Abstract
We have developed a microfabricated biochip that enumerates CD4+ T lymphocytes from leukocyte populations obtained from human blood samples using electrical impedance sensing and immunoaffinity chromatography. T cell counts are found by obtaining the difference between the number of leukocytes before and after depleting CD4+ T cells with immobilized antibodies in a capture chamber. This differential counting technique has been validated to analyze physiological concentrations of leukocytes with an accuracy of ~9 cells per µL by passivating the capture chamber with bovine serum albumin. In addition, the counter provided T cell counts which correlated closely with an optical control (R2 = 0.997) for CD4 cell concentrations ranging from approximately 100 to 700 cells per µL. We believe that this approach can be a promising method for bringing quantitative HIV/AIDS diagnostics to resource-poor regions in the world.
Introduction
HIV/AIDS affects more than 33 million people throughout the world, and is an especially critical problem in resource-poor regions in sub-Saharan Africa, where 67% of all HIV/AIDS patients live.1 Antiretroviral therapy (ART) increases the longevity and quality of life for HIV patients, and global efforts increased the accessibility of such treatment 30-fold in sub-Saharan Africa between 2003 and 2008.2–4 However, the lack of objective diagnostic tests to determine when to start ART and to monitor its success hinders the effective use of treatment. In the absence of appropriate diagnostic tests, clinicians may rely on less accurate assessments, ranging from self-described symptoms or insensitive laboratory tests.
An important diagnostic procedure is to obtain a patient’s CD4+ T lymphocyte count.5–7 Flow cytometers using laser light scattering and laser-induced fluorescence principles are the current gold standard to provide accurate CD4+ T cell counts at high throughputs. However, the cost and technical requirements of such instruments are too taxing for the debilitated healthcare infrastructures of resource-poor regions.
As an answer to this problem, much effort has been placed on creating highly specialized microfabricated point-of-care (PoC) biochips for HIV/AIDS analysis. Such a technology may use electrical and/or optical sensing methods, and would greatly reduce initial and operating costs, increase portability, and be simpler to use by more healthcare workers.
Many efforts have used fluorescent tagging and subsequent image processing to automatically enumerate CD4+ T lymphocytes in microchambers. Some designs relied on the even distribution of cells in a plastic chamber to produce accurate counts.8 Others have used a microfabricated membrane to filter out erythrocytes, leaving leukocytes, which were fluorescently labeled.9 Fluorescence detection was later enhanced by using quantum dots.10 Immunospecific paramagnetic beads were used to bring fluorescently labeled CD4+ T cells into the focusing plane for analysis, reducing counting error.11,12 Cheng et al. have investigated CD4+ T cell capture by controlling shear stresses at the chamber walls and enumerating cells using a cocktail of fluorescently labeled antibodies.13,14 They improved their design by including a monocyte depletion chamber to reduce the positive bias created at lower CD4+ T cell concentrations.15
The aforementioned optical methods require the use of lenses and focusing to analyze samples, but this can increase the cost and decrease the portability of the device. Gohring and Fan quantified the antibody-mediated attachment of T cells by the amount of shift in the whispering gallery mode of an optofluidic ring resonator.16 Moon et al. counted immobilized helper T cells by their shadows cast over a charge coupled device (CCD) by a white light source.17 Wang et al. further simplified the optics by not requiring an external light source: immobilized CD4+ T cells were labeled with CD3-conjugated horseradish peroxidase to facilitate a chemiluminescent reaction, which was amplified and quantified by a photodetector.18
Electrical methods for enumerating CD4+ T lymphocytes prove to be promising due to the fact that they would not require optical components such as lenses, filters, light sources, photo-detectors, and CCDs, which can be expensive, bulky, fragile, and require periodic maintenance. An electrical PoC solution could require only solid state components to electrically interrogate a sensing geometry, process sensor output, and provide input from and results to the user.
Recent advancements have used the Coulter principle to electrically analyze cells individually within a population.19,20 Adams et al. enumerated low concentrations of circulating tumor cells in whole blood samples after specifically capturing and releasing them.21 Holmes et al. have used impedance analysis at multiple frequencies as a label-free method to differentiate between different leukocyte subsets, and were able to further enhance electrical differentiation by specifically attaching latex beads to CD4+ T cells.22,23 Lee et al. have developed a method to find the concentration of erythrocytes by subtracting the total number of cells that are entering a control volume from the total number of cells that are exiting the control volume. The concentration was found in real time by dividing this difference by the control volume.24
Instead of electrically analyzing cells one-by-one, others have interrogated entire populations to estimate the helper T cell concentration. Mishra et al. used three electrode cyclic voltammetry to estimate the number of CD4+ T cells that were selectively captured on a working electrode.25 Jiang and Spencer have used this design as a building block to create an array of 200 of these electrochemical sensing regions, or “pixels”, conjugated with CD4 antibody. A pixel would be considered “on” when a CD4+ T cell was captured, and a total cell count was the total number of “on” pixels.26 In an earlier report, we have correlated the number of selectively captured CD4+ T cells with the impedance of their lysate, achieving a detection threshold of 20 cells per µL.27
We have also previously shown that we could obtain accurate counts for homogenous populations of CD4+ T lymphocytes using electrical impedance sensing in a microfabricated biochip.28 However, a next major step toward a practical PoC was needed to selectively enumerate the T cells among the various cell types found in blood.
In this paper, we present a novel device and analysis method to accurately enumerate CD4+ T lymphocytes from healthy human subject blood samples. We lyse the erythrocytes off-chip using standard lysing protocols and then use electrical impedance sensing coupled with immunoaffinity chromotography to electrically differentiate between CD4+ cells and other leukocytes. This method incorporates the inherent accuracy of individually analyzing cells one-by-one, identifies CD4+ T cells without the need for labeling or optical detection, provides results that do not require further analysis or gating by a healthcare worker, and comprises a streamlined and inexpensive sensing geometry amenable to low-cost fabrication.
Principle
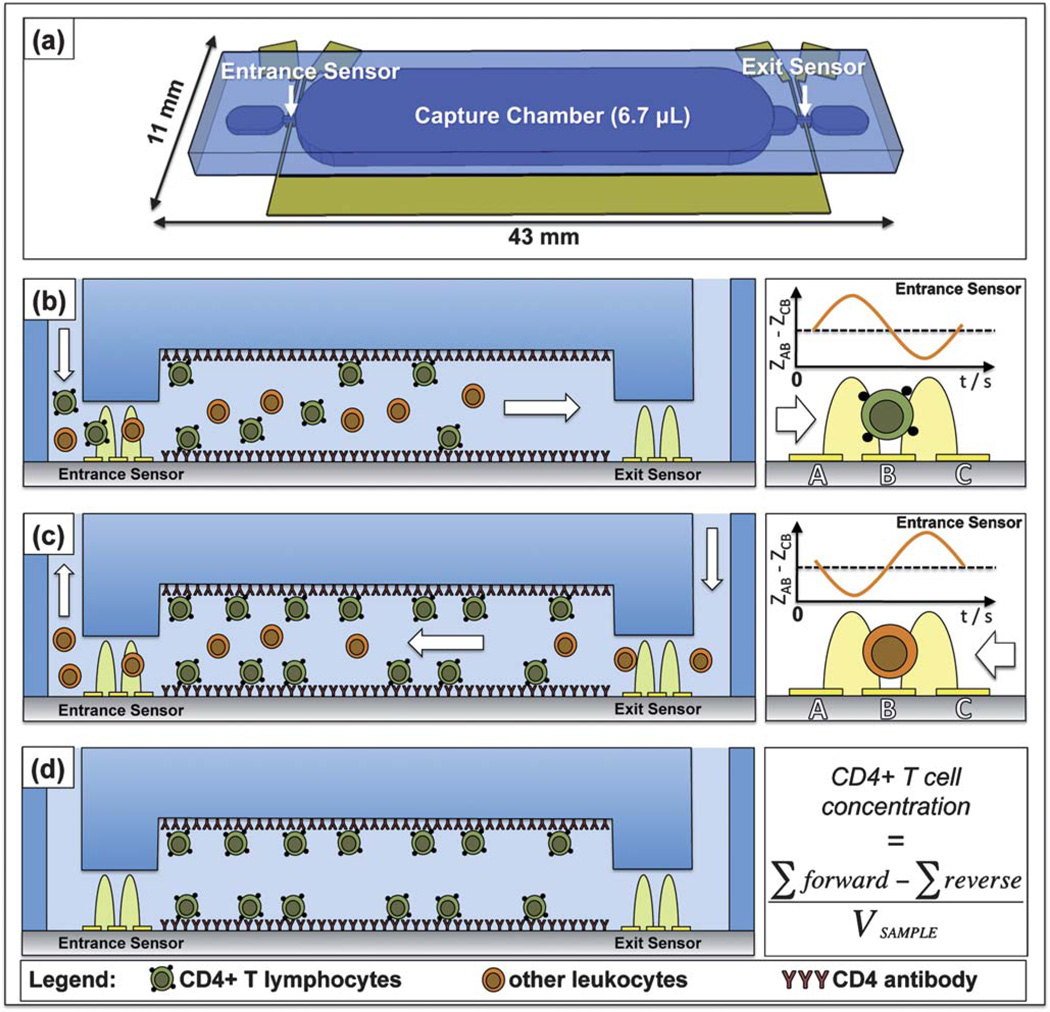
Fig. 1(a) depicts the microfabricated differential counting device that uses affinity chromatography, electrical impedance sensing, and a reverse-flow method to selectively enumerate CD4+ T cells. The main feature of the chip is its large capture chamber, which is conjugated with anti-human CD4 antibody to selectively isolate and deplete all CD4+ T cells from the heterogeneous leukocyte population, which we have described previously.13 Monocytes also express the CD4 antigen and bind to the capture region, but can be removed by the application of a controlled shear stress, as monocytes contain an order of magnitude less CD4 surface antigen than T cells.13,29,30 The electrical sensors are designed to count individual cells using the Coulter principle, where the passage of a cell perturbs the electrical current within an orifice, creating a distinct impedance pulse. The chip contains two such counters, both configured to measure the relative impedance change caused by the passage of a cell. Specifically, each counter is comprised of three electrodes, configured so that the impedance signal between electrode pairs B and C is subtracted from the signal between pairs A and B (Fig. 1(b), right panel). This self-referencing design is valuable in impedance spectroscopy in microfluidics31,32—and is especially useful in our case, as it identifies which direction cells are flowing within the chip. A single cell entering the chip through the entrance port creates an up-down pulse pair, while cells exiting back out of the entrance port create down-up pulse pairs (Fig. 1(b and c), right panel).
