Abstract
Cells must regulate the synthesis and degradation of their proteins to maintain a balance that is appropriate for their specific growth conditions. Here we present the results of an investigation of the balance between protein folding and degradation for mammalian chaperone Hsp90-dependent client proteins. The central players are the molecular chaperones Hsp70 and Hsp90, the co-chaperone HOP, and ubiquitin ligase, CHIP. Hsp70 and Hsp90 bind to HOP thus forming a ternary folding complex whereas the binding of CHIP to the chaperones has previously been shown to lead to ubiquitination and ultimately to degradation of the client proteins as well as the chaperones. To understand the folding/degradation balance in more detail, we characterized the stoichiometries of the CHIP-Hsp70 and CHIP-Hsp90 complexes and measured the corresponding dissociation constants to be ~ 1 µM and ~ 4.5 µM respectively. We quantified the rate of ubiquitination of various substrates by CHIP in vitro. We further determined that the folding and degradation machineries cannot coexist in one complex. Lastly, we measured the in vivo concentrations of Hsp70, Hsp90, HOP, and CHIP under normal conditions and when client proteins are being degraded due to inhibition of the folding pathway. These in vivo measurements along with the in vitro data allowed us to calculate the approximate cellular concentrations of the folding and degradation complexes under both conditions and formulate a quantitative model for the balance between protein folding and degradation as well as an explanation for the shift to client protein degradation when the folding pathway is inhibited.
"La fixité du milieu intérieur est la condition d'une vie libre et indépendante" Claude Bernard. Living cells maintain a constant milieu intérieur in part by regulating the synthesis and degradation of proteins. An appropriate balance is essential for normal cellular growth and function. If this balance is upset, the result can be aberrant growth and disease. Here we present the results of an investigation of the balance between protein folding and degradation for mammalian chaperone Hsp90-dependent proteins. An understanding of these processes is not only of intrinsic interest, but is also of substantial biomedical importance. Many cancer-associated proteins are ‘clients’ of Hsp90 and intervention to tilt the balance from folding to degradation is an ongoing therapeutic strategy (1).
A complex array of chaperones and co-chaperones play a role in Hsp90 associated protein folding and degradation. The central players, on which we focus, are Hsp70, Hsp90, HOP, and CHIP:
Hsp70 is a heat shock induced monomeric molecular chaperone (see experimental procedures and Figure S1). Heat shock cognate 70 protein (Hsc70) is a constitutively expressed homolog of Hsp70. Hsp70 and Hsc70 are ~ 95 % identical in sequence. Other than the difference in expression profiles, Hsp70 and Hsc70 appear to be functionally identical and are considered here to be interchangeable. Hsp70 contains two domains: an N-terminal domain with ATPase activity and a C-terminal domain, which binds short hydrophobic sequences on its client proteins (2). A hinge region in the C-terminal domain allows the domain to ‘open’, exposing a client protein binding site. This change in Hsp70 conformation is coupled to the binding of ATP to the N-terminal domain. The ATP bound state is ‘open’ – a client protein can access the site, but can also leave it readily: kon/koff rates are high and affinity is low (3). By contrast, the ADP bound state has the hinge closed so that the substrate is enveloped. Here, kon/koff rates are slow and affinity is high. Certain mutations in Hsp70 can shift the equilibrium between the closed and open states (4). At the very C-terminus of Hsp70 is the sequence PTIEEVD, which binds to the TPR1 domain of the co-chaperone Hsp70/Hsp90 organizing protein (HOP)1 (5), see below.
Hsp90 is a heat shock induced homo-dimeric molecular chaperone (6). Each monomer has three domains: an N-terminal ATP-binding domain, a middle domain that regulates the ATPase activity of the N-terminal domain, and a C-terminal dimerization domain. At the very C-terminus of each monomer is the sequence MEEVD, which binds to the TPR2A domain of HOP (5).
