Abstract
Solid materials, such as silicon, glass, and polymers, dominate as structural elements in microsystems including microfluidics. Porous elements have been limited to membranes sandwiched between microchannel layers or polymer monoliths. This paper reports the use of micropatterned carbon-nanotube forests confined inside microfluidic channels for mechanically and/or chemically capturing particles ranging over three orders of magnitude in size. Nanoparticles below the internanotube spacing (80 nm) of the forest can penetrate inside the forest and interact with the large surface area created by individual nanotubes. For larger particles (>80 nm), the ultrahigh porosity of the nanotube elements reduces the fluid boundary layer and enhances particle–structure interactions on the outer surface of the patterned nanoporous elements. Specific biomolecular recognition is demonstrated using cells (≈10 µm), bacteria (≈1 µm), and viral-sized particles (≈40 nm) using both effects. This technology can provide unprecedented control of bioseparation processes to access bioparticles of interest, opening new pathways for both research and point-of-care diagnostics.
1. Introduction
Most clinical diagnostics and basic research studies aimed at understanding the causes underlying disease require isolation of specific biomolecules or cells from complex samples such as blood, saliva, and cell-culture supernatant. Unfortunately, many bioparticles of interest are present in the samples in very small quantities. This is the case, for example, of antigen-specific T-cells, circulating tumor cells (CTCs), and HIV viral particles for monitoring immune response, cancer, and AIDS progression, respectively. As miniaturized detection methods are becoming increasingly sensitive, capable of single-cell[1,2] and even single-virus detection,[3,4] efficient capture of specific bioparticles is now the rate-limiting step to achieving complete lab-on-a-chip assays for particle identification. There is huge diversity in current microfluidic methods for bioparticle isolation.[5–7] Whereas some commonly used techniques differentiate particles based on their physical characteristics alone,[8–12] higher specificity is often achieved by methods that make use of surface chemistry and biomolecular recognition to target the particle of interest. These include solid-phase techniques, such as affinity chromatography,[13,14] and solution-phase techniques, such as immunomagnetic beads[15] and on-chip flow cytometry.[16] Although solid-phase techniques generally offer higher detection sensitivity and more flexibility for further interrogation, a significant challenge is to achieve sufficient physical interaction between the bioparticles and surfaces to promote binding.[17,18] Difficulties arise from the formation of boundary layers at solid–fluid interfaces and the low diffusivity for all but the smallest (<10 nm) particles, so that even microscale systems become diffusion limited and particles in solution are prevented from contacting the solid surfaces, thus resulting in poor isolation yields.
In this Full Paper, we show that particle–surface interactions can be greatly enhanced by triggering changes in the boundary layer as well as increasing the particle–solid-surface interaction area. Specifically, we describe the use of patterned, aligned, and chemically functionalized carbon nanotube (CNT) forests fully integrated into microfluidic devices for the isolation of bio-particles with sizes spanning over several orders of magnitude. While previous efforts on combining CNTs with microfluidics have focused on making use of the electrical properties of disorganized CNTs for sensing applications,[19–22] we take advantage of the physical characteristics, such as high nanoscale porosity and surface area, and surface functionalizability of organized nanoporous CNT-forest elements for bioseparation. Our CNT forests are grown with a Fe/Al2O3 catalyst using atmospheric-pressure thermal chemical vapor deposition (CVD) and have been previously characterized as multiwalled CNTs with an average diameter of 8 nm (5–8 walls).[23] The CNTs can be grown as an aligned forest morphology, sometimes referred to as vertically-aligned CNTs (VACNTs) up to several millimeters in height, and with an average spacing of 80 nm, which results in an overall forest porosity of 99% (1 v/v% CNTs). The forest modulus along the CNT axis has been previously characterized to have an elastic modulus of ≈2 MPa.[25] The CNT-forest structures are patterned on silicon by using photolithography to control the area of catalyst coverage on the wafer,[24] thereby creating nanoporous bulk elements that are introduced into the microfluidic-channel flow field. Until now, the inclusion of porous elements in microfluidics has been limited to membranes sandwiched between microchannel layers[26] or polymer monoliths that fill the inside of channels.[27] With membranes, geometrical control of the porous region is restricted to two dimensions, defined by the overlap between channels on either side of the membrane. Their applications are mostly in size-exclusion filtration and dialysis.[26,28,29] For polymer monoliths, examples have been limited to simple porous plugs in the channel that are commonly utilized for chromatography and solid-phase extraction applications.[30,31] A fundamental limitation to both of these techniques is the lack of geometrical control other than that provided by the nonporous channel walls. This has limited their application to simple size-based separation and does not enable them to take full advantage of the exquisite control of fluid dynamics possible within microfluidics. In contrast, the nanoporous CNT elements allow for three-dimensional geometrical control of the porous elements independently from the channel boundaries: photolithography of the catalyst layer allows definition of the microscale geometry, and varying the time of growth can finely control the forest height. With these added levels of control and flexibility, this approach transforms the capabilities of porous elements in microfluidics to enable manipulation of fluid flow, particle flow, and molecular diffusion in ways that have not been previously feasible, opening up new capabilities and applications.