Fig. 1.
Principle of electrical differential counting of CD4+ T cells. (a) Graphically-rendered image depicting the chip’s geometry, specifically its two electrical counters and capture chamber. Cross-sectional views of the chip during an experiment: (b) forward flow direction to obtain total leukocyte count at the entrance counter, (c) reverse flow direction and enumeration of uncaptured cells after leukocytes reach the exit sensor, and (d) finished experiment after all unbound leukocytes are washed from the capture chamber. The concentration of CD4+ cells can be obtained simply by normalizing the differential count by the known sample volume ((d), right panel). The right panels of (b) and (c) illustrate how cell flow direction at the entrance counter can be determined by the change in pulse signature polarity, with the pulse signature changing from up-down (b) to down-up (c) in time.
Fig. 1(b–d) illustrates the reverse-flow differential counting method used to obtain accurate and objective cell counts. Red blood cells from whole blood samples are lysed off-chip, and a pure leukocyte population is injected into the chip’s entrance. All white blood cells are counted as they pass through the entrance counter and enter a chamber functionalized with CD4 antibody. This capture chamber depletes CD4+ T cells from the leukocyte population (Fig. 1 (b)). Flow direction is reversed once cells are detected at the exit counter, and uncaptured or nonspecifically bound cells are washed back out of the chip’s entrance after being enumerated again by the entrance counter (Fig. 1(c)). Fig. 1(d) depicts a completed experiment in which the capture chamber contains only specifically bound CD4+ cells. The differential count depicts the number of captured CD4+ T lymphocytes by obtaining the difference between the forward and reverse counts. Actual helper T cell concentrations can simply be found by normalizing this difference to the volume of sample flowed into the chip.
Although this application is focused on CD4+ T cell enumeration for HIV/AIDS diagnostics, it can be expanded to analyze other cell types simply by choosing different capture antibodies for the particular cell surface antigen of interest.
Materials and methods
Channel and sensor design
Each counter was comprised of a 15 µm-wide and 15 µm-high microchannel that funneled the cells over three 10 µm-wide platinum electrodes (with 10 µm spacings). The counting channel dimensions were chosen to be large enough to facilitate leukocyte passage without clogging (leukocytes have an average diameter of ~13 µm),33 but small enough to ensure the cells flowed past the electrodes in a single file manner with small variation in passage height to ensure consistent enumeration of cells.28,31 The counting electrodes’ dimensions were chosen to provide adequate spatial resolution (50 µm) and to create a sensing region volume of ~11 pL (50 µm × 15 µm × 15 µm), both to ensure coincident events would not occur for leukocyte concentrations up to ~9 × 104 cells per µL, well above the 4–11 × 103 cells per µL physiological range found in humans.34
The 50 µm height of the capture region was chosen to provide a large enough sample volume of ~7 µL for an accurate depiction of a patient’s health and to ensure proper shear stresses at the fluid–wall interface for optimal CD4+ T cell capture. According to Cheng et al., a shear stress greater than 3 dyn cm−2 resulted in less effective CD4+ T cell capture.13 The equation
can be used to estimate the shear stress at the walls of a rectangular microfluidic channel of a constant width, ω1, where μ is the dynamic viscosity of the fluid, Q is the volumetric flow rate, and h is the height of the channel.35 This shows the sensitive, inverse-squared relationship between the channel height and the shear stress at the chamber’s floor and ceiling. If the capture chamber remained the same height as the counter channels (15 µm), the capture chamber would give a shear stress of over 5.5 dyn cm−2 at Q = 5 µL min−1, which would be over the aforementioned 3 dyn cm−2 upper boundary. However, a 50 µm capture chamber height significantly reduces the average shear stress to 0.5 dyn cm−2, greatly increasing the interactions between the cells’ surface antigens and the immobilized Ab, which facilitate cell capture. The 34 mm capture chamber length ensured sufficient interaction time (~80 s at sample flow rate of 5 µL min−1 for flowing in a single direction).
Differential counter fabrication
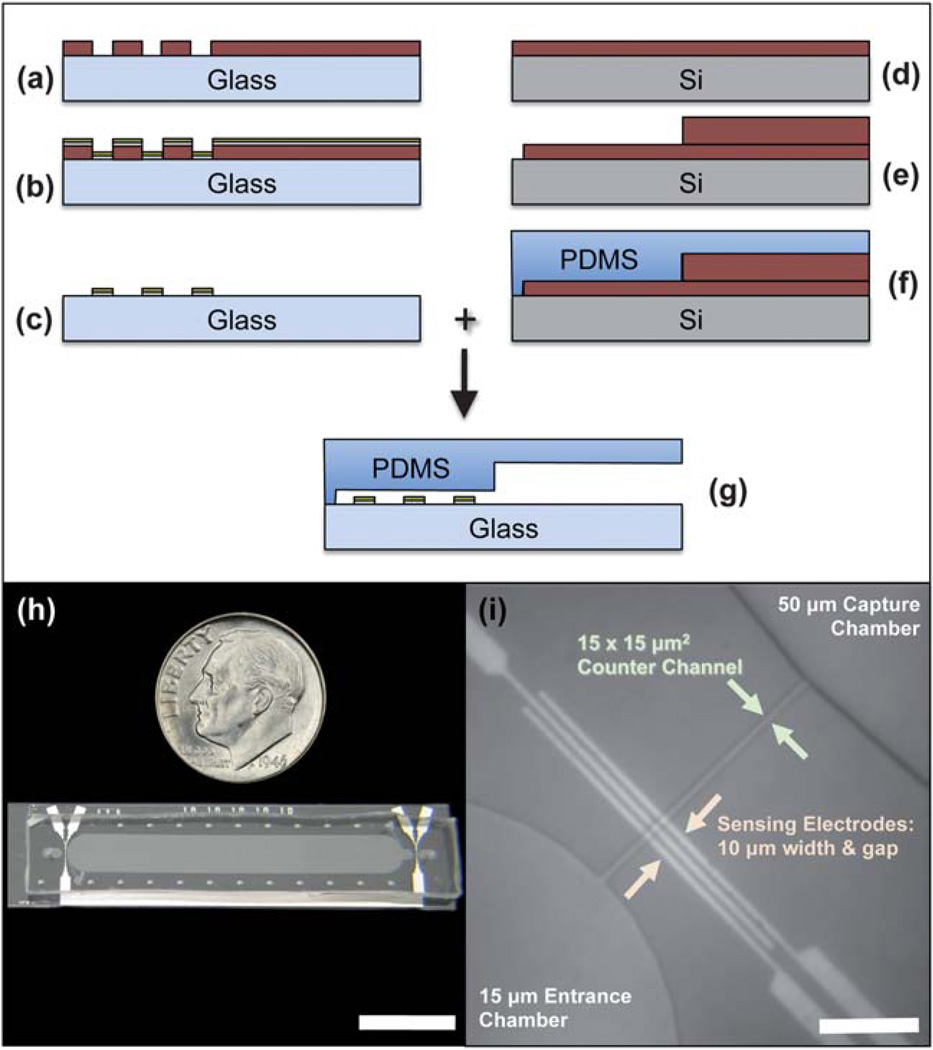
Fabrication of the differential counter is illustrated for one counter region in Fig. 2(a–g).