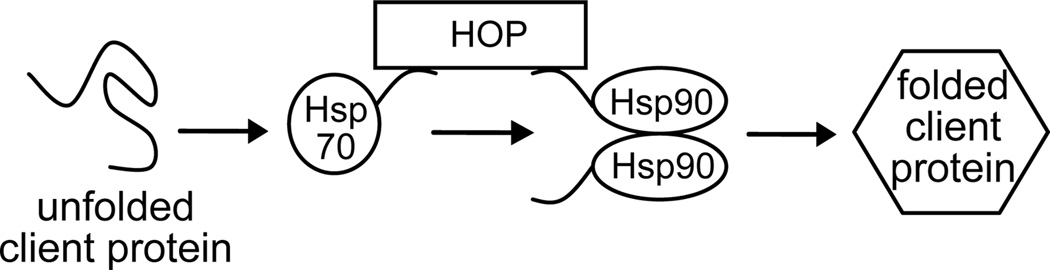
HOP is a modular protein that contains independent tetratricopeptide repeat (TPR) domains. TPR1 specifically recognizes the C-terminus of Hsp70 and TPR2A specifically recognizes the C-terminus of Hsp90. The simultaneous binding of both Hsp70 and Hsp90 to HOP brings them together into an active complex, in which client proteins are passed from Hsp70 to Hsp90 to complete their folding and maturation (Figure 1) (7). Although previously some researchers proposed that HOP is a dimer (8–10), recent conclusive data indicated that HOP is a monomer (11).
FIGURE 1.
Schematic of the Hsp70/Hsp90 folding pathway. Hsp70 and Hsp90 are brought into spatial proximity by binding to separate HOP domains. An unfolded client protein first interacts with Hsp70 and partially folded, is then passed to Hsp90, where its folding process is completed.
Carboxyl terminus of Hsc70-interacting protein (CHIP) was first identified as a ubiquitin ligase which binds to Hsc70 (12) and has since been implicated as being the quality control regulator of the Hsp70/Hsp90 folding pathway. CHIP is a homo-dimer (see experimental procedures) (13, 14) and each monomer contains an N-terminal TPR domain, a central helical domain, and a C-terminal U-box ubiquitin ligase domain. The TPR domain of CHIP has been reported to interact not only with the C-terminus of Hsc70 or Hsp70, but also with the C-terminus of Hsp90 (13, 15). CHIP has been proposed to ubiquitinate Hsp70/Hsp90 client proteins, thus targeting them to the proteasome for degradation (15, 16). Inhibition of Hsp90 by benzoquinone ansamycin geldanamycin or its derivative 17-AAG, for example, results in rapid degradation of the Hsp70/Hsp90 client protein HER2/neu in a CHIP-dependent fashion (17). CHIP, thus, links the protein folding with the protein degradation pathway. It has also recently been proposed that after CHIP ubiquitinates Hsp70/Hsp90 client proteins, it ubiquitinates Hsp70 itself (18). This chaperone ubiquitination has been suggested to be a mechanism to decrease the high levels of Hsp70 after a heat shock.
The CHIP-dependent quality control mechanism of the Hsp70/Hsp90 folding pathway is not well understood. Here we investigate the interplay between Hsp70, Hsp90, HOP, and CHIP and how they influence the folding/degradation balance. In order to perform these studies, it was essential that we fully characterize the properties of all the components and their interactions in vitro. We then used these quantitative measurements, in combination with estimates of protein concentrations in vivo, to propose a regulatory mechanism of the balance between Hsp90-dependent client protein folding and degradation.
EXPERIMENTAL PROCEDURES
DNA constructs
Full-length human CHIP was cloned as a BamHI/NcoI restriction fragment from pcDNA3-CHIP (a gift from Cam Patterson) (15) into pET11a (Stratagene) which had been modified to include a TEV protease cleavable His6-tag between the promoter and cloning cassette to create pET11a-His-TEV-hCHIP. GST-CHIP fusion construct in pGEX-KG (pGST-hCHIP (1–303)) was a gift from Cam Patterson (12). His6-Hsp90β in pET-14b (19) and His6-Hsp70 in pET28 were gifts from Sophie Jackson. The T13G mutation was introduced into Hsp70 using site-directed mutagenesis to create pET28-His6-Hsp70-T13G. Full-length human HOP in pACT2 was a gift from Chung Wang (20). It was sub-cloned into pProEX-HTa (Invitrogen) using SpeI and XhoI restriction sites to construct pProEX-HTa-His6-TEV-HOP. Full-length human UbcH5c in pET28a (without a tag) was a gift from Rachel Klevit (21).