2. Results and Discussion
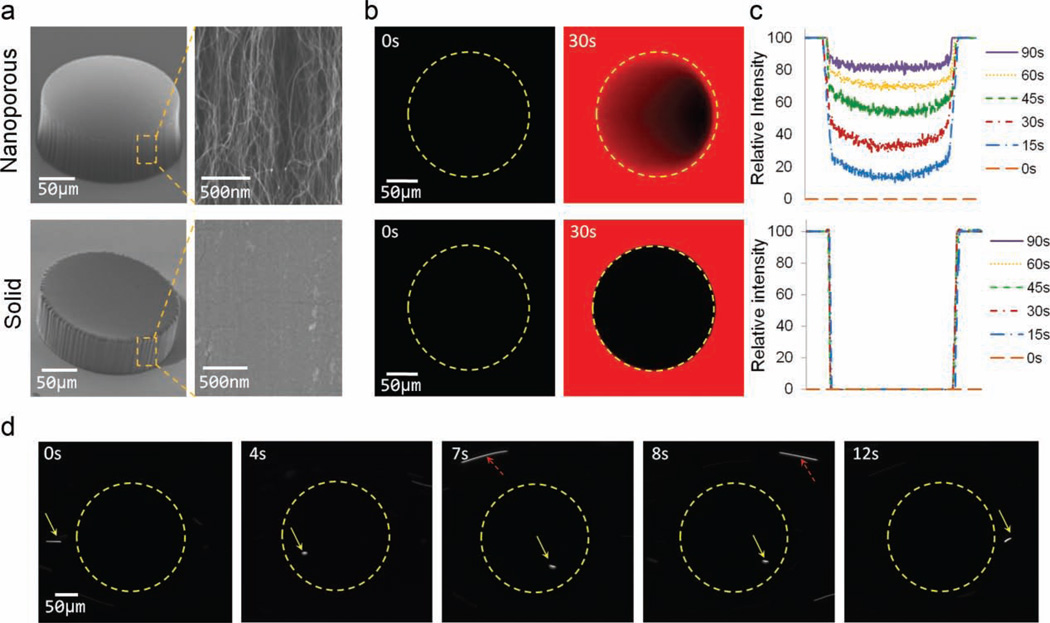
Figure 1a highlights the porosity of a circular post-shaped element made from a nanoporous CNT forest in comparison to a nonporous (solid) post made from polydimethylsiloxane (PDMS). To test the nanoporous structures under fluid flow, we sealed microfabricated PDMS channels to the silicon surface around the CNTs using oxygen-plasma bonding. Channels are made several micrometers shorter than the height of the as-grown CNT elements so that the ceiling of the channel slightly compresses the top of the CNT elements to produce an interference-fit seal. The surfaces of CNTs, which are hydrophobic, can be made hydrophilic by the addition of Tween-20 (wet functionalization) to facilitate infiltration of fluid inside the nanostructures. Fluid was injected by pressure-driven flow using a syringe pump. We do not observe any deformation of the CNT features upon wetting or under fluid flow in the regimes studied. Figure 1b demonstrates the penetration of water and small-molecule dyes (rhodamine; molecular weight = 479 Da) inside a nanoporous CNT post over 90 s; by comparison, and as expected, there is no dye penetration inside the solid PDMS post. Fluorescence-intensity analysis of each type of post as the dye penetrates over time is quantified in Figure 1c. To characterize flow of nanoparticles inside the nanoporous structures, we injected a dilute suspension of fluorescent quantum dots (QDs) 10–20 nm in size and observed their flow paths inside and outside of a 200-µm-diameter nanoporous post via confocal imaging. Figure 1d follows the path of a single QD as it travels through a nanoporous post (shown with yellow arrows) and compares it with another QD (indicated by dashed red arrows) that passes around the outside of the same post. Due to increased fluid resistance inside the nanoporous structure, the QDs are observed to travel inside the CNT post at a significantly reduced velocity, only 8% of its counterpart traveling outside the post. These experiments clearly demonstrate the ability of aligned CNT structures to allow pressure-driven flow of both molecular and particulate species inside the nanoporous elements.
Figure 1.
Nanoporous micropatterned elements are permeable to nanometer-scale molecules and particle flow. a) Comparison between a nanoporous CNT post and a solid PDMS post. b) Confocal micrographs of 200-µm-diameter nanoporous and solid posts as a fluorescent dye solution flows through the microchannel. c) Relative intensity plots of dye infiltration with time inside nanoporous and solid posts. Intensity values were evaluated along the vertical center line across each post. d) Fluorescent QDs (10–20 nm) passing through a 200-µm-diameter nanoporous post (outlined via dotted lines) having 80-nm CNT spacing. Yellow arrows follow the passage of one QD through the post; red arrows follow another passing around the outside of the post.
