains an SNC gene that is orthologous to X. laevis XNC10.
One notable feature of the XNC10 protein was the uniqueness of its predicted peptide-binding residues (pPBRs) (13). To explore further the orthologous relationship between X. laevis XNC10 and S. tropicalis SNC10 and obtain indirect evidence of a critical, conserved immune function, we analyzed molecular features of the pPBRs of SNC10. As previously done for X. laevis class Ia and XNC genes (11, 12), we based our analysis on the amino acid alignment of these genes with HLA-A (26). Specifically, we compared SNC10 amino acid residues that align with invariant amino acids of the peptide binding domain of human HLA-A (27) to analyze any conservation of residues potentially involved in peptide binding. In mammalian class Ia, side chains of these invariant residues point into the MHC class I cleft and are essential for peptide anchoring through hydrogen or ionic bonding to backbone atoms of peptide N and C termini (Table III). The putative peptide N terminus of XNC10 contains two XNC10-specific residues, Asp/Asn59 and His171 (13), which are not conserved with SNC10. The putative peptide C terminus docking residues of SNC10, in contrast, are conserved with those of XNC10 (Table III). For example, the peptide C terminus docking residues of XNC10 are very different compared with residues in the same position of other XNC molecules. In XNC10, Lys146 is conserved with class Ia and Tyr/Arg84 is conservatively substituted for Ser, a residue that is still capable of forming hydrogen bonds. Both of these C terminus docking residues are conserved with SNC10. The XNC10-specific residue Ala143 is also conserved in SNC10. Of interest, the only peptide C terminus residue that is not conserved between XNC10 and SNC10 is Trp147. Notably, this residue is identical to that found in the same position in class Ia. Therefore, although N terminus residues of SNC10 pPBRs are not very similar to those of XNC10 (they are actually more conserved with XNC1–9 pPBRs), putative peptide C terminus docking residues of SNC10 are remarkably similar to those of XNC10 (Table III). These results are consistent with the interesting possibility that the XNC/SNC10 amphibian class Ib subfamily binds a common or conserved antigenic motif with its putative peptide C terminus docking residues while simultaneously building flexibility into the system at the peptide N terminus binding residues. Whether XNC/SNC10 does indeed bind an Ag and the nature of this Ag (e.g., peptide, glycolipid, or other), however, remains to be determined.
Table III.
Comparison of putative SNC10 peptide N and C termini docking residues of the pPBRs between SNC10, XNC10, and XNC1–9
| Docking Residuesa | XNC1–9b | XNC10 | SNC10 |
|---|---|---|---|
| Peptide N terminus | |||
| Tyr7 | 6 Tyr, 2 Phe | Tyrc | Tyrc |
| Tyr59 | 1 Tyr, 8 His | Asp/Asn | His |
| Tyr159 | 4 Tyr, 4 Phe | Tyrc | Tyrc |
| Trp167 | 2 Gln, 6 His | Gln | His |
| Tyr171 | 7 Tyr | His | Tyr |
| Peptide C terminus | |||
| Tyr84 or Arg84 | 3 Tyr, 1 Arg, 3 Phe, 2 Val | Serc | Serc |
| Thr143 | 4 Val, 3 Met, 1 Leu | Alac | Alac |
| Lys146 | 3 Gln, 5Leu | Lysc | Lysc |
| Trp147 | 7 Trp, 1 Leu | Arg | Trp |
pPBRs of Xenopodinae class Ia and class Ib were predicted by alignment with human HLA-A as previously published (12, 13, 28).
Numbering based on HLA-A2.
Not all residue positions sum up to 9 because transcripts originally obtained for XNC5, XNC7, and XNC9 are incomplete and therefore data are not available for every position.
Residue conserved between XNC10 and SNC10.
One hallmark of class Ib genes, which contrasts with class Ia, is their limited polymorphism (29). This is thought to reflect differences in selective pressures that drive distinct functions of class Ib compared with class Ia (30). To address the degree of SNC10 gene polymorphism, we amplified, analyzed, and aligned 13 α1 domain and 10 α2 domain nucleotide sequences for SNC10 that were obtained by 5′ RACE-PCR of a pool of 20 outbred S. tropicalis larvae. This analysis reveals that, like XNC10 (13), the α1 and α2 domains of SNC10 are not very polymorphic. Only six sites in the α1 domain, of 258 total sites, have a single-nucleotide polymorphism in 1 of the 14 sequences analyzed (Supplemental Fig. 4). Similarly, only two sites of 170 analyzed are mutated in the α2 domain. A larger sample size would be necessary to infer any more from this analysis, therefore we will simply conclude that as is expected for a typical class Ib gene, the coding region of SNC10 is not very polymorphic.