Recombinant protein expression and purification
All recombinant proteins were expressed in BL21(DE3) E. coli cells induced with 0.8 mM IPTG. Proteins were purified using either Ni-NTA agarose (Qiagen) for those with a His6-tag or glutathione agarose (Sigma) for GST-CHIP according to manufacturer’s instructions. All proteins with His6-tag were further purified over Superdex S200 16/60 gel filtration column (Amersham) in 50 mM Tris pH 7.4, 10 mM NaCl, 5 mM β-mercaptoethanol (βME) buffer. GST-CHIP was dialyzed into the same buffer directly after purification with glutathione agarose. Proteins were used without removing the tags, unless noted otherwise.
UbcH5c purification was performed as described in (21) with slight modifications. Briefly, UbcH5c in cell lysate was loaded onto UNO S1 ion exchange column (Bio-Rad) equilibrated in 30 mM MES pH 6, 1 mM EDTA, 2 mM βME and eluted using 0–1 M NaCl gradient. Further purification was achieved using a Superdex S75 16/60 gel filtration column (Amersham), equilibrated in 25 mM Na phosphate pH 7, 150 mM NaCl, 1 mM βME. Final protein stocks were dialyzed into the same buffer but with lower βME concentration of 0.1 mM. Protein concentrations were determined using absorbance at 280 nm.
All peptides were synthesized by the W. M. Keck Foundation (Yale School of Medicine) and their concentrations determined by amino acid analysis at the W. M. Keck Foundation or by absorbance at 280 nm.
Oligomeric states of Hsp70 and CHIP. Although predominantly a monomer, recombinant Hsp70 has been observed by us and others to also associate into higher order structures (22). These Hsp70 higher order states however are not biologically relevant to Hsp70 chaperone function, as they cannot bind substrate peptides and are not observed in Hsp70-HOP-Hsp90 folding complexes (23, 24). Because these higher order states do not exist when Hsp70 is in the open conformation (Figure S1 in Supporting Information), they most likely form when an Hsp70 molecule via its substrate binding site associates with an unfolded region or a more specific site on another Hsp70 molecule (25). Here, we used purified Hsp70 monomers for our biochemical analyses.
Our experiments where CHIP was chromatographed over a size-exclusion column indicate (data not shown) that the protein is in equilibrium between two oligomeric states. Because in the two available crystal structures (13, 14) CHIP is a homodimer with an extensive dimerization interface, we presume that the protein is in a dimer-tetramer equilibrium. The two oligomeric species of CHIP can be separated on a gel-filtration column and the inter-conversion rate is on the order of days at 4°C. ITC experiments indicate (data not shown) that both oligomeric species of CHIP bind Hsp70 with the same binding affinity and stoichiometry and are therefore functional. CHIP dimer was used in characterizing CHIP-Hsp70 and CHIP-Hsp90 complexes (see below).
Characterization of protein complexes
To monitor CHIP-Hsp70 and CHIP-Hsp90 complex formation, equimolar mixtures of CHIP dimer with Hsp70 monomer or CHIP dimer with Hsp90 dimer were chromatographed on a Superdex S200 10/30 size-exclusion column (Amersham) equilibrated in 50 mM Tris pH 7.4, 10 mM NaCl, 5 mM βME at 4°C. The protein composition of each fraction was determined by Coomassie-stained SDS-polyacrylamide gels. To determine the stoichiometry of the complexes, the relative amount of protein in each band in fractions 25, 26, 27, 28, and 30 when CHIP + Hsp70 were chromatographed and in fractions 30 and 32 when CHIP + Hsp90 were chromatographed was determined by calculating band staining intensities (sum of all pixel intensities in a band) using Kodak 1D image analysis software v 3.5. We corrected for inherent differences in the staining between CHIP, Hsp70, and Hsp90 by determining the intensities of each band in standard solutions.