Fig. 2.
(a–g) Microfabrication process flow for the electrical differential counter chip. (h) Fabricated chip compared to the size of a US dime (scale bar length is 10 mm). (i) Micrograph of an entrance counter region, depicting the three-electrodes setup, narrow counter channel, and 50 µm-high capture chamber (scale bar length is 100 µm).
The electrical sensing layer was fabricated using the standard metal lift-off process. A 4″ glass wafer (Pyrex 7740) was first spin-coated with LOR2A liftoff resist (Microchem Corp., Newton, MA), soft-baked at 183 °C for 5 minutes, and then coated with Microposit S-1805 photoresist (Rohm and Haas Electronic Materials, Marlborough, MA). After another soft-bake at 110 °C for 90 s, the wafer was aligned to the electrodes mask on a Quintel (Neutronix-Quintel, Morgan Hill, CA) Q7000 mask aligner and exposed for a total dose of 2.8 mJ cm−2. The wafer was then placed on a 110 °C hotplate for a 60 s post-exposure bake before being immersed into Microposit MF CD-26 developer (Rohm and Haas Electronic Materials) for 80 s and rinsed with DI water for 2 minutes (Fig. 2(a)). The wafer was then de-scummed in an O2 plasma system for 20 s before being placed in a CHA SEC-6000 evaporator (CHA Industries, Fremont, CA) for the deposition of 25 nm of Ti adhesion layer, followed by a 75 nm Pt conduction layer (Fig. 2(b)). The undesired metal was lifted off by placing the wafer in a 70 °C bath of Microchem Remover PG for 15 min, creating the necessary conduction paths for the counters (Fig. 2(c)).
The multi-height fluidics layer was created by fabricating a negative image of the desired channels using Microchem SU-8 25 photoresist. SU-8 25 was spun on a 4″ Si wafer to a height of 15 µm, and was pre-baked in two steps for 2 minutes at 65 °C and then 95 °C for 5 minutes. The wafer was then aligned and exposed to a mask defining all of the fluidic channels, including the capture region, counters, sample inlet and outlet, and antibody functionalization ports (Fig. 2(d)). A second layer of SU-8 was spun on to obtain a total thickness of 50 µm for the entire wafer, and was pre-baked first at 65 °C for 5 minutes and then 95 °C for 15 minutes. The wafer was then exposed to a second mask that only defined the capture chamber, allowing it to have a height of 50 µm, compared to the other fluidic regions of 15 µm in height. The wafer was developed in Microchem SU-8 developer for 2 minutes at room temperature, rinsed with isopropyl alcohol, and hard-baked at 125 °C for 15 minutes (Fig. 2(e)). Poly(dimethylsiloxane) (PDMS), 1 : 10 ratio of curing agent to base, was poured over the negative mold and allowed to cure overnight at 65 °C (Fig. 2(f)). The polymerized mold was peeled off, and ports were punched for all inlets and outlets using a blunt syringe needle.
A sealed fluidic chip was completed by aligning and bonding the electrode sensing layer to the fluidics layer after oxygen plasma activation in a barrel etcher (Fig. 2(g)). Teflon microbore tubing was used to make fluidic connections among the chip, valves, and syringe pumps.
Fig. 2(h) compares the footprint of the completed chip to a US dime, showing how the device could be manufactured as a single-use, disposable chip. Fig. 2(i) is a micrograph of the entrance counter region.
Obtaining purified leukocytes from whole blood
Red blood cells were lysed before flowing samples through the differential counter chip, as the concentration of erythrocytes would be too high to analyze at the desired flow rates and could also introduce additional counting error. On the day of the experiment, 4 mL samples of whole blood from healthy donors were collected in EDTA-coated Vacutainer tubes (BD Biosciences, Franklin Lakes, NJ). Twelve mL of lysis solution (0.12% (v/v) formic acid and 0.05% (w/v) saponin in DI) was added to 1 mL of whole blood and incubated for 6 s with agitation. Lysis was immediately stopped by the addition of quenching solution (5.3 mL of 0.6% (w/v) sodium carbonate and 3% (w/v) sodium chloride in DI).22 The solution was centrifuged for 5 minutes at 200× gravity at room temperature, supernatant was aspirated, and the leukocyte pellet was resuspended in 5 mL PBS + 1% (w/v) bovine serum albumin (BSA). The suspension was centrifuged again and resuspended in 1 mL PBS + 1% BSA, giving the physiological concentration of white blood cells. The volume of resuspension PBS was also varied to provide different leukocyte concentrations.
The saponin/formic acid lysing method was chosen because it proved to be the most rapid and effective erythrocyte lysis technique and ensured cell sample concentrations were low enough to be successfully analyzed by the electrical counter (see Maximum pulse density sub-section, below). However, this aggressive lysing method has been known to differentially damage the membranes of leukocyte subtypes with possible loss of viability.22 Flow analysis showed that the saponin/formic acid lysis agent did decrease the total leukocyte concentration when compared to a commercially available solution (BD Pharm Lyse™, BD Biosciences), but T lymphocyte and monocyte populations were not affected. This held true even for varying the duration of incubation in the lysis solution (3 s to 18 s). Leukocyte vitality was preserved by resuspending the cells in a PBS + 1% BSA solution immediately after halting the lysis process with the quenching solution.
Experimental setup and reverse-flow differential counting technique
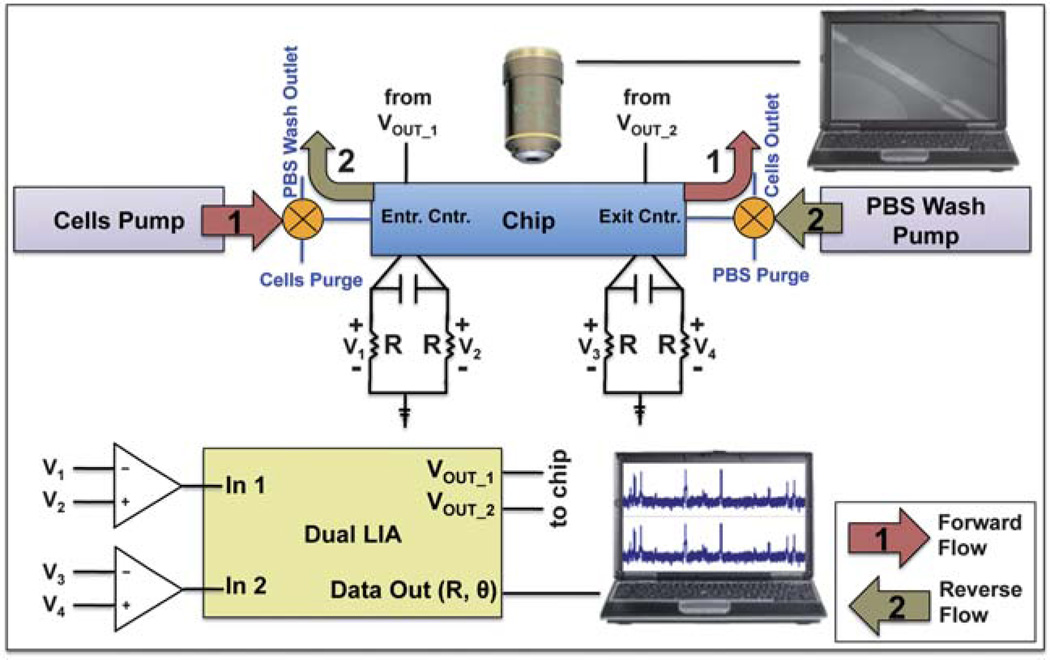
Fig. 3 illustrates the experimental setup and process used to differentially count CD4+ T cells. The process can be split into two segments: forward flow (Fig. 3(1)) and reverse-flow analysis (Fig. 3(2)). A Harvard Apparatus (Holliston, MA) PicoPlus syringe pump injected a constant stream of leukocytes into the chip through the entrance counter at 5 µL min−1 to create a forward count of all leukocytes. The enumerated cells entered the CD4 capture chamber and were depleted of CD4+ T cells. Forward flow continued until cells were detected at the exit counter. At this time, flow direction was reversed by switching the entrance and exit valves and by initiating the PBS+ 1% BSA solution wash pump at 5 µL min−1. The unattached leukocytes were washed back out the capture chamber and entrance counter to be enumerated, creating a reverse count. The total number of captured CD4+ T lymphocytes could be simply calculated by subtracting the reverse count from the forward count.