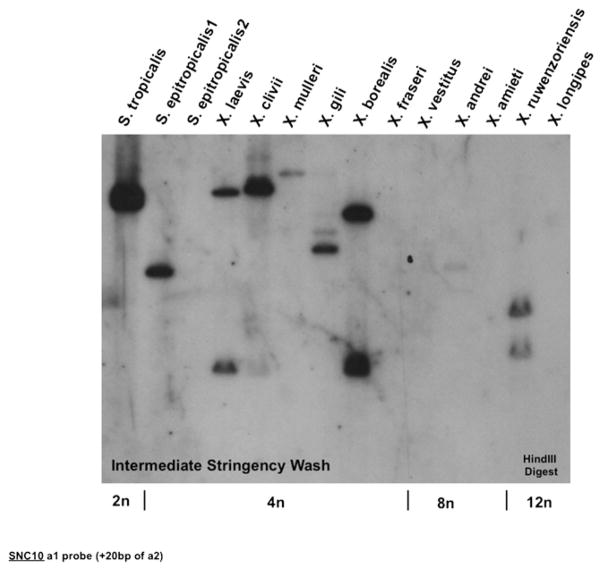
Southern blot hybridization with an XNC consensus α3 domain probe has shown that all Xenopus and Silurana species maintain a large number of class Ib genes (12). Furthermore, we have established that the XNC10 gene is conserved in all species tested of the genus Xenopus (13). To address whether other species of the genera Silurana and/or Xenopus maintain the XNC/SNC10 gene lineage, hybridization of a genomic Southern blot of representative polyploid species from these two genera was performed with an SNC10 α1 domain probe. Results show that SNC10 is conserved in S. tropicalis, as expected, and in S. epitropicalis1, but not S. epitropicalis2, and in some species of the Xenopus genus (Fig. 4). In addition, the gene is diploidized in all species that retain a copy. Because a similar amount of genomic DNA is present, the most likely explanation for the absence of an SNC10 hybridizing band in the S. epitropicalis2 lane is that the SNC10 gene has been lost in this individual. Unfortunately, no more DNA from this individual is available for genomic PCR. Regardless of this sample, we can conclude that SNC10 is conserved and diploidized among multiple species of the genera Silurana and Xenopus.
FIGURE 4.
Conservation and silencing of the SNC10 gene in multiple Xenopodinae species. Equivalent amounts of genomic DNA (adjusted per ploidy) from a representative of each species within the genera Xenopus and Silurana digested with HindIII, hybridized at 42°C with a 32P-labeled 278-bp (258-bp α1 exon plus 20 bp of α2 exon) SNC10 cDNA fragment, and washed under intermediate stringency. Ploidy of each species is indicated at the bottom of the blot.
There is some evidence for the idea that SNC10 and XNC10 represent two distinct sublineages of the XNC/SNC10 subfamily that are differentially retained in different species of Xenopodinae. Whereas the blot from Fig. 4 demonstrates that an SNC10-specific probe hybridizes to species from the genus Silurana, and to some, but not all, species from the genus Xenopus, probing this same blot with an XNC10-specific probe results in hybridization to all species from the genus Xenopus but not to any from the genus Silurana (13). The Xenopus genus has been categorized into various “subgroups” (10), and the SNC10 probe primarily hybridizes to species from the “laevis” (including X. laevis and X. gilli) and “muelleri” (including X. muelleri, X. clivii, and X. borealis) subgroups. These results suggest that there may be two divergent lineages of class Ib genes from the XNC/SNC10 subfamily that are selectively maintained in individual species of the amphibian subfamily Xenopodinae. This is perhaps not unexpected because the different species are thought to have arisen by allopolyploidization.
Early expression of SNC10 in the radiosensitive thymocytes of S. tropicalis larval thymus
In X. laevis larvae, the XNC10 gene is primarily expressed by thymocytes (13). The striking molecular features and expression profile of XNC10 are suggestive of a function in T cell development in X. laevis larvae. If the orthology of SNC10 and XNC10 is indicative of a similar function that has been conserved over evolutionary time, then SNC10 expression should also preferentially occur in S. tropicalis larval thymus. To explore this possibility, we determined the expression pattern of SNC10 at adult (Fig. 5A) and early developmental stages (Fig. 5B, 5C) of S. tropicalis, focusing especially on the thymus. RT-PCR was performed on various organs pooled from 20 individual tadpoles (stage 56) using SNC10-specific primers. Like XNC10, SNC10 expression was found primarily in adult and larval thymus and to a lesser extent in the spleen (Fig. 5A, 5B). SNC10 was also markedly expressed in the gills and at low levels in the pancreas. All RT-minus controls were negative, and each amplified band was cloned and sequenced to verify its authenticity.