Surface Plasmon Resonance
Surface plasmon resonance (SPR) experiments were performed on a Biacore 3000 system (Biacore). Typically, ~3000 RU of NeutrAvidin (Thermo Fisher Scientific) was immobilized on CM5 sensor chip through amine linkage, according to manufacturer’s instructions. The entire immobilization procedure was performed in 10 mM HEPES pH 7.5, 150 mM NaCl, 3 mM EDTA, 0.005% [vol/vol] polysorbate 20 (P20) buffer. In the experiments shown, 343 RU of Hsp90 C-terminal peptide (biotin-SAAVTEEMPPLEGDDDTSRMEEVD-COO−), 168 RU of Hsp70 C-terminal peptide (biotin-GGFPGGGAPPSGGASSGPIEEVD-COO−), and 722 RU or 789 RU of model client protein (FYQLALTGGGGKGKGKGKGKGKGKK-biotin) were immobilized. To measure the affinity of the CHIP-Hsp70 and CHIP-Hsp90 interactions, 240 µl of various concentrations of CHIP were injected over immobilized Hsp70 and Hsp90 C-terminal peptides at 30 µl/min in the above immobilization buffer. To measure the binding affinity between the model client protein and Hsp70, Hsp70 T13G, and CHIP, various concentrations of Hsp70 (85 µl at 40 µl/min), Hsp70 T13G (200 µl at 30 µl/min), and CHIP (180 µl at 30 µl/min) were injected over immobilized model client protein in 50 mM Tris pH 7.4, 10 mM NaCl, 150 mM KCl, 3 mM MgCl2, 5 mM ATP, 5 mM βME, 0.005 % [vol/vol] P20 buffer. Sensor chip surface was regenerated between injections with 1 M NaCl. The average difference values in response units at steady-state equilibrium (RUeq) of the reference channel subtracted sensograms were plotted as a function of protein concentration ([P]). The data were fit to a one-site binding model (eq 1).
| (1) |
where Rmax is the response value at saturation and Kd is the dissociation constant.
Isothermal Titration Calorimetry
Shown ITC experiments were performed using VP ITC (Microcal) in 50 mM Tris pH 8, 10 mM NaCl, 10 mM βME buffer at 25°C. 19.7 µM CHIP was titrated with 95 – 3 µl injections of 0.322 mM Hsp70 C-terminal peptide GGGAPPSGGASSGPTIEEVD-COO−. 96 – 3µl injections of 0.4 mM Hsp90 C-terminal peptide GSSAAWTEEMPPLEGDDDTSRMEEVD-COO− were also added to 22.4 µM CHIP. The average integration value of the last 20 or 10 heats of dilutions (for Hsp70 and Hsp90 respectively) in each experiment were subtracted from all injection heats. Data were fit to one set of sites binding model with stoichiometry, association constant and binding enthalpy as floating variables.
In-vitro ubiquitination assays
To compare rates of Hsp70, Hsp70 T13G, and Hsp90 ubiquitination, 0.091 µM human ubiquitin activating enzyme (UBE1) (BostonBiochem), 20 µM UbcH5c, 20 µM CHIP, and either Hsp70, Hsp70T13G, or Hsp90 (all 20 µM) were combined in 50 mM Tris pH 7.4, 10 mM NaCl, 3 mM MgCl2, 150 mM KCl, 10 mM ATP, 0.1 mM βME buffer. For model client protein ubiquitination experiments, 0.091 µM UBE1, 20 µM UbcH5c, 40 µM CHIP, 40 µM model client protein (FYQLALTGGGGKGKGKGKGKGKGKK-biotin), and either 40 µM Hsp70, 40 µM Hsp70 T13G, or no Hsp70 were combined in the above buffer. To compare Hsp70 T13G ubiquitination with and without the presence of the model client protein, three independent experiments were performed for each condition where 0.091 µM UBE1, 20 µM UbcH5c, 20 µM CHIP, 100 µM Hsp70 T13G, and +/− 188 µM model client protein were combined in the above buffer. All ubiquitination reactions were pre-incubated at 30°C for ~10 minutes, initiated by the addition of 0.5 mM ubiquitin (Sigma), and kept at 30°C for the duration of the reaction. Aliquots were quenched with SDS sample buffer and analyzed by Coomassie-stained SDS-polyacrylamide gels.