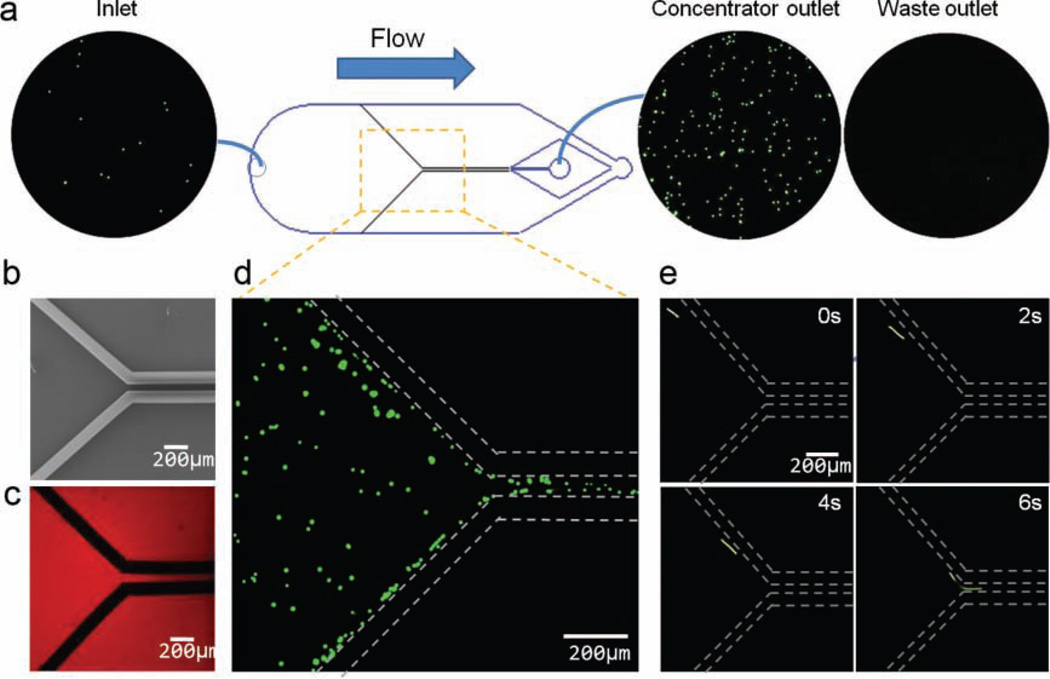
A straight forward yet powerful application that takes advantage of the high porosity and ease of patterning of the nanoporous CNT structures is mechanical filtration. In Figure 2, we demonstrate filtration and concentration by micropatterning a Y-shaped nanoporous filter. The filter sidewalls are made from 100-µm-wide CNT forests. It narrows from a 3-mm-wide section to a 100-x000B5;m-wide section. A schematic of the filter-chip design is shown in Figure 2a and a scanning electron microscopy (SEM) image of the CNT structure is shown in Figure 2b. In this configuration, large particles (10-µm fluorescent-green beads), which cannot pass through the 80-nm spaces between CNTs in the nanoporous forest, are guided by the barrier into the narrow neck of the Y-filter (Figure 2d,e), whereas fluid and particles smaller than 80 nm efficiently pass through the barrier, such as, for example, the ≈10-nm red bovine serum albumin (BSA) molecules in Figure 2c, resulting in an 11-fold concentration of the larger particles in the sample (inlet and outlet circles, Figure 2a). Higher-concentration factors could be achieved by adjusting the ratio of the nanoporous-passage width to the overall PDMS-channel width by cascading several filters or altering the nanoporosity (e.g.,via the average spacing of CNTs) in the elements.
Figure 2.
Nanoporous Y-filter. a) Device schematic. Overall dimensions 3 mm (W)× 20 mm (L) × 100 µm (H). The inside width of the nanoporous central channel is 100 µm. Blue lines are PDMS-channel boundaries; black lines show filter structure. Circular insets show micrographs of the inlet, concentrator outlet, and waste outlet, showing selective concentration of 10-µm fluorescent beads. b) SEM image of the nanoporous filter. c) Fluorescent micrograph showing that red fluorescent BSA molecules have passed through the nanoporous Y-filter walls. d) 10-µm polymer beads (green) cannot pass through the nanoporous filter sidewalls (dotted lines) and are directed to the central channel. e) Streak images of a single 10-µm bead (green) as it enters the constricted section of the filter.
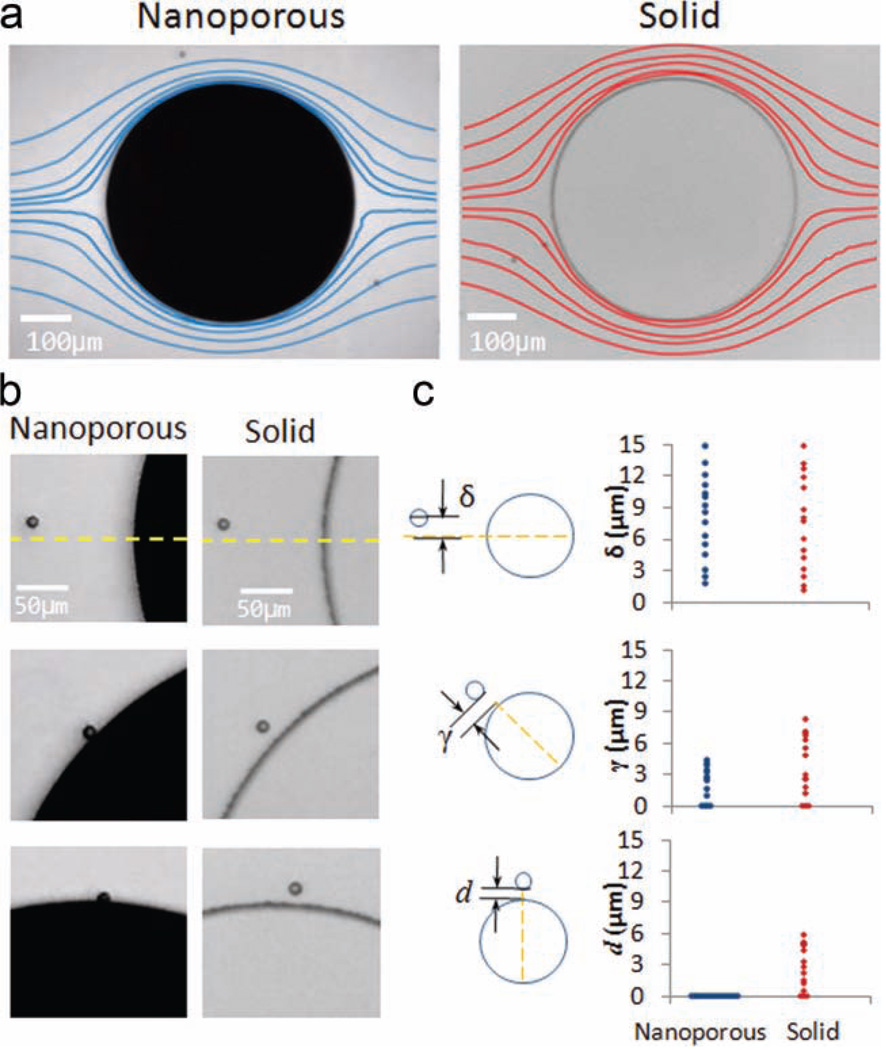
We expected that fluid flow through a porous structure could alter the flow characteristics of the fluid path in the vicinity of the structure and change the boundary-layer behavior. We investigated this phenomenon by flowing suspensions of 10-µm-diameter beads through a microchannel containing a 500-µm-diameter nanoporous-post element and tracking the beads’ flow trajectories. In Figure 3a (left panel), we show superimposed particle tracks from multiple beads flowing around the nanoporous post. For comparison, flow trajectories around a 500-µm solid PDMS post are also shown (Figure 3a, right panel). We observed that the tracks around the nanoporous post are concentrated in an area that is closer to the post surface than the tracks around the solid post. A comparison of several snapshots of two single particles approaching each type of post from the same starting position demonstrates that some particle trajectories can only be captured with the nanoporous post (Figure 3b). Analysis of the trajectories of multiple beads shows that, when solid posts are used, only particles approaching at <2.5 µm away from the center line contact the post, whereas nanoporous posts ensure 100% interception of bioparticles approaching from as far as 17 µm away from the post center line, as shown in Figure 3c and Figure S2 of the Supporting Information (SI). Interactions between particles and microstructure surfaces can therefore be enhanced sevenfold. This improvement is achieved through the ability of porous surfaces to modulate boundary-layer flow dynamics (and therefore streamlines) by allowing fluid transport through the nanoporous element. For solid features, an area of stagnation occurs at the solid–fluid interface due to a zero-slip condition at the boundary. This boundary-layer effect results in streamlines developing away from the solid surfaces, such that only the fluid streamlines aligned with the center of a circular solid post will bring particles to interact with the post feature. However, with porous features, some streamlines pass through the outer boundary of the element/feature, hence reducing the overall boundary-layer thickness by avoiding flow stagnation. A greater percentage of particles traveling along streamlines in the bulk flow will therefore reach the outer surface of a porous feature than for a solid feature of identical element geometry. We expect that this beneficial effect of porous elements holds both at low (studied here) and high (future work) Reynolds flow regimes, hence making nanoporous elements attractive for a variety of microfluidic applications.
Figure 3.
Nanoporous elements alter particle flow paths. a) Particle streamline tracks of 10-µm beads around a nanoporous post and a solid post, showing tightening of the particle tracks towards the element surfaces due to the nanoporous post altering the flow. b) Snapshots of single particles approaching a nanoporous circular post element and a solid post from the same start position. The particle approaching the nanoporous post eventually touches the post and the one approaching the solid post never does. c) Positional data of multiple beads as they approach both posts. All beads approaching the nanoporous post from 17 µm or less away from the center line contact the post (γ or d = 0), with most never reaching the solid post.