FIGURE 5.
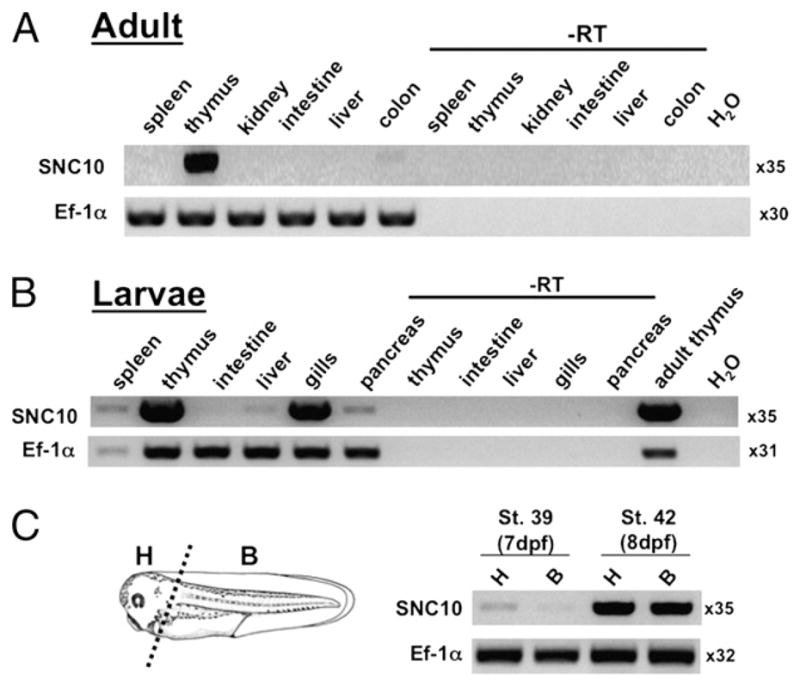
SNC10 expression primarily in thymus and larval gills from early in ontogeny. A and B, RT-PCR with primers specific for SNC10 was performed on cDNA generated from total RNA from a pool of 20 individual adult (A) and larval (B) premetamorphic (stage 56) S. tropicalis tissues. C, RT-PCR of a pool of 10 larvae stage 39 or 42 head or body tissues using primers specific for SNC10 and EF-1α as control. Number of cycles used is listed next to each gel picture. B, body; H, head.
Another key feature of XNC10 expression in X. laevis tadpoles is its early onset of expression, at developmental stage 39 (13), which corresponds with the onset of thymic organogenesis (31). To address whether SNC10 is also expressed early during ontogeny in S. tropicalis thymus, its expression was assessed by RT- PCR on anterior and posterior regions of larvae at developmental stage 39 (7 dpf) and stage 42 (8 dpf). Because S. tropicalis develops faster and may be slightly different [i.e., the detailed developmental characterization by Nieuwkoop and Faber (31) was done with X. laevis], we focused on precise comparable developmental morphological criteria for an accurate comparison between S. tropicalis and X. laevis developmental stages. In several experiments, expression of SNC10 was first detected at stage 39 and its intensity increased through stage 42 (Fig. 5C). Therefore, similar to XNC10, SNC10 is expressed in S. tropicalis larval thymus from the onset of thymic organogenesis.
In mammals, unlike class Ia molecules that are expressed by thymic stromal and epithelial cells, class Ib molecules involved in the education of T cells are primarily expressed by the thymocytes themselves, and this expression pattern is required for the selection of class Ib-restricted T cells (32). We therefore asked if SNC10 is preferentially expressed by larval thymocytes, an expression pattern that would mirror that of XNC10. Premetamorphic tadpoles (three per group) were sublethally gamma-irradiated (10 Gy), and three were left untreated as controls. Thymuses were harvested and prepared for RT-PCR analysis at 1, 2, and 3 d after gamma-irradiation or untreated. As in X. laevis with XNC10 (13), a significant decrease of SNC10 expression was already detectable 1 d after gamma-irradiation and became more pronounced in the following 2 d after treatment, in parallel with other thymocyte markers such as CTX, Rag1, CD8β, and CD3ε. Notably, expression of Aire also decreases upon gamma-irradiation. This was an unexpected result because in mammals it is expressed in radio-resistant thymic epithelial cells (33). Frog Aire is divergent from human and mouse, both in sequence and domain/region composition (34), and therefore suggests that the cells expressing Aire in frog thymus might differ from that in mammals. Other markers, such as the housekeeping gene EF-1α, did not change upon gamma-irradiation (Fig. 6). These results strongly suggest that SNC10 is preferentially expressed by radiosensitive thymocytes of S. tropicalis larvae, identical to XNC10 expression in X. laevis larval thymus. Such an expression pattern is consistent with a specialized role of genes from the XNC/SNC10 MHC class Ib lineage in the development of T cells.