To compare the rates of ubiquitination of Hsp70, Hsp70 T13G, and Hsp90, the intensity of staining of the band corresponding to unmodified species was determined with Kodak 1D image analysis software v 3.5 and the amount of ubiquitinated protein calculated. nmole of ubiquitinated protein (y) was graphed as a function of time (t) and the data fit to equation 2:
| (2) |
where a is the maximum amount of ubiquitinated protein and b is the rate of ubiquitination. Because in vitro ubiquitination of Hsp70, Hsp70 T13G, and Hsp90 were performed at the same time with the same stocks of UBE1, UbcH5c, CHIP, ubiquitin, and Mg-ATP, discrepancies in the rate of ubiquitination of various target proteins due to differences in protein stocks were eliminated. To analyze the rates of Hsp70 T13G ubiquitination with and without the model client protein, the amount of Hsp70 T13G or ubiquitin in each Hsp70 T13G-ubiquitin conjugate species and in unmodified Hsp70 species was determined using band staining intensity data. The number of ubiquitin molecules attached to Hsp70 T13G per CHIP monomer with standard deviation error bars determined from three independent experiments was graphed as a function of time and the beginning of the curve fit to a linear equation where the slope is the rate of ubiquitination.
Pull-down experiments
Competition pull-down experiments were performed in 50 mM Tris pH 7.4, 10 mM NaCl, 5 mM βME buffer. 3, 6, 10, 15, 20, or 30 µM His6-HOP, 10 µM GST-CHIP, and 10 µM His6-Hsp90 or various combinations of the proteins for control experiments were combined as 20 µl protein mixtures and incubated on ice for ~10 minutes. The mixtures were added to 10 µl of glutathione agarose (Sigma) and incubated rotating at 4°C for 1 hour. The resin was washed 4 times with 200 µl aliquots of buffer. Proteins remaining on the resin were eluted with SDS sample buffer and analyzed on a Coomassie-stained SDS-polyacrylamide gel.
To confirm that Hsp90 is able to pull down HOP, a control experiment was performed in the above buffer supplemented with 150 mM KCl and 3 mM MgCl2. 20 µM HOP without the His6-tag and +/− 20 µM His6-Hsp90 were combined as 20 µl protein mixtures and incubated on ice for ~ 10 minutes. The mixtures were added to 10 µl of washed Ni-NTA agarose (Qiagen) and incubated with rotation at 4°C for 1 hour. The resin was washed 3 times with 150 µl aliquots of buffer. Proteins remaining on the resin were eluted with 50 mM Tris pH 7.4, 150 mM NaCl, 2 mM βME, 300 mM imidazole elution buffer and resolved on a Coomassie-stained SDS-polyacrylamide gel.
Quantitative Western blot analysis
Approximately 2.5×106 BT474 breast cancer cells (ATCC) were seeded (25 cm2 flask) and allowed to adhere overnight. They were then treated with 0.178 µM 17-AAG or left untreated. 17-AAG treated cells were harvested at 7 and 14 hours and untreated cells were harvested at 15 hours after treatment. Cells were detached with 0.25% Trypsin-EDTA (Gibco) and their concentration determined by counting at least 1500 cells on a hemacytometer. Cells were washed with PBS and finally resuspended in 50 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% SDS, 1 ‘Complete’ protease inhibitor cocktail tablet (Roche Molecular Biochemicals) lysis buffer. DNA was sheared with a 30G needle. Cell concentration was kept constant during the PBS wash and in the lysis buffer. To qualitatively assess the amounts of GAPDH and HER2, total protein extract from 35,000 lysed cells from each treatment condition were separated on an SDS-polyacrylamide gel and Western blotted as in (26). Membranes were probed with HRP-conjugated anti-GAPDH antibody (Abcam) or with rabbit polyclonal anti-Neu antibody (Santa Cruz Biotechnology, C-18). To determine the amount of Hsp70, Hsp90, CHIP, and HOP in a cell, total protein extract from either 35,000 cells (for Hsp70, Hsp90, and HOP blots) or 70,000 cells (for CHIP blots) was separated on an SDS-polyacrylamide gel. A concentration series of purified protein was run along side for concentration calibration. Membranes were probed with mouse monoclonal anti-Hsp70 (Stressgen, SPA-810), rabbit polyclonal anti-Hsp90α/β (Santa Cruz Biotechnology, H-114), rabbit polyclonal anti-CHIP (Santa Cruz Biotechnology, H-231), or mouse monoclonal anti-HOP (gift from David Smith, F5) (27) antibodies. Blots were further incubated with anti-mouse or anti-rabbit HRP-conjugated secondary antibodies (both from Amersham) and developed using enhanced chemiluminescence (Amersham). The intensity of staining (determined using Kodak 1D image analysis software v 3.5) of bands corresponding to a purified recombinant protein was plotted as a function of the known protein amount and the data fit to the linear equation to create a concentration calibration graph. The calibration curve was then used to convert the intensity of staining of bands corresponding to the endogenous protein to the amount of the protein in the cells. Cellular concentrations of Hsp70, Hsp90, CHIP, and HOP were calculated based on the known number of cells from which protein extracts were made and loaded on a gel, amount of protein in those cells and taking the average BT474 cell volume of 3.5×10−6 µl as reported (28).