Fig. 3.
Experimental setup and process for electrical differential counting of CD4+ T cells. A dual lock-in amplifier is used to inject an AC signal and monitor its change through the chip’s counter regions, which are balanced by a Wheatstone configuration. Process: (1) the cells pump injects leukocytes into the chip (forward direction) to be enumerated at the entrance counter; (2) after cells are sensed at the exit counter, both valves are switched to reverse flow direction and the PBS wash pump purges the capture chamber of uncaptured cells, which are enumerated again by the entrance counter.
Counting was performed using a Zurich Instruments (Zurich, Switzerland) HF2LI dual lock-in amplifier which injected a 5 V (rms) 1.1 MHz AC signal into the entrance and exit counters (VOUT_1 and VOUT_2, respectively). Relative impedance was measured using three microelectrodes setup in a Wheatstone bridge configuration, balanced with 10 kΩ resistors (R) and a 68 pF capacitor. For each counter, the voltage signal across each resistor was subtracted (e.g., V2 − V1, or V4 − V3) using a differential preamplifier before entering the lock-in’s input. An increased sensitivity and stable impedance signal were some of the benefits of this balancing bridge method, but the major advantage of the three-electrode configuration was that one could precisely and accurately determine when the forward counting stopped and reverse counting started, as already described.
The bridge potential difference signals for the entrance and exit counters were input into the HF2LI, and the impedance magnitude and phase angle (R and θ, respectively) were output to a computer for real-time observation and recording of data at a 200 kHz sampling rate using a National Instruments (Austin, TX) PCI-6351 DAQ card and Lab-VIEW software. The data were imported into and analyzed with Clampfit software. Impedance pulses were counted using a threshold detection method, and forward and reverse counts were compared.
Another computer connected to a digital camera on a Nikon Eclipse E600FN microscope (Nikon Instruments, Inc., Melville, NY) was used to observe cell passage through the channels as well as cellular interactions with the capture chamber’s walls.
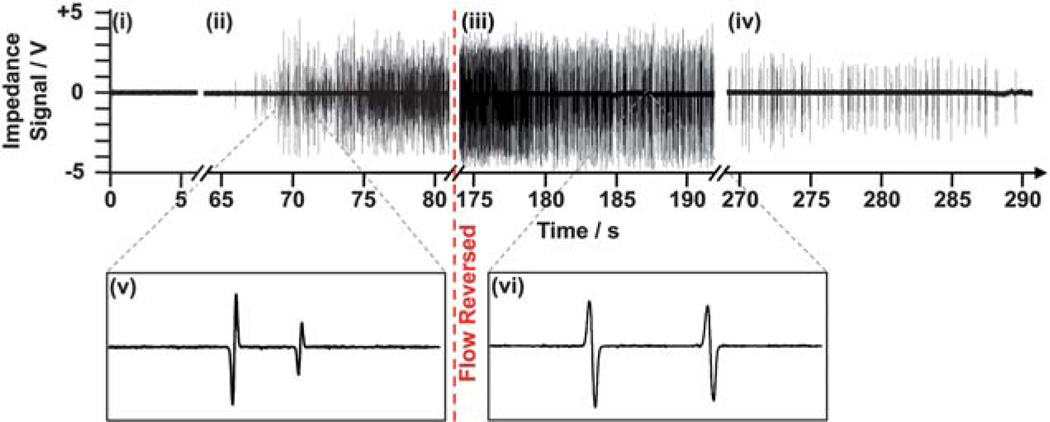
Fig. 4 shows an example of the reverse-flow differential counting method from actual experimental data. Flow reversal caused impedance pulses to invert polarity from down-up (v) to up-down (vi), exhibiting a straightforward method to differentiate between cells entering and exiting the chip.
Fig. 4.
Impedance data of the entrance counter for the entire duration of a reverse-flow differential counting experiment. The data can be separated into four regimes: (i) no cells present, (ii) cells beginning to enter the chip’s entrance, (iii) just after reversing flow and counting cells exiting back out the entrance, and (iv) near the end of purging the capture chamber of uncaptured cells. Magnifications of the impedance signal during forward (v) and reverse flow (vi) show how pulse signature polarity can be used to ensure when reverse counting begins.
Maximum pulse density limit
It is imperative that the differential counter can analyze physiological concentrations of white blood cells (~1 × 104 cells per µL) flowing at the desired rate of 5–10 µL min−1. This flow rate range would provide as rapid sample analysis as possible, given the limitations of (1) creating a capture chamber shear stress <3 dyn cm−2, (2) ensuring the capture chamber volume is large enough to contain an adequate sample volume (≥5 µL), (3) creating a capture chamber (and chip) footprint that would be amenable to PoC applications (smaller than a microscopy slide, ~75 mm × ~25 mm), and (4) ensuring the pulses are not attenuated by the lock-in amplifier’s low pass filter (in our case of AC analysis). In the last case, as flow rate is increased, impedance pulses become shorter in duration and contain mostly frequency components that exceed the filter’s bandwidth. These components are attenuated, resulting in diminished pulses that cannot be detected above the baseline noise level.
A maximum pulse density (pulses per s) must be found to test whether the electrical counters can accurately count physiological concentrations of leukocytes at the aforementioned flow rate range. Some limiting factors for accurate counting in any electrical cytometer are (1) the concentration of cells in the sample, (2) the speed in which they flow through the sensor, and (3) the measurement system’s sampling frequency. As the concentration of cells increases at a constant flow rate, the duration between passage events decreases. Eventually, the concentration becomes high enough where two cells will be in the same sensing region, creating coincidence events that cause two impedance pulses to overlap. For a finite sampling frequency—even if the cells are not coincident within the sensing region—a high enough velocity will eventually cause overlap of the pulses from two subsequent cell passages.
Hence, we found the maximum pulse density by flowing increasing concentrations of blood cells into the chip until noticeable pulse overlap occurred (i.e., an up-down or down-up pulse signature was not completed before another cell passed through the sensing region).
Reverse-flow differential technique validation
The described reverse-flow differential counting technique was implemented in response to problems found using a simpler flow-through method, where flow was constant in one direction and both entrance and exit sensors were used to quantify the number leukocytes before and after depletion of CD4+ T cells, respectively. The difference in the electrical characteristics between each counter and its respective external circuitry made it impossible to objectively compare the counting results between both counters. Although both counters could detect cell passage, their sensitivities differed enough to make subsequent analysis subjective. For example, the average impedance pulse height at the entrance counter may be lower than that found at the exit counter for the same population of cells or beads with the capture chamber passivated from specific and non-specific adsorption. Ideally, both counters would have the same sensitivity and produce the same average impedance pulse heights for identical populations, resulting in a differential count of zero. However, using the same threshold trigger level (determines at what height an impedance pulse is a cell) would result in the non-real result of a larger exit count than entrance count—a negative differential count. Scaling the trigger level based on average pulse height, noise level, and other variables proved unsuccessful and subjective, precluding further application.
The reverse-flow differential counting method would remove the ambiguity in electrical analysis and lessen the counting error by using the entrance counter alone to enumerate cells entering and exiting the chip. Because only one sensor is being used, the same threshold trigger level can be used for both the forward and reverse counts, resulting in a more straightforward and objective method to obtain differential counts. The exit counter would only be used as a qualitative means to determine when to reverse fluid flow direction to prevent cells from being lost through the chip’s exit and to initiate reverse counting.
Experiments were designed to evaluate the plausibility of the reverse-flow differential counting technique and to quantify its inherent error. This can be found by passivating the capture chamber from any cellular interactions—specific and nonspecific—and performing differential counts of leukocyte populations. Ideally, a passivated capture chamber would result in equal forward and reverse counts (a differential count of zero), but error may come from imperfections in the passivation layer, cell disruption inside the capture chamber, and the electrical analysis itself.