FIGURE 6.
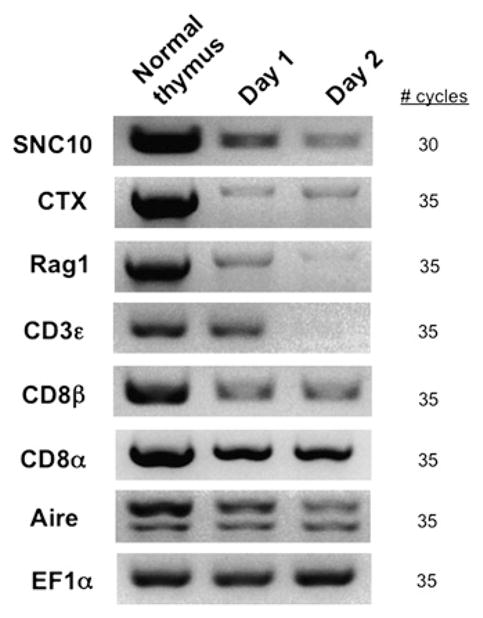
SNC10 is mainly expressed by thymic hematopoietic cells in larvae. RT-PCR analysis of total thymocytes from premetamorphic tadpoles untreated or 1 and 2 d after sublethal gamma-irradiation (10 Gy). Number of cycles used is listed next to each gel picture.
Discussion
In the current study, we have conducted an extensive genomic, molecular, and phylogenetic characterization of class Ib genes in two amphibian species of two different genera that diverged around 65 MYA. Our results reveal that in contrast to the unsteadiness of class Ib genes in most vertebrates analyzed to date, class Ib genes of two divergent species from the Xenopodinae subfamily display an unusual degree of conservation of most of their class Ib gene lineages. This striking conservation at the gene level includes a monogenic subfamily (XNC/SNC10) that is at least as ancient as the divergence between the two species from which they were characterized and whose expression pattern is consistent with a specialized function in early T cell development.
Conservation of amphibian class Ib gene lineages over a long evolutionary time
Our comparative study reveals an unusual conservation of multiple class Ib gene lineages among two divergent amphibian species. Although each lineage varies in SNC gene copy number, the majority of the lineages themselves are conserved with X. laevis. The conservation of these gene lineages in S. tropicalis does not, however, imply that the SNC genes themselves are static. Because our sequence analysis demonstrates that some SNC gene lineages contain many more genes than others, SNC genes in different lineages have differentially undergone gene duplication and are thus plastic, having experienced some lineage-specific expansion. Therefore, it is surprising that within the amphibian subfamily of Xenopodinae, class Ib gene lineages are preserved despite the plasticity of individual class Ib genes. Thirty-five percent of the genome of S. tropicalis is composed of transposable elements (14), and this could predispose certain genomic regions to reshuffling. The preservation of multiple distinct class Ib gene lineages over a relatively long evolutionary time is suggestive of the selection of these molecules for performing important functions.
This pattern of class Ib evolution within the Xenopodinae subfamily is in stark contrast to what is seen in mammals where different families or orders usually do not have orthologous genes. In primates, for example, MHC class I genes diverged so recently that even humans and New World monkeys, which diverged only ~33–35 MYA, do not share functional genes (35, 36). Similarly, MHC class I genes from two marsupial species, which separated ~48 MYA from a common ancestor, show no orthologous relationships (37). The turnover rate of MHC class I genes in mammals is therefore very high. Class Ib gene evolution in the Xenopodinae subfamily of amphibians appears to have taken a different route compared with that of mammals. Various attempts to date the origin of Xenopodinae using extant species have provided an estimate that ranges from 50.4 to 81.3 MYA (9, 38), which is why we decided to use the average time of 65 MYA. Although the evolutionary relationships of some individual class Ib genes are established within mammals (e.g., CD1, HLA-E/Qa1), the conservation of 9 of 14 S. tropicalis and X. laevis class Ib lineages over this relatively long period of evolution is to our knowledge the first case in vertebrates of such preservation of a group of class Ib genes.