RESULTS
Quantification of the affinity and stoichiometry of the CHIP-Hsp70 and CHIP-Hsp90 interactions
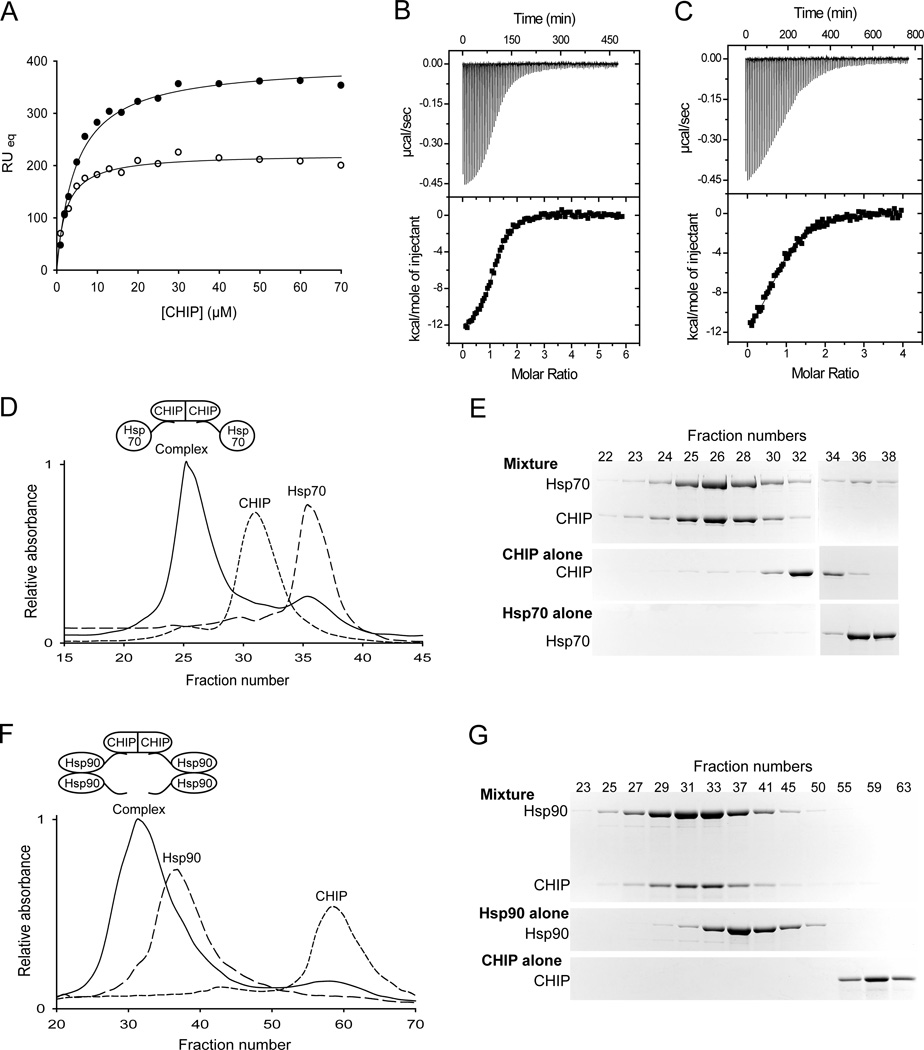
We measured the affinity of the interaction of CHIP with the C-terminal peptides of Hsp70 and Hsp90 using surface plasmon resonance (SPR) and determined the dissociation constants to be ~ 2 µM and ~ 5 µM, respectively (Figure 2A). We also used isothermal titration calorimetry (ITC) to confirm these dissociation constants and to determine the stoichiometry of the interactions. By ITC the dissociation constant for CHIP-Hsp70 is ~ 1 µM and stoichiometry of the interaction is ~ 0.8 C-terminal Hsp70 peptide per CHIP monomer (Figure 2B). The dissociation constant for CHIP-Hsp90 is ~ 4.4 µM and the stoichiometry of the interaction is ~ 0.9 Hsp90 C-terminal peptide per CHIP monomer (Figure 2C).