The capture chamber was passivated by flowing in phosphate buffered saline (PBS, pH 7.4), with 1% BSA (A3059-50G; Sigma–Aldrich, St Louis, MO) and incubating for at least one hour at room temperature to prevent the non-specific adsorption of cells. BSA is a well-known protein for surface passivation, and readily binds to the hydrophilic glass substrate at pH 7.4.36 The passivated chip was then evaluated using the reverse differential counting technique, described above, for various leukocyte dilutions.
Capture chamber modification for CD4+ T lymphocytes
Other experiments were performed to enumerate CD4+ T lymphocytes in the heterogeneous leukocyte populations by selectively immobilizing them in the capture chamber, which was functionalized with CD4 antibody.
Purified mouse antibody to human CD4 (MHCD0400) from Invitrogen, diluted (1 : 10) in PBS, was flowed through a newly fabricated chip until the entire capture chamber was filled. Flow was stopped, and the solution was allowed to incubate at room temperature for 30 minutes, followed by two more filling plus incubation cycles. A solution of PBS + 1% BSA was used to wash away unbound CD4 antibody and block any unconjugated sites from the nonspecific binding of cells. Chamber functionalization was performed either immediately before flowing leukocyte samples or the day before with the functionalized chip stored overnight at 4 °C.
CD4+ T-lymphocyte count comparison with optical control
An optical counting method using image processing was implemented to evaluate the electrical microcytometer chip’s differential counting technique. Immediately after electrical differential counting was finished, the chip’s entire capture region was sequentially imaged using an Olympus (Tokyo, Japan) IX81 inverted microscope with phase contrast under a total magnification of 64. Images were merged using standard imaging software, and cells were automatically enumerated in ImageJ to find the actual number of cells that were captured. This count was then compared with the electrical differential count.
Cell capture efficiency
A BD Biosciences LSRII flow cytometer was used to quantify the efficiency of the chip in capturing CD4+ T lymphocytes. Two leukocyte populations were analyzed: cells before flowing into the chip and cells that have exited the chip after a differential counting experiment. Alexa Fluor® 488-conjugated mouse antibody to human CD4 (MHCD0420) was purchased from Invitrogen. Phycoerythrin-Cy7 (PE-Cy7™)-conjugated mouse antibody to human CD14 (25-0149-42) and peridinin chlorophyll protein-cyanine 5.5 (PerCP-Cy5.5™)-conjugated mouse antibody to human CD3 (45-0037-42) were purchased from eBioscience (San Diego, CA). Both populations were stained with a cocktail of the above labels by following the manufacturer’s suggested protocols, were rinsed with centrifugation (200× g, 5 minutes), and resuspended in PBS for flow analysis to compare the percentages of CD4+ T lymphocytes (CD3+ CD4+ CD14−) in both populations.
Cell capture purity
After optical control micrographs were obtained, the captured cells were fluorescently labeled to examine their phenotype and to understand the capture purity of the system (i.e., the ratio between CD4+ T lymphocytes captured and total cells captured). Alexa Fluor® 488-conjugated mouse antibody to human CD4 (MHCD0420), allophycocyanin (APC)-conjugated mouse antibody to human CD14 (MHCD1405), and 4′,6-diamidino-2-phenylindole (DAPI, D21490) were purchased from Invitrogen (Carlsbard, CA). The CD14 antibody was used to determine which CD4+ cells were monocytes, which express both the CD4 and CD14 surface proteins. DAPI was used to determine which cells were nucleated, in the case un-lysed erythrocytes were nonspecifically captured. A 1 : 20 cocktail of the above three fluorophores was created in PBS and flowed into the chip at 2.5 µL min−1 for a total volume of 50 µL. Flow was stopped and the mixture was allowed to incubate for 30 additional minutes. This was followed by a 5 µL min−1 wash with PBS + 1% BSA to ensure unbound labels were purged from the chip. Fluorescent micrographs of selected areas of the chip were taken to create a representative of the entire population using the Olympus IX81 with appropriate fluorescent filter sets at a total magnification of 320.
Results and discussion
Maximum pulse density limit
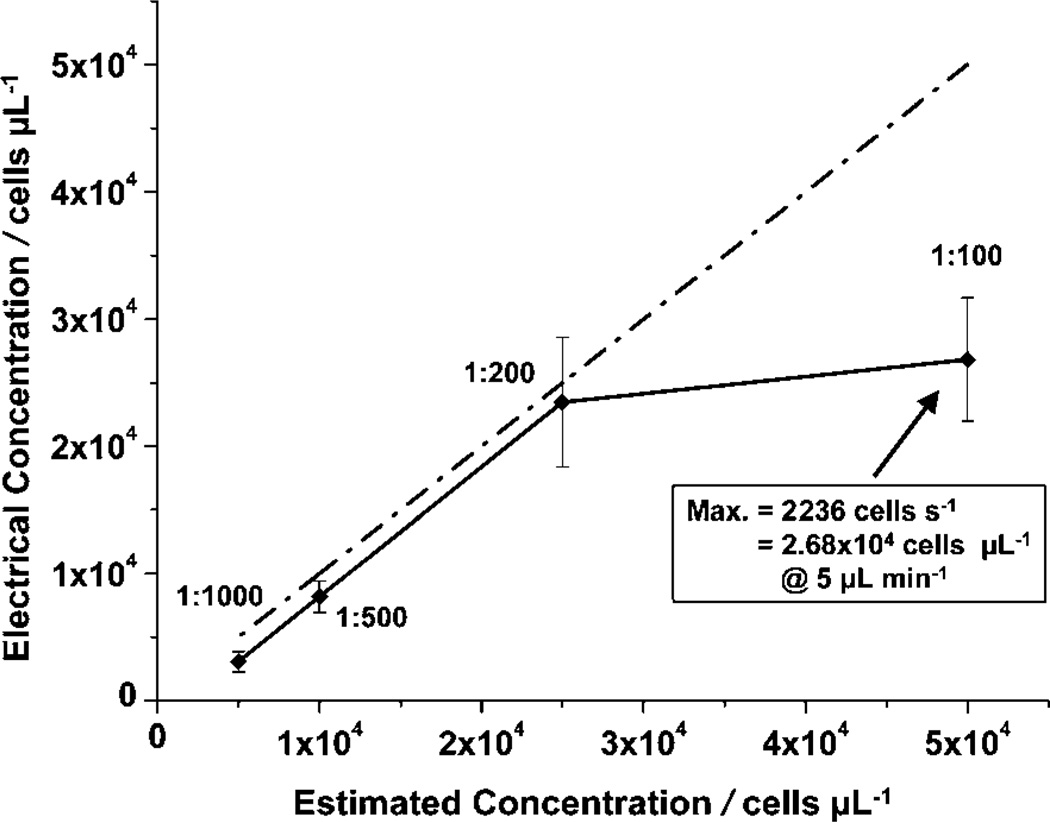
Varying dilutions (1 : 1000 to 1 : 100) of whole blood were injected into a chip at a constant flow rate of 5 µL min−1 to test the pulse density limits (pulses per s) of the differential counter. Flow direction was not reversed (kept in forward direction), and pulses were only analyzed for the entrance counter. Pulse density was calculated by normalizing the total number of pulses found by the number of fixed time windows that were applied at random points in the raw data.
Fig. 5 compares the cell concentration found using the chip (calculated by the number of pulses for a known volume flowed) versus the calculated concentration of each sample dilution, assuming the concentration of whole blood was 5 × 106 cells per µL. At a 5 µL min−1 flow rate, the chip could handle the 1 : 200 dilution of whole blood (~2.5 × 104 cells per µL), but failed to count every pulse individually for the 1 : 100 dilution (~5 × 104 cells per µL) because of pulse merging. The maximum pulse density the chip could handle was 2236 cells per s, equivalent to a concentration of 2.68 × 104 cells per µL at a flow rate of 5 µL min−1. While in some pathologic conditions physiological leukocyte counts can exceed this threshold, the vast majority of leukocyte counts will be lower. This maximum pulse density also confirms that effective lysis of erythrocytes is necessary before sample entry to avoid cell counting error.
Fig. 5.
Maximum pulse density characterization. Concentration calculations were based on the number of pulses per s for a known flow rate. Error bars represent the standard deviations obtained from measurements of 9 (1 : 1000), 14 (1 : 500), 25 (1 : 200), and 23 (1 : 100) duration windows.