The morphological resemblance of S. tropicalis and X. laevis may suggest a corresponding genetic stability that would explain a higher overall conservation of class Ib locus compared with that of mammals. However, we think this is unlikely. Even though there is a small degree of morphological difference between these two amphibian species, they differ far more from each other in their protein sequences than do organisms with large morphological differences, such as humans and chimpanzees (39, 40). Although S. tropicalis and X. laevis have many morphological similarities, numerous studies of their mitochondrial DNA have demonstrated the large degree of divergence between species of the Silurana and Xenopus genera (9, 38, 41). This divergence between the Xenopodinae subfamily does not appear to be due to drastic differences in rates of nucleotide substitution between amphibians and mammals because when rates of nucleotide substitution have been compared between pairs of X. laevis duplicated genes and the orthologous loci of humans and rodents, it has been determined that, at the amino acid level, the X. laevis genes and the mammalian genes are under similar constraints (41). Collectively, these results suggest that morphological evolution and protein sequence evolution can proceed at contrasting rates.
All extant species of frogs from the Xenopodinae subfamily are confined to the continent of Africa. The antiquity of the Xenopodinae subfamily of amphibians is highlighted by the discovery of a fossil identified as a species from the Xenopus genus in South America (42). This indicates that evolutionary radiation with the Xenopodinae subfamily of amphibians must have preceded the separation of the African and South American continents more than 100 MYA (10). The substantial degree of divergence between X. laevis and S. tropicalis (43) is therefore one of the reasons why the maintenance of multiple and distinct class Ib lineages in these two species of frogs is suggestive of important conserved functions. One possible driving force behind the conservation of multiple class Ib lineages in two species of the Xenopodinae subfamily of amphibians might be their exposure to relatively similar pathogens. There is some overlap of the natural geographical niches that S. tropicalis and X. laevis occupy in the continent of Africa (9). These organisms may have therefore needed to develop a way to defend themselves against a common set of pathogens for which certain class Ib lineages may be specialized to recognize. Furthermore, in contrast to most mammals where there are three class Ia genes per haplotype, a large number of class Ib gene lineages in species of the amphibian subfamily Xenopodinae might serve to compensate for the presence of only one class Ia gene.
Nonfunctionalization of S. tropicalis class Ib gene lineage
The particular expression profile of the XNC11 gene (e.g., high expression by three different lymphoid tumors derived from independent spontaneous thymic tumors of genetically unrelated animals, very weak expression by thymocytes, and no expression elsewhere) suggests an association with tumorigenesis in X. laevis (13). In case XNC11 would, for example, provide some survival signal during thymocyte differentiation as some class Ib molecules do in mammals (44), one can conceive that a deregulated over-expression of the gene would be an advantageous step in neoplastic transformation. Alternatively, the aberrant expression of XNC11 in thymic tumors may just be related to aberrant regulation or the stage of thymic differentiation from which the tumor developed. Notably, no XNC11 homologue can be found in S. tropicalis, which makes us speculate that genes of the X. laevis XNC11 subfamily may not have been conserved in the Silurana lineage because of their potential association with tumors.
In X. laevis, transcripts of the XNC11 subfamily are most similar to the XNC5 subfamily (13). The S. tropicalis orthologue of the X. laevis XNC5 subfamily is the SNC5 gene. It is interesting to note that the SNC5 subfamily is monogenic. In fact, most class Ib gene lineages in S. tropicalis are monogenic (Fig. 2). Although the loss of duplicated genes over time does not need to be the result of a selection, the maintenance of many monogenic Xenopodinae class Ib gene lineages together with a few multigenic ones, all located in the same genetic region, suggests to us that some class Ib genes are under pressure to remain a single copy in the genome. In this regard, the targeted diploidization of the class Ib locus in polyploid species as a control of gene copy number is consistent with the idea that, as for class Ia, some class Ib genes cannot tolerate gene duplication because of a critical non-redundant dose-dependent function. For example, because there is only one copy of SNC5 in the S. tropicalis genome, it is possible that in X. laevis, XNC11 originally duplicated from XNC5, and the consequence of having more than one XNC5-like gene was a predisposition to tumors. This interesting possibility remains to be explored.
Stability of the divergent XNC/SNC10 Xenopodinae class Ib gene lineage
The XNC/SNC10 subfamily forms an independent phylogenetic clade that is intermediate between S. tropicalis and X. laevis class Ib and class Ia sequences. This topology suggests an independent evolutionary origin of XNC/SNC10 relative to other Xenopodinae class Ib lineages. SNC10 and XNC10 might have thus become fixed to serve a critical function early during Xenopodinae evolution. Notably, in both S. tropicalis and X. laevis there is only one gene representing this class Ib lineage, and genomic Southern analysis strongly suggests that the monogenic and diploidized state of this subfamily has also been maintained in other species, including polyploids. The fact that this subfamily is old and the gene dosage has been maintained to one is a strong argument for an important, nonredundant function.