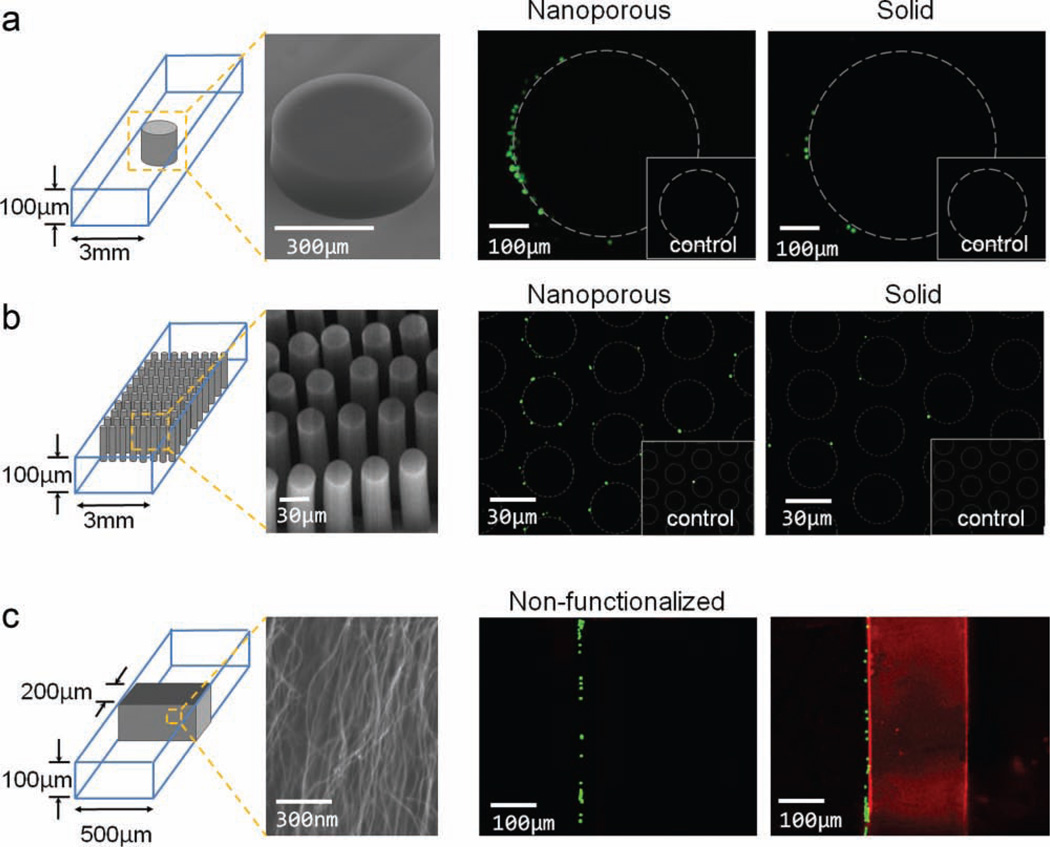
To demonstrate the capability of nanoporous structures to separate bioparticles, we integrated nanoporous elements into microfludic channels to capture bioparticles ranging over three orders of magnitude in size. We designed both a 500-µm-diameter post element to capture CD4+ T-cells (≈10 µm) and an array of circular posts to capture Escherichia coli bacteria (≈1 µm) (Figure 4a,b). Posts were then wet functionalized with anti-CD4 and anti-E. coli antibodies, respectively.[32] Capture data from four different devices in each case is shown in Figure S4 of the SI and the accompanying discussion. For the single-post geometry, as in Figure 4a, we observed a 5.5-fold enhancement of capture on the nanoporous posts compared to the solid posts, with a standard deviation of 12.7% in capture across 4 devices. For the post-array geometry, as in Figure 4b, we observed a 6.3-fold enhancement of capture on the nanoporous posts compared to the solid posts, with a standard deviation of 15.9% in capture across 4 devices. Nonspecific binding was low in all cases (insets of Figure 4a,b). These experiments confirm our expectation that the changes in the boundary layer described above (Figure 3) would enhance interactions between bioparticles and post surfaces, ultimately resulting in increased particle-capture efficiency.
Figure 4.
Bioparticles of three size orders are captured with functionalized nanoporous elements. a) 10-µm-size fluorescently labeled CD4 T-cells are captured on 500-µm-diameter posts. Comparison in capture between nanoporous and solid posts of identical geometry is shown. Inset boxes show capture on nonfunctionalized control devices. b) 1-µm-size Streptococcus pneumoniae capture. Nanoporous- and solid-post arrays with identical geometries are functionalized with anti-S. pneumoniae antibody and fluorescently labeled Pneumoniae S. pneumoniae samples are flowed through the device. Inset boxes show capture on nonfunctionalized control devices. c) 40-nm-size virus-like particles captured within a functionalized nanoporous filter. A mixture of nonfunctionalized 1-µm beads (green) and 40-nm avidin-coated beads (red) are flowed through nonfunctionalized and biotin-functionalized nanoporous filters, respectively. The 1-µm beads are physically trapped in front of both filters. The 40-nm beads are trapped inside the functionalized filter and the nonfunctionalized filter does not capture the 40-nm beads.
Finally, we combined the mechanical-filtration capabilities of the CNT posts with surface chemistry to design a device that can efficiently capture virus-sized particles. Using 40-nm fluorescent beads with an avidin-coated surface, we show that virus-size particles can be captured inside a nanoporous filter containing biotin-functionalized aligned CNTs (Figure 4c). The filter mechanically excludes 1-µm-size particles, which are larger than the 80-nm spacing between individual nanotubes in the forest, while chemically trapping the 40-nm beads, which can flow through the nanoporous structure. Since there are ≈108 individual CNTs per mm2 of forest, a 400× increase in surface area inside a 100-µm-height channel is achieved and particles traveling through the forest will encounter a high degree of contact with the functionalized CNT sidewalls.
Thus, the nanoporous elements enhance surface interactions for particle capture via two mechanisms: the first is through boundary-layer modulation and benefits particles larger than the inter-CNT spacing (Figure 3 and Figure 4a,b); the second is through the high internal surface area of the CNT forests and benefits particles smaller than the inter-CNT spacing (Figure 4c). As a consequence, a single device can be optimized to simultaneously separate particles of different size orders.