A possible explanation for the discrepancy between the theoretical (9 × 104 cells per µL) and the observed concentration maxima (2.68 × 104 cells per µL) is the theoretical calculations assumed an even distribution of cells in the sample volume. However, cells may not be evenly distributed at the microscale, as they could travel in packets that result in coincidence events (multiple cells in the sensing region). This is likely with the higher concentrations (i.e., 1 : 100) and effectively reduced the maximum cell concentration that the electrical counters could handle.
The sampling frequency (115.2 kHz was used in this particular experiment) was not a limitation, as cell velocity through the sensing region (370 mm s−1 for Q = 5 µL min−1) was not high enough to cause two pulse signatures to overlap from consecutive cell passages. Cell translocation time through the sensing region was 135 µs, or ~15 data samples. This was well above our estimated minimum of 5 samples needed to resolve the passage of a cell and to distinguish between two different cells—as long as they were not simultaneously within the sensing region. (This estimated minimum sample number was determined by assuming distinct cell passage could be resolved if impedance signal values were recorded at the (1) baseline immediately before cell passage, (2) first peak (positive- or negative-going), (3) baseline between peaks, (4) second peak (opposite the polarity of the first peak), and (5) baseline after both peaks.)
Reverse-flow differential technique validation
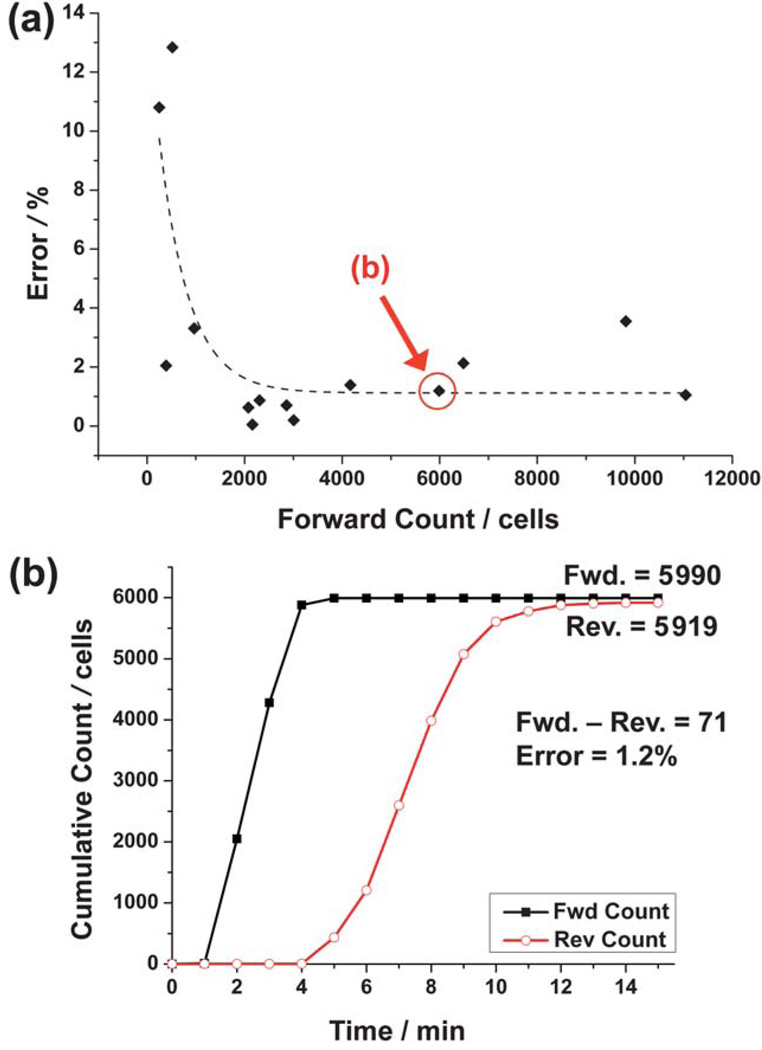
Fig. 6(a) illustrates the results for 14 experiments on the analysis of leukocyte populations using the reverse differential counting method with a passivated capture chamber. Leukocyte sample dilutions ranged from 0.25 to 2 times the physiological concentration to understand the inherent error of the device and technique for a range of leukocyte counts. The percent error decreased for larger forward counts (i.e., leukocytes entering the chip), which is desirable for clinical situations where samples will contain physiological concentrations of leukocytes. The main reason for this was that the absolute counting error remained roughly constant for the entire forward count range. (The percent error here is the absolute counting error normalized by the forward count, while the absolute counting error is the absolute difference from the ideal differential count of zero.)
Fig. 6.
Analysis of reverse differential counting for a passivated capture chamber. (a) Percent error decreases exponentially (dashed line, R2 = 0.57) as the number of leukocytes entering the chip (forward count) increases. (b) Cumulative counts of forward and reverse-flow regimes for the particular experiment highlighted in (a). Error is calculated by normalizing the differential count (absolute difference between the forward and reverse counts) to the forward count.
Fig. 6(b) illustrates the accumulated forward and reverse counts during the experiment highlighted in Fig. 6(a). This demonstrates how the reverse count eventually leveled off and became close to the forward count, resulting in an absolute counting error of 71 cells that equates to a 1.2% error.
Table 1 quantifies the data in Fig. 6(a), and affirms that larger leukocyte counts (>2000 cells) resulted in a smaller percent error (~1.2%). The absolute counting error for this range of forward counts was ~46 cells. The significance of this small error can best be expressed by normalizing it to the estimated 5 µL injected sample volume, resulting in a concentration accuracy of ~9 cells per µL. This not only validates the differential counter as a plausible CD4+ T cell enumeration method, but opens the possibility of analyzing patients with advanced AIDS, who may have CD4+ T cell concentrations <50 cells per µL.
Table 1.
Error analysis of electrical differential counting of leukocytes using a passivated capture chamber
| Error (%) | Abs. counting error (cells) |
Est. conc. accuracy (cells per µL) |
||||
|---|---|---|---|---|---|---|
| SD | SD | SD | ||||
| All WBC | 2.91 | 3.93 | 44.2 | 31.3 | 8.84 | 6.26 |
| WBC < 2000 | 7.25 | 5.37 | 38.8 | 25.0 | 7.76 | 5.00 |
| WBC > 2000 | 1.18 | 1.02 | 46.4 | 34.5 | 9.28 | 6.90 |
Although the capture chamber volume was ~7 µL (Fig. 1(a)), we can only estimate that approximately 5 µL of sample entered the chamber before flow was reversed to prevent the loss of cells. This is because cell sample diffusion resulted in a lower concentration of cells reaching the exit counter before the more concentrated cell population completely filled the chamber. Estimation was made by observing the portion of the capture chamber filled with the concentrated cell population.
The main source of counting error was caused by non-specific adsorption of cells onto the chamber surface, despite passivating the capture chamber with BSA. Passivation using more incubation time and/or PBS with a pH closer to BSA’s isoelectric point of 5 would substantially decrease this error and further emphasize the accuracy of this enumeration technique.37 Another possible source of error may be dead/dying cells rupturing under the high shear rates found in the counter channel during forward counting, and not having the opportunity to be captured or counted again during the reverse counting phase.