Expression of XNC10 and SNC10 is associated with thymocyte differentiation during early larval development at the onset of thymic organogenesis. This is of particular interest given the intriguing issue, which is not yet fully elucidated, that X. laevis larvae have thymus-dependent circulating mature CD8 T cells but lack consistent class Ia surface expression, especially in the thymus, until metamorphosis (45). One can postulate that the relatively high level of SNC10 and XNC10 expressed in thymus, from early developmental stages, compensates for the lack or low level of class Ia expression. As such, it may be the case that in larvae, certain class Ib molecules, such as XNC10 and/or SNC10, are primarily and critically involved in regulating the differentiation of CD8 T cells needed for rapid protection during early development, considering that tadpoles are free swimming by 3 dpf. Larval sublethal gamma-irradiation experiments have shown that in the thymus, XNC10 and SNC10 are predominately expressed by the radiosensitive hematopoietic population. In mice, there are several examples of thymic T cells being selected on class Ib molecules expressed by hematopoietic cells, in the absence of class Ia. For example, the class Ib molecules TL and CD1d are expressed on cortical double positive thymocytes and seem to be the key to the development of a subset of γ/δ as well as NKT cells, respectively (46). These findings are consistent with a convergence of function within amphibian and mammalian class Ib. It will be fascinating to explore the possibility that because space and time are limited in Xenopus and Silurana larvae, they may preferentially generate class Ib-restricted “innate” T cells, which have the intrinsic property of an activated phenotype thus requiring much less expansion and time to carry out effector functions (47). This may constitute a critical defense strategy early in life when an organism’s adaptive immune system is not yet fully competent. Having cells around with an accelerated ability to detect and control danger might serve to manage a potential infection before it is allowed to establish. Furthermore, one of the useful features of X. laevis larvae as a potential model to study class Ib function is that this model is a natural system that has minimal contribution from class Ia.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants F31-AI068610 (to A.G.), AI27877 (to Y.O.), R03-HD-06167-01, and R24-AI-059830.
We thank Dr. Martin Flajnik for invaluable discussions. We also thank an anonymous reviewer for helpful suggestions and Hristina Nedelkovska for critical reading of the manuscript and useful suggestions. The expert animal husbandry of David Albright and Tina Martin are greatly appreciated.
Abbreviations used in this paper
- Ψ
pseudogene
- AN
accession number
- B
body
- class Ia
classical MHC class Ia
- class Ib
nonclassical MHC class Ib
- EST
expressed sequence tag
- H
head
- β2m
β2-microglobulin
- MYA
million years ago
- pPBR
predicted peptide-binding residue
- RACE
rapid amplification of cDNA ends
- RACE-PCR
rapid amplification of cDNA ends-PCR
- RT
reverse transcriptase
- SNC
S. tropicalis nonclassical MHC class Ib
- XNC
X. laevis nonclassical MHC class Ib
Footnotes
Disclosures
The authors have no financial conflicts of interest.
The online version of this article contains supplemental material.