3. Conclusion
In this work, we have shown several device concepts that combine nanoporous CNT elements within microfluidics and have demonstrated their ability to both mechanically and chemically manipulate bioparticles of varying sizes. In contrast to previously described microfluidic platforms for particle isolation, which are highly tailored to perform optimally for particles of a single size,[33–35] our porous elements provide a more universal platform, capable of high-efficiency separation of bioparticles across multiple size scales, ranging from viruses to bacteria and cells. Furthermore, the fabrication process is simple with a fast turnaround for prototyping. In addition to optimal design of the microstuctured nanoporous elements via simple patterning, there is potential for additional versatility and utility by altering the spacing between individual nanotubes, that is, changing the degree of nanoporosity, such as by modulating CNT-growth conditions.[36,37] This would allow different size particles to penetrate into the nanoporous structure, as well as change the fluidic resistance for liquid passing through the structures and therefore their surrounding flow field. Another important direction for future work would be to integrate bioseparation with sensing, taking advantage of the body of work already performed on CNT-based miniature biosensors that focuses on using nonmechanical aspects of the CNTs.[38,39] We envisage the ability to create new families of devices using this technique for a very broad range of applications, including lab-on-a-chip devices for blood analysis to monitor patients at the point-of-care, ultrarapid cell sorters to detect rare cells in circulation for diagnostics (e.g., cancer, prenatal, infections), high-throughput filters for pathogen depletion, and isolation of bacteria and viruses for diagnosing infectious diseases.
4. Experimental Section
CNT Patterning and Growth
Figure S1 of the SI shows a schematic version of the fabrication process used in this work. Plain <100> 6-inch silicon wafers (p-type, 1–10 Ω cm, Silicon Quest International), which have been cleaned using a standard “piranha” (3:1 H2SO4:H2O2) solution, are patterned by photolithography using a 1.25-µm layer of image-reversal photoresist (AZ-5214E, Figure S1a). A catalyst film of 0.1-nm Fe/Al2O3 is then deposited by electron-beam evaporation in a single pump-down cycle using a Temescal VES-2550 with a FDC-8000 film-deposition controller. Film thickness is monitored during deposition using a quartz crystal monitor. Catalyst areas for patterning are removed by photoresist lift-off, soaking the wafer in acetone for 10 min with mild sonication (Figure S1b).
CNT growth is performed in a 4-inch inner diameter quartz-tube CVD furnace (G. Finkenbeiner, Inc.) at atmospheric pressure using reactant gases of C2H4, H2, and He (Airgas, 400/1040/1900 sccm). Catalyst annealing is carried out in a reducing He/H2 environment at 650 °C, leading to the formation of catalyst nanoparticles ≈10 nm in diameter (Figure S1c). C2H4 is then introduced into the furnace to initiate CNT growth, occurring at a rate of approximately 100 µm min−1 until the flow of C2H4 is terminated (Figure S1d). The nanotubes grown using this method are multiwalled (2–3 concentric walls) with a diameter of ≈8 nm. The CNTs are spaced by approximately 80 nm and their morphology is characterized by very good vertical alignment.
Microfluidic Channel Fabrication and Bonding
Microfluidic channels were generated using standard soft lithography.[40] SU-8 photoresist (Microchem) was patterned on a silicon wafer by photolithography to form a negative mold. A 10:1 mixture of PDMS prepolymer and curing agent (Sylgard 184, Dow Corning) was poured onto the mold and baked at 75 °C until cured. The PDMS channels were then bonded to the silicon wafers containing the CNT structures after oxygen-plasma treatment. Channels are made several micrometers shorter than the height of the CNT structures so that the ceiling of the channel slightly compresses the top of the CNTs to produce a seal. For the ‘solid’ devices used as controls, PDMS channels with cylindrical posts were fabricated using soft lithography and bonded to 1-inch × 3-inch glass microscope slides after oxygen-plasma treatment.
Fluorescent-Dye and Quantum-Dot Infiltration
The devices used for the fluorescent-dye and quantum-dot infiltration experiments in Figure 1b and c were single posts 200 µm in diameter and 100 µm in height, sealed inside a 3-mm × 20-mm × 100-µm PDMS channel. 0.5% Tween-20 in deionized (DI) water was used to treat the devices after fabrication to make the surfaces hydrophilic and to block nonspecific binding. Rhodamine B (Sigma Aldrich) was injected into the channel using a syringe pump (Harvard Apparatus) set to a flow rate of 5 uL min−1. Imaging was performed using a confocal microscope (Zeiss) and intensity plots shown in Figure S2 of the SI were made using ImageJ software.
Y-Filter Experiments
Y-filter devices were blocked against nonspecific binding with 0.5% Tween-20 in DI water. 10-µm green fluorescent polymer beads (Duke Scientific) were injected at 10 µL min−1 using a syringe pump. Flow from both outlets were collected with tubing and imaged under a fluorescent microscope.
Particle Tracing
Devices used for particle-tracing experiments in Figure 2 were single posts 500 µm in diameter and 100 µm in height, sealed inside a 3-mm × 20-mm × 100-µm PDMS channel. 0.5% Tween-20 in DI water was used to block nonspecific binding. Particles used were 10-µm-diameter green fluorescent polymer particles (Duke Scientific) suspended in water with 0.1% Tween-20. Flow injection was via a syringe pump set to 20 µL min−1. Video images were recorded using NIS elements software. Particle paths were traced using the manual tracking plug-in in ImageJ.