CD4+ T-lymphocyte count comparison: optical vs. electrical (ESI†)
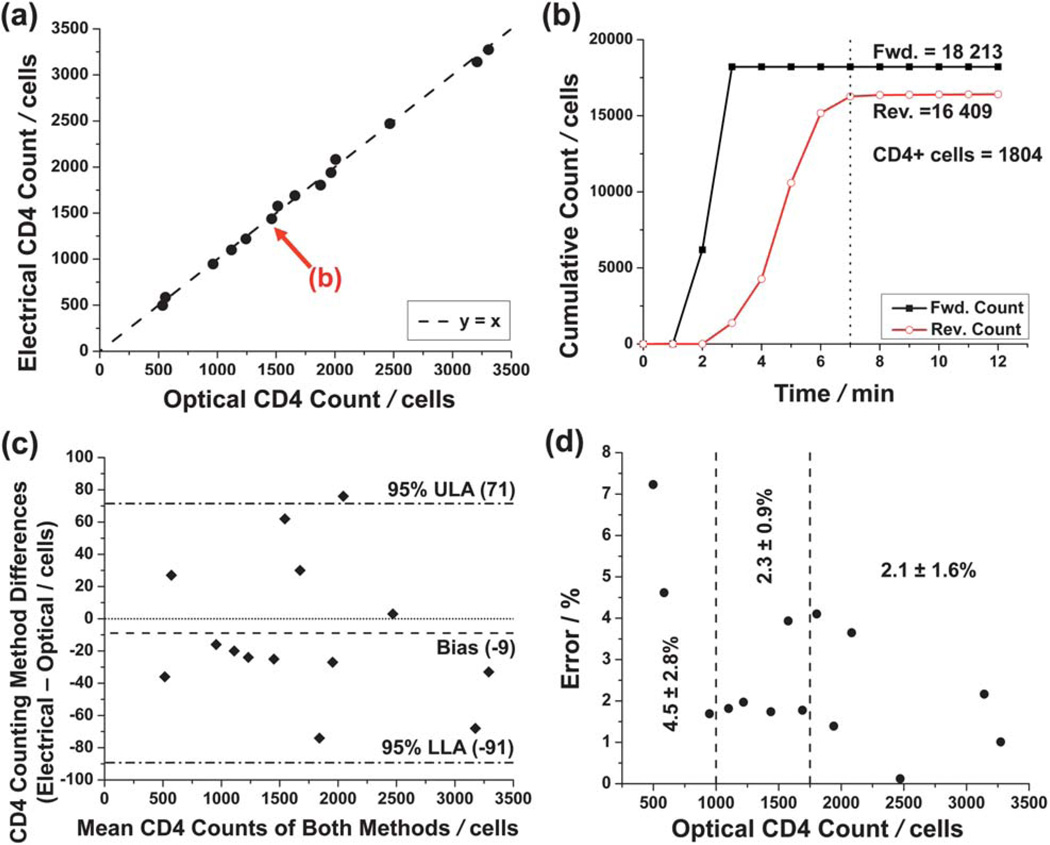
Fourteen purified leukocyte samples of varying dilution (0.5 to 2 times the physiological leukocyte concentration) were analyzed using a capture chamber conjugated with CD4 antibody. Fig. 7(a) shows the results of the CD4+ T cell counting experiments and the close correlation (y = 0.994x, R2 = 0.997) between the electrical differential method and the optical control experiments.
Fig. 7.
Reverse-flow differential counting of CD4+ T lymphocytes. (a) Comparison of electrical differential counting method and the optical controls. A linear fit (R2 = 0.997) is not shown, as it is would be indistinguishable from the y = x correlation standard. (b) Cumulative counts of forward and reverse-flow regimes for the particular experiment highlighted in (a). The absolute number of captured CD4+ T cells was found simply by subtracting the reverse count from the forward count. The dotted line denotes a completed analysis (where reverse counting levels off) duration of approximately 7 minutes. (c) Bland–Altman analysis of the data in (a). The dash-dot lines denote upper and lower levels of agreement (ULA and LLA, respectively). The dashed line shows a bias of 9 cells toward the optical method. (d) Counting error decreases as the total number of captured CD4+ T cells increases. Listed statistics are for three regimes demarcated by dashed lines: 0 to 1000 captured helper T cells (0 to 200 cells per µL, assuming 5 µL of sample flowed into chip), n = 3; 1000 to 1750 (200 to 350 cells per µL), n = 5; and greater than 1750 (>350 cells per µL), n = 6.
Fig. 7(b) illustrates the cumulative forward and reverse counts during an experiment, showing how the absolute number of helper T cells was calculated. This particular experiment was finished within 7 minutes, as the reverse count levels off at this point. The average counting time for 12 experiments was 10 ± 1.8 minutes, which can easily be shortened by using higher flow rates. The two other experiments were not included in the average because their average flow rates differed from the majority, affecting the analysis time.
Fig. 7(c) shows Bland-Altman comparison analysis between the electrical differential and optical counting methods. A bias of only −9 cells confirms the accuracy of the electrical differential counting method for the entire range of enumerated CD4+ T cells. One reason for the negative bias (larger optical counts than electrical counts) could be that leukocytes traveling in aggregates were counted as one entity during the forward count, were then separated by the high shear forces of the entrance counter, and then were counted individually during reverse counting. This would result in a lower electrical CD4+ T cell count—and a negative bias when compared to the optical count.
Fig. 7(d) shows how the percent error relates to the total number of CD4+ T cells counted. (The percent error here is defined as the absolute difference in optical and electrical counts, normalized by the optical count.) Assuming that 5 µL of sample flowed into the chip, the data can be separated into three important CD4+ T cell concentration regimes for AIDS diagnostics. For less than 1000 cells captured on the chip (5 µL sample at 200 cells per µL, the clinical definition of AIDS), the average error was 4.5% (n = 3). This shows to have high accuracy; for example, a patient with an actual CD4+ T cell concentration of 100 cells per µL would have a counting error of only ±4.5 cells per µL. The device has also shown to be even more accurate for the regimes surrounding the clinically critical threshold of 350 cells per µL (1750 cells captured), which has been shown to be an optimal condition to start ART.3
CD4+ capture efficiency
Flow cytometry analysis for 8 chips showed that 60.2 ± 18% of CD4+ T cells were immobilized in the capture chamber. A major reason for this low capture efficiency may be that cells entering into the chip later do not have enough time to interact with the antibody before flow reversal. In addition, the low shear stress used during the experiments (0.5 dyn cm−2) may also cause a loss in capture efficiency. Cheng et al. have shown that shear stresses between 1 and 3 dyn cm−2 will ensure adequate CD4+ T cell capture (~95%), while efficiency sharply drops off outside of this zone.13 However, they relied on erythrocytes flowing in the center of the capture chamber to effectively push the leukocytes toward the chamber walls, facilitating more leukocyte-antibody interactions. In our case, erythrocytes must be lysed to accurately enumerate leukocytes, but this removes the leukocyte margination mechanism. Although most leukocytes were able to come into contact with the chamber’s floor by sedimentation, other leukocytes traveled near the center of the 50 µm-high channel at higher velocities and away from interactions with its walls. Simply reducing the capture chamber’s height would not only increase the shear stress to within the desired range, but also would ensure cells would interact with the chamber’s walls. In addition, a covalent surface chemistry using linkers could be used to ensure proper CD4 antibody fragment antigen-binding (Fab) region orientation for optimal cell capture.
Additional error may have been introduced simply by the fact that a small sample volume (5 µL) was collected for flow analysis. Some practical issues are cells lost by non-specific adsorption to tubing during collection and cells lost during flow cytometry analysis preparation (fluorescent staining and washing).
CD4+ capture purity (ESI†)
Three chips were analyzed for CD4+ T cell capture purity after completing electrical differential counting experiments. In all chips, it was found that all captured cells were leukocytes (DAPI+) expressing the CD4 antigen (AF488+), confirming the high specificity of the surface chemistry for CD4+ cells. It was found that 81.7% of captured cells were CD4+ T lymphocytes, with monocytes comprising the remaining portion. According to Cheng et al., monocyte capture increased for shear stresses <0.7 dyn cm−2, a regime in which our chip was operating (0.5 dyn cm−2).13 One of the chips had a CD4+ T cell capture purity of 56.5% because flow was temporarily stopped before reversing flow direction. This effectively decreased shear stress even further, allowing more monocytes to be captured. The other two chips had no such stoppage, and showed an average capture purity of 94.3%, which agrees with our previous work.13 Although relatively small, the percentage of captured monocytes can still cause a positive bias for patients CD4+ T cell counts below 200 cells per µL. One solution for this problem is to create a monocyte depletion chamber conjugated with CD14 Ab immediately before the entrance to the entrance counter and main capture chamber.15
Conclusions
We have demonstrated as a proof of concept that a differential cell counter device using the reverse-flow technique and cell immunoaffinity depletion is a viable method to electrically enumerate CD4+ T lymphocytes from purified leukocyte populations from donor blood samples. Our microfabricated device and detection approach resulted in a short testing time of 10 minutes, had an accuracy (9 cells per µL) that allows for lower detection limits, and correlated closely with an optical standard (R2 = 0.997), demonstrating its capability to analyze patients in all stages of HIV infection. Its microfabricated nature suggests it may be an inexpensive, simple, and portable alternative to current flow cytometric practices that would enhance the penetration of CD4+ T cell tests into resource-poor regions.
Several advancements to the current design are necessary to create a complete PoC AIDS diagnostics device for whole blood samples. An on-chip erythrocyte removal/lysis module is needed to eliminate off-chip sample preparation steps. Also, an on-chip sample metering system is necessary to accurately ensure a known amount of blood is injected into the chip and for comparison to other CD4+ T cell counting methods. Furthermore, an optimized capture chamber design and surface chemistry would provide greater CD4+ T cell capture efficiency.