References
- 1.Gleimer M, Parham P. Stress management: MHC class I and class I-like molecules as reporters of cellular stress. Immunity. 2003;19:469–477. doi: 10.1016/s1074-7613(03)00272-3. [DOI] [PubMed] [Google Scholar]
- 2.Gomes AQ, Correia DV, Silva-Santos B. Non-classical major histocompatibility complex proteins as determinants of tumour immuno-surveillance. EMBO Rep. 2007;8:1024–1030. doi: 10.1038/sj.embor.7401090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahram S. MIC genes: from genetics to biology. Adv Immunol. 2000;76:1–60. doi: 10.1016/s0065-2776(01)76018-x. [DOI] [PubMed] [Google Scholar]
- 4.Howcroft TK, Singer DS. Expression of nonclassical MHC class Ib genes: comparison of regulatory elements. Immunol Res. 2003;27:1–30. doi: 10.1385/IR:27:1:1. [DOI] [PubMed] [Google Scholar]
- 5.Adams EJ, Parham P. Species-specific evolution of MHC class I genes in the higher primates. Immunol Rev. 2001;183:41–64. doi: 10.1034/j.1600-065x.2001.1830104.x. [DOI] [PubMed] [Google Scholar]
- 6.Nei M, Gu X, Sitnikova T. Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc Natl Acad Sci USA. 1997;94:7799–7806. doi: 10.1073/pnas.94.15.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasahara M, Flajnik MF, Ishibashi T, Natori T. Evolution of the major histocompatibility complex: a current overview. Transpl Immunol. 1995;3:1–20. doi: 10.1016/0966-3274(95)80001-8. [DOI] [PubMed] [Google Scholar]
- 8.Kobel HR, Du Pasquier L. Genetics of polyploidy. Xenopus Trends Genet. 1986;2:310–315. [Google Scholar]
- 9.Evans BJ, Kelley DB, Tinsley RC, Melnick DJ, Cannatella DC. A mitochondrial DNA phylogeny of African clawed frogs: phylogeography and implications for polyploid evolution. Mol Phylogenet Evol. 2004;33:197–213. doi: 10.1016/j.ympev.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 10.Evans BJ. Genome evolution and speciation genetics of clawed frogs (Xenopus and Silurana) Front Biosci. 2008;13:4687–4706. doi: 10.2741/3033. [DOI] [PubMed] [Google Scholar]
- 11.Robert J, Ohta Y. Comparative and developmental study of the immune system in. Xenopus Dev Dyn. 2009;238:1249–1270. doi: 10.1002/dvdy.21891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flajnik MF, Kasahara M, Shum BP, Salter-Cid L, Taylor E, Du Pasquier L. A novel type of class I gene organization in vertebrates: a large family of non-MHC-linked class I genes is expressed at the RNA level in the amphibian. Xenopus EMBO J. 1993;12:4385–4396. doi: 10.1002/j.1460-2075.1993.tb06123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goyos A, Ohta Y, Guselnikov S, Robert J. Novel nonclassical MHC class Ib genes associated with CD8 T cell development and thymic tumors. Mol Immunol. 2009;46:1775–1786. doi: 10.1016/j.molimm.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, et al. The genome of the western clawed frog. Xenopus tropicalis Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohta Y, Goetz W, Hossain MZ, Nonaka M, Flajnik MF. Ancestral organization of the MHC revealed in the amphibian. Xenopus J Immunol. 2006;176:3674–3685. doi: 10.4049/jimmunol.176.6.3674. [DOI] [PubMed] [Google Scholar]
- 16.Wong WM, Au DM, Lam VM, Tam JW, Cheng LY. A simplified and improved method for the efficient double-stranded sequencing of mini-prep plasmid DNA. Nucleic Acids Res. 1990;18:5573. doi: 10.1093/nar/18.18.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 18.Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 19.Roelants K, Gower DJ, Wilkinson M, Loader SP, Biju SD, Guillaume K, Moriau L, Bossuyt F. Global patterns of diversification in the history of modern amphibians. Proc Natl Acad Sci USA. 2007;104:887–892. doi: 10.1073/pnas.0608378104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson JS, Reisz RR, Scott D, Fröbisch NB, Sumida SS. A stem batrachian from the Early Permian of Texas and the origin of frogs and salamanders. Nature. 2008;453:515–518. doi: 10.1038/nature06865. [DOI] [PubMed] [Google Scholar]
- 21.Shum BP, Avila D, Du Pasquier L, Kasahara M, Flajnik MF. Isolation of a classical MHC class I cDNA from an amphibian. Evidence for only one class I locus in the Xenopus MHC. J Immunol. 1993;151:5376–5386. [PubMed] [Google Scholar]
- 22.Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salter-Cid L, Nonaka M, Flajnik MF. Expression of MHC class Ia and class Ib during ontogeny: high expression in epithelia and coregulation of class Ia and lmp7 genes. J Immunol. 1998;160:2853–2861. [PubMed] [Google Scholar]