CNT-Device Functionalization
We used the noncovalent functionalization method by Chen et al.[32] using 1,1-carbonyldiimidazole (CDI)-activated Tween. Tween-20 was reacted with CDI (Sigma Aldrich) under dimethyl sulfoxide (DMSO) for 2 h at 40 °C then dried using a Rotovap. Devices were treated with 1% CDI-activated Tween for 30 min then flushed with DI water. For the biotin-functionalized devices in Figure 4c, 50 µg mL−1 biotin (Pierce) in PBS was then injected and incubated at room temperature for 1 h before use. For the antibody-functionalized devices in Figure 4a and b, 50 µg mL−1 NeutrAvidin (Thermo Scientific) in phosphate buffered saline (PBS) was injected and incubated at room temperature for 30 min, followed by 30 ug mL−1 of biotinylated antibody in PBS for 30 min. The devices were then washed and blocked for nonspecific binding with 1% BSA in PBS.
PDMS-Device Functionalization
For the PDMS solid-structure comparison experiments in Figure 3b and c, the all-PDMS devices were functionalized using previously described methods.[41] Freshly bonded devices were pretreated with a 4 v/v% solution of 3-mercaptopropyltrimethoxysilane in ethanol for 30 min at room temperature, followed with incubation with 0.01 µmol mL−1 4-maleimidobutyric acid N-hydroxysuccinimide ester (GMBS in ethanol for 15 min at room temperature. NeutrAvidin and biotinylated antibodies was then added in the same way as for the CNT devices.
Bioparticle Capture
The geometries of the three devices used are as annotated in Figure 4. Particles used in Figure 4c were 40-nm strepavidin-coated red fluorescent beads (Invitrogen), and 1-µm uncoated green fluorescent beads (Duke Scientific) suspended in DI water. The experiments in Figure 4b used fluorescently labeled S. pneumoniae bacteria, and devices were coated with anti-S. pneumoniae antibody (rabbit polyclonal, Abcam). In the cell-capture experiment shown in Figure 4a, human leukocytes were obtained from healthy volunteer blood and devices were coated with anti-CD4 antibody (clone 13B8.2 Beckman Coulter). All control devices were treated with 0.5% Tween-20. All flow rates were set at 10 µL min−1.
Supplementary Material
Acknowledgements
We thank Octavio Hurtado (MGH) and Namiko Yamamoto (MIT) for technical support with nano- and microfabrication procedures. This work was supported by the National Institute of Biomedical Imaging and Bioengineering under Grant P41 EB002503 (BioMEMS Resource Center) and U.S. Department of State’s Fulbright Science and Technology Award.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Grace D. Chen, BioMEMS Resource Center, Massachusetts General Hospital, 114 16th Street, Charlestown, MA 02139, USA.
Fabio Fachin, Department of Aeronautics and Astronautics, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139, USA.
Marta Fernandez-Suarez, BioMEMS Resource Center, Massachusetts General Hospital, 114 16th Street, Charlestown, MA 02139, USA.
Brian L. Wardle, Department of Aeronautics and Astronautics, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139, USA.
Mehmet Toner, Email: mehmet_toner@hms.harvard.edu, BioMEMS Resource Center, Massachusetts General Hospital, 114 16th Street, Charlestown, MA 02139, USA.
References
- 1.Burg TP, Godin M, Knudsen SM, Shen W, Carlson G, Foster JS, Babcock K, Manalis SR. Nature. 2007;446:1066–1069. doi: 10.1038/nature05741. [DOI] [PubMed] [Google Scholar]
- 2.Lee H, Sun E, Ham D, Weissleder R. Nat. Med. 2008;14:869–874. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patolsky F, Zheng GF, Hayden O, Lakadamyali M, Zhuang XW, Lieber CM. Proc. Natl. Acad. Sci. USA. 2004;101:14017–14022. doi: 10.1073/pnas.0406159101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uram JD, Ke K, Hunt AJ, Mayer M. Small. 2006;2:967–972. doi: 10.1002/smll.200600006. [DOI] [PubMed] [Google Scholar]
- 5.Meldrum DR, Holl MR. Science. 2002;297:1197–1198. doi: 10.1126/science.297.5584.1197. [DOI] [PubMed] [Google Scholar]
- 6.Dittrich PS, Tachikawa K, Manz A. Anal. Chem. 2006;78:3887–3907. doi: 10.1021/ac0605602. [DOI] [PubMed] [Google Scholar]
- 7.Whitesides GM. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 8.Gascoyne PRC, Vykoukal J. Electrophoresis. 2002;23:1973–1983. doi: 10.1002/1522-2683(200207)23:13<1973::AID-ELPS1973>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacDonald MP, Spalding GC, Dholakia K. Nature. 2003;426:421–424. doi: 10.1038/nature02144. [DOI] [PubMed] [Google Scholar]