Supplementary Material
Acknowledgements
The authors would like to thank Adil Ghafoor (Purdue University) for his help in image analysis. Partial support was provided by the United States Department of Homeland Security graduate student fellowship (NNW), by the NSF NSEC at OSU EEC-0914790, and by the University of Illinois at Urbana-Champaign. The devices were fabricated and tested at the Micro-and Nanotechnology laboratory (www.mntl.illinois.edu).
Footnotes
Electronic supplementary information (ESI) available: Phase contrast imaging of captured cells (Fig. S1) and cell capture purity analysis using fluorescent labels (Fig. S2). See DOI: 10.1039/c0lc00556h
References
- 1.UNAIDS and WHO. [accessed 7 October, 2010];AIDS Epidemic Update. 2009 http://data.unaids.org/pub/Report/2009/jc1700_epi_update_2009_en.pdf.
- 2.Kitahata M, Gange S, Abraham A, Merriman B, Saag M, Justice A, Hogg R, Deeks S, Eron J, Brooks J, Rourke S, Gill M, Bosch R, Martin J, Klein M, Jacobson L, Rodriguez B, Sterling T, Kirk G, Napravnik S, Rachlis A, Calzavara L, Horgerg M, Silverberg M, Gebo K, Goedert J, Benson C, Collier A, Van Rompaey S, Crane H, McKaig R, Lau B, Freeman A, Moore R. N. Engl. J. Med. 2009;360:1815. doi: 10.1056/NEJMoa0807252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Severe P, Juste M, Ambroise A, Eliacin L, Marchand C, Apollon S, Edwards A, Bang H, Nicotera J, Godfrey C, Gulick M, Johnson W, Jr, Pape J, Fitzgerald D. N. Engl. J. Med. 2010;363:257. doi: 10.1056/NEJMoa0910370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO, UNICEF, UNAIDS. [accessed 7 October, 2010];Towards Universal Access: Scaling Up Priority HIV/AIDS Interventions in the Health Sector. http://www.who.int/entity/hiv/pub/tuapr_2009_en.pdf.
- 5.Phillips A, Lee C, Elford J, Janossy G, Timms A, Bofill M, Kernoff P. Lancet. 1991;337:389. doi: 10.1016/0140-6736(91)91166-r. [DOI] [PubMed] [Google Scholar]
- 6.Miller V, Staszewski S, Sabin C, Carlebach A, Rottmann C, Weidmann E, Rabenau H, Hill A, Lepri A, Phillips A. J. Infect. Dis. 1999;180:530. doi: 10.1086/314890. [DOI] [PubMed] [Google Scholar]
- 7.Lepri A, Phillips A, Monforte A, Castelli F, Antinori A, de Luca A, Pezzotti P, Alberici F, Cargnel A, Grima P, Piscopo R, Prestileo T, Scalise G, Vigevani M, Moroni M. AIDS. 2001;15:983. doi: 10.1097/00002030-200105250-00006. [DOI] [PubMed] [Google Scholar]
- 8.Bae S, Park H, Oh J, Yoon S-Y, Park D, Choi I, Kim J, Oh J, Hur D, Chung C, Chang J, Robinson J, Lim C. Cytometry, Part B. 2009;76:354. doi: 10.1002/cyto.b.20470. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez W, Christodoulides N, Floriano P, Graham S, Mohanty S, Dixon M, Hsiang M, Peter T, Zavahir S, Thior I, Romanovicz D, Bernard B, Goodey AP, Walker BD, McDevitt J. PLoS Med. 2005;2:663. doi: 10.1371/journal.pmed.0020182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jokerst J, Floriano P, Christodoulides N, Simmons G, McDevitt J. Lab Chip. 2008;8:2079. doi: 10.1039/b817116e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Ymeti A, Lunter B, Breukers C, Tibbe A, Terstappen L, Greve J. Cytometry, Part B. 2007;72B:397. doi: 10.1002/cyto.b.20165. [DOI] [PubMed] [Google Scholar]
- 12.Ymeti A, Li X, Lunter B, Breukers C, Tibbe A, Terstappen L, Greve J. Cytometry, Part A. 2007;71A:132. doi: 10.1002/cyto.a.20375. [DOI] [PubMed] [Google Scholar]
- 13.Cheng X, Irimia D, Dixon M, Sekine K, Demirci U, Zamir L, Tompkins R, Rodriguez W, Toner M. Lab Chip. 2007;7:170. doi: 10.1039/b612966h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng X, Irimia D, Dixon M, Ziperstein J, Demirci U, Zamir L, Tompkins R, Toner M, Rodriguez W. JAIDS, J. Acquired Immune Defic. Syndr. 2007;45:257. doi: 10.1097/QAI.0b013e3180500303. [DOI] [PubMed] [Google Scholar]
- 15.Cheng X, Gupta A, Chen C, Tompkins R, Rodriguez W, Toner M. Lab Chip. 2009;9:1357. doi: 10.1039/b818813k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gohring J, Fan X. Sensors. 2010;10:5798. doi: 10.3390/s100605798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moon S, Keles J, Ozcan A, Khademhosseini A, Hæggstrom E, Kuritzkes D, Demirci U. Biosens. Bioelectron. 2009;24:3208. doi: 10.1016/j.bios.2009.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Chin S, Chin C, Sarik J, Harper M, Justman J, Sia S. Anal. Chem. 2010;82:36. doi: 10.1021/ac902144w. [DOI] [PubMed] [Google Scholar]
- 19.Coulter WH. 2656508. US Pat. 1953
- 20.Coulter W. Proceedings of National Electronics Conference; Chicago. 1956. p. 1034. [Google Scholar]
- 21.Adams A, Okagbare P, Feng J, Hupert M, Patterson D, Gottert J, McCarley R, Nikitopoulos D, Murphy M, Soper S. J. Am. Chem. Soc. 2008;130:8633. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes D, Pettigrew D, Reccius C, Gwyer J, van Berkel C, Holloway J, Davies D, Morgan H. Lab Chip. 2009;9:2881. doi: 10.1039/b910053a. [DOI] [PubMed] [Google Scholar]
- 23.Holmes D, Morgan H. Anal. Chem. 2010;82:1455. doi: 10.1021/ac902568p. [DOI] [PubMed] [Google Scholar]
- 24.Lee D, Yi S, Cho Y-H. J. Microelectromech. Syst. 2008;17:139. [Google Scholar]
- 25.Mishra N, Retterer S, Zieziulewicz T, Isaacson M, Szarowski D, Mousseau D, Mousseau D, Lawrence D, Turner J. Biosens. Bioelectron. 2005;21:696. doi: 10.1016/j.bios.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 26.Jiang X, Spencer M. Biosens. Bioelectron. 2010;25:1622. doi: 10.1016/j.bios.2009.11.024. [DOI] [PubMed] [Google Scholar]
- 27.Cheng X, Liu Y-S, Irimia D, Demirci U, Yang L, Zamir L, Rodriguez W, Toner M, Bashir R. Lab Chip. 2007;7:746. doi: 10.1039/b705082h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watkins N, Venkatesan B, Toner M, Rodriguez W, Bashir R. Lab Chip. 2009;9:3177. doi: 10.1039/b912214a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis K, Abrams B, Iyer S, Hoffman R, Bishop J. Cytometry. 1998;33:197. doi: 10.1002/(sici)1097-0320(19981001)33:2<197::aid-cyto14>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 30.Lee B, Sharron M, Montaner L, Weissman D, Doms R. Proc. Natl. Acad. Sci. U. S. A. 1999;96:5215. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gawad S, Schild L, Renaud Ph. Lab Chip. 2001;1:76. doi: 10.1039/b103933b. [DOI] [PubMed] [Google Scholar]
- 32.Morgan H, Holmes D, Green N. Curr. Appl. Phys. 2006;6:367. [Google Scholar]
- 33.Young B, Heath J. Wheater’s Functional Histology: A Text and Colour Atlas. 4th edn. Edinburgh: Churchill Livingstone; 2000. [Google Scholar]
- 34.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 4th edn. New York: Garland Science; 2002. [Google Scholar]
- 35.Usami S, Chen H-H, Zhao Y, Chien S, Skalak R. Ann. Biomed. Eng. 1993;21:77. doi: 10.1007/BF02368167. [DOI] [PubMed] [Google Scholar]
- 36.Sweryda-Krawiec B, Devaraj H, Jacob G, Hickman J. Langmuir. 2004;20:2054. doi: 10.1021/la034870g. [DOI] [PubMed] [Google Scholar]
- 37.Freeman N, Peel L, Swann M, Cross G, Reeves A, Brand S, Lu J. J. Phys.: Condens. Matter. 2004;16:S2493. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.