- 24.Braud VM, Allan DS, McMichael AJ. Functions of nonclassical MHC and non-MHC-encoded class I molecules. Curr Opin Immunol. 1999;11:100–108. doi: 10.1016/s0952-7915(99)80018-1. [DOI] [PubMed] [Google Scholar]
- 25.Rodgers JR, Cook RG. MHC class Ib molecules bridge innate and acquired immunity. Nat Rev Immunol. 2005;5:459–471. doi: 10.1038/nri1635. [DOI] [PubMed] [Google Scholar]
- 26.Saper MA, Bjorkman PJ, Wiley DC. Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 A resolution. J Mol Biol. 1991;219:277–319. doi: 10.1016/0022-2836(91)90567-p. [DOI] [PubMed] [Google Scholar]
- 27.Koller BH, Orr HT. Cloning and complete sequence of an HLA-A2 gene: analysis of two HLA-A alleles at the nucleotide level. J Immunol. 1985;134:2727–2733. [PubMed] [Google Scholar]
- 28.Madden DR, Gorga JC, Strominger JL, Wiley DC. The structure of HLA-B27 reveals nonamer self-peptides bound in an extended conformation. Nature. 1991;353:321–325. doi: 10.1038/353321a0. [DOI] [PubMed] [Google Scholar]
- 29.Shawar SM, Vyas JM, Rodgers JR, Rich RR. Antigen presentation by major histocompatibility complex class I-B molecules. Annu Rev Immunol. 1994;12:839–880. doi: 10.1146/annurev.iy.12.040194.004203. [DOI] [PubMed] [Google Scholar]
- 30.Sullivan LC, Hoare HL, McCluskey J, Rossjohn J, Brooks AG. A structural perspective on MHC class Ib molecules in adaptive immunity. Trends Immunol. 2006;27:413–420. doi: 10.1016/j.it.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 31.Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin) North Holland; Amsterdam: 1967. [Google Scholar]
- 32.Berg LJ. Signalling through TEC kinases regulates conventional versus innate CD8(+) T-cell development. Nat Rev Immunol. 2007;7:479–485. doi: 10.1038/nri2091. [DOI] [PubMed] [Google Scholar]
- 33.Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 34.Saltis M, Criscitiello MF, Ohta Y, Keefe M, Trede NS, Goitsuka R, Flajnik MF. Evolutionarily conserved and divergent regions of the autoimmune regulator (Aire) gene: a comparative analysis. Immunogenetics. 2008;60:105–114. doi: 10.1007/s00251-007-0268-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watkins DI, Chen ZW, Hughes AL, Evans MG, Tedder TF, Letvin NL. Evolution of the MHC class I genes of a New World primate from ancestral homologues of human non-classical genes. Nature. 1990;346:60–63. doi: 10.1038/346060a0. [DOI] [PubMed] [Google Scholar]
- 36.Cadavid LF, Shufflebotham C, Ruiz FJ, Yeager M, Hughes AL, Watkins DI. Evolutionary instability of the major histocompatibility complex class I loci in New World primates. Proc Natl Acad Sci USA. 1997;94:14536–14541. doi: 10.1073/pnas.94.26.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Houlden BA, Greville WD, Sherwin WB. Evolution of MHC class I loci in marsupials: characterization of sequences from koala (Phascolarctos cinereus) Mol Biol Evol. 1996;13:1119–1127. doi: 10.1093/oxfordjournals.molbev.a025674. [DOI] [PubMed] [Google Scholar]
- 38.Kobel HR, Barandun B, Thiebaud CH. Mitochondrial rDNA phylogeny in. Xenopus Herpetol J. 1998;8:13–17. [Google Scholar]
- 39.Cherty LM, Case SM, Wilson AC. Frog perspective on the morphological difference between humans and chimpanzees. Science. 1978;200:209–211. doi: 10.1126/science.635583. [DOI] [PubMed] [Google Scholar]
- 40.Cherry LM, Case SM, Kunkel JG, Wilson AC. Comparisons of frogs, humans, and chimpanzees. Science. 1979;204:435. doi: 10.1126/science.204.4391.435. [DOI] [PubMed] [Google Scholar]
- 41.Hughes MK, Hughes AL. Evolution of duplicate genes in a tetraploid animal. Xenopus laevis Mol Biol Evol. 1993;10:1360–1369. doi: 10.1093/oxfordjournals.molbev.a040080. [DOI] [PubMed] [Google Scholar]
- 42.Estes R. Xenopus from the Palaeocene of Brazil and its zoogeographic importance. Nature. 1975;254:48–50. [Google Scholar]
- 43.Cannatella DC, de Sa RO. Xenopus laevis as a model organism. Syst Biol. 1993;42:476–507. [Google Scholar]
- 44.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 45.Flajnik MF, Du Pasquier L. The major histocompatibility complex of frogs. Immunol Rev. 1990;113:47–63. doi: 10.1111/j.1600-065x.1990.tb00036.x. [DOI] [PubMed] [Google Scholar]
- 46.Ladi E, Yin X, Chtanova T, Robey EA. Thymic microenvironments for T cell differentiation and selection. Nat Immunol. 2006;7:338–343. doi: 10.1038/ni1323. [DOI] [PubMed] [Google Scholar]
- 47.Urdahl KB, Sun JC, Bevan MJ. Positive selection of MHC class Ib-restricted CD8(+) T cells on hematopoietic cells. Nat Immunol. 2002;3:772–779. doi: 10.1038/ni814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.