- 10.Petersson F, Nilsson A, Holm C, Jonsson H, Laurell T. Lab Chip. 2005;5:20–22. doi: 10.1039/b405748c. [DOI] [PubMed] [Google Scholar]
- 11.Long ZC, Liu DY, Ye NN, Qin JH, Lin BC. Electrophoresis. 2006;27:4927–4934. doi: 10.1002/elps.200600252. [DOI] [PubMed] [Google Scholar]
- 12.Davis JA, Inglis DW, Morton KJ, Lawrence DA, Huang LR, Chou SY, Sturm JC, Austin RH. Proc. Natl. Acad. Sci. USA. 2006;103:14779–14784. doi: 10.1073/pnas.0605967103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malmstadt N, Yager P, Hoffman AS, Stayton PS. Anal. Chem. 2003;75:2943–2949. doi: 10.1021/ac034274r. [DOI] [PubMed] [Google Scholar]
- 14.Cheng XH, Irimia D, Dixon M, Sekine K, Demirci U, Zamir L, Tompkins RG, Rodriguez W, Toner M. Lab Chip. 2007;7:170–178. doi: 10.1039/b612966h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gijs MAM. Microfluid. Nanofluid. 2004;1:22–40. [Google Scholar]
- 16.Fu AY, Spence C, Scherer A, Arnold FH, Quake SR. Nat. Biotechnol. 1999;17:1109–1111. doi: 10.1038/15095. [DOI] [PubMed] [Google Scholar]
- 17.Squires TM, Messinger RJ, Manalis SR. Nat. Biotechnol. 2008;26:417–426. doi: 10.1038/nbt1388. [DOI] [PubMed] [Google Scholar]
- 18.Yanik AA, Huang M, Artar A, Chang TY, Altug H. Appl. Phys. Lett. 2010;96 [Google Scholar]
- 19.Ghosh S, Sood AK, Kumar N. Science. 2003;299:1042–1044. doi: 10.1126/science.1079080. [DOI] [PubMed] [Google Scholar]
- 20.Crevillén AG, Ávila M, Pumera M, González MC, Escarpa A. Anal. Chem. 2007;79:7408–7415. doi: 10.1021/ac071247i. [DOI] [PubMed] [Google Scholar]
- 21.Jeng ES, Nelson JD, Prather KLJ, Strano MS. Small. 2010;6:40–43. doi: 10.1002/smll.200900944. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Chen G, Chatrathi MP, Musameh M. Anal. Chem. 2004;76:298–302. doi: 10.1021/ac035130f. [DOI] [PubMed] [Google Scholar]
- 23.Wardle BL, Saito DS, Garcia EJ, Hart AJ, de Villoria RG, Verploegen EA. Adv. Mater. 2008;20:2707–+. doi: 10.1002/adma.200800295. [DOI] [PubMed] [Google Scholar]
- 24.Garcia EJ, Hart AJ, Wardle BL, Slocum AH. Nanotechnology. 2007;18 [Google Scholar]
- 25.Yaglioglu O, Martens R, Hart A, Slocum A. Adv. Mater. 2008;20:357–362. [Google Scholar]
- 26.Wang PC, DeVoe DL, Lee CS. Electrophoresis. 2001;22:3857–3867. doi: 10.1002/1522-2683(200110)22:18<3857::AID-ELPS3857>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 27.Yu C, Xu MC, Svec F, Frechet JMJ. J. Polym. Sci. A. 2002;40:755–769. [Google Scholar]
- 28.de Jong J, Lammertink RGH, Wessling M. Lab Chip. 2006;6:1125–1139. doi: 10.1039/b603275c. [DOI] [PubMed] [Google Scholar]
- 29.Xiang F, Lin YH, Wen J, Matson DW, Smith RD. Anal. Chem. 1999;71:1485–1490. doi: 10.1021/ac981400w. [DOI] [PubMed] [Google Scholar]
- 30.Chon K, Moon J, Kim S, Kim SD, Cho J. Desalination. 2007;202:215–223. [Google Scholar]
- 31.Throckmorton DJ, Shepodd TJ, Singh AK. Anal. Chem. 2002;74:784–789. doi: 10.1021/ac011077o. [DOI] [PubMed] [Google Scholar]
- 32.Chen RJ, Bangsaruntip S, Drouvalakis KA, Kam NWS, Shim M, Li YM, Kim W, Utz PJ, Dai HJ. Proc. Natl. Acad. Sci. USA. 2003;100:4984–4989. doi: 10.1073/pnas.0837064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–U1210. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dharmasiri U, Witek MA, Adams AA, Osiri JK, Hupert ML, Bianchi TS, Roelke DL, Soper SA. Anal. Chem. 2010;82:2844–2849. doi: 10.1021/ac100323k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huber DL, Manginell RP, Samara MA, Kim BI, Bunker BC. Science. 2003;301:352–354. doi: 10.1126/science.1080759. [DOI] [PubMed] [Google Scholar]
- 36.Chhowalla M, Teo KBK, Ducati C, Rupesinghe NL, Amaratunga GAJ, Ferrari AC, Roy D, Robertson J, Milne WI. J. Appl. Phys. 2001;90:5308–5317. [Google Scholar]
- 37.Nessim GD, Hart AJ, Kim JS, Acquaviva D, Oh JH, Morgan CD, Seita M, Leib JS, Thompson CV. Nano Lett. 2008;8:3587–3593. doi: 10.1021/nl801437c. [DOI] [PubMed] [Google Scholar]
- 38.Li J, Ng HT, Cassell A, Fan W, Chen H, Ye Q, Koehne J, Han J, Meyyappan M. Nano Lett. 2003;3:597–602. [Google Scholar]
- 39.Crevillen AG, Pumera M, Gonzalez MC, Escarpa A. Lab Chip. 2009;9:346–353. doi: 10.1039/b809963d. [DOI] [PubMed] [Google Scholar]
- 40.McDonald JC, Whitesides GM. Acc. Chem. Res. 2002;35:491–499. doi: 10.1021/ar010110q. [DOI] [PubMed] [Google Scholar]
- 41.Murthy SK, Sin A, Tompkins RG, Toner M. Langmuir. 2004;20:11649–11655. doi: 10.1021/la048047b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.