FIGURE 2.
Complex formation between Hsp70 or Hsp90 and CHIP. (A) CHIP-peptide interactions monitored by surface plasmon resonance (SPR). Plot of the average response at equilibrium (RUeq) versus concentration of CHIP. Open circles show binding to immobilized C-terminal peptide of Hsp70, solid circles show binding to immobilized C-terminal peptide of Hsp90. The solid lines show fits to a one-site binding equation, with dissociation constants of ~ 2 µM and ~ 5 µM, for CHIP binding to Hsp70 and Hsp90, respectively. (B and C) Isothermal Titration Calorimetry (ITC). Upper panels: thermogram of the titration of the C-terminal peptide of Hsp70 (B) or Hsp90 (C) into a solution of CHIP. Lower panels: plot of integrated areas under the peaks of heat in the upper panel as a function of molar ratio. Fitting the data gives a dissociation constant of ~ 1 µM with a stoichiometry of ~ 0.8 C-terminal Hsp70 peptide per CHIP monomer and a dissociation constant of ~ 4.4 µM with a stoichiometry of ~ 0.9 Hsp90 C-terminal peptide per CHIP monomer. (D – G) Complex formation between CHIP and Hsp70 or CHIP and Hsp90, all full-length. Size-exclusion chromatograms of purified CHIP dimer alone, purified Hsp70 monomer alone and an equimolar mixture of the two proteins (D). Coomassie-stained SDS-polyacrylamide gels of the indicated fractions (E). Size-exclusion chromatography analysis of purified CHIP dimer alone, purified Hsp90 dimer alone and an equimolar mixture of the two proteins (F). Coomassie-stained SDS-polyacrylamide gels of the indicated fractions (G). Cartoons of the proposed CHIP-Hsp70 and CHIP-Hsp90 complexes, based on the analysis of the composition of each fraction (see experimental procedures), are shown in (D) and (F), respectively. CHIP does not elute in the same fractions in (D) and (F), because the volume of the fractions is different in the two experiments.
We used size-exclusion chromatography to analyze the nature of the complexes between full-length Hsp70 (Figure 2D and 2E) or full-length Hsp90 (Figure 2F and 2G) and CHIP. Complexes between Hsp70 and CHIP or Hsp90 and CHIP formed readily and were clearly identifiable as new peaks in the gel filtration profile. By analyzing the composition of these peaks, we determined the stoichiometry of the CHIP-Hsp70 complex to be ~ 0.8 Hsp70 monomer per CHIP monomer, in other words, a CHIP dimer binds to two Hsp70 monomers. The stoichiometry of the CHIP-Hsp90 complex was determined to be ~ 2.1 Hsp90 monomers per CHIP monomer. In other words, a CHIP dimer binds to two Hsp90 dimers.
Stoichiometries for CHIP-Hsp70 and CHIP-Hsp90 complexes obtained using either C-terminal peptides or full-length proteins, thus, agree well with each other. These measured dissociation constants and stoichiometries for the CHIP and Hsp90 C-terminal peptide interaction also agree well with the previously reported affinity between CHIP and full length Hsp90 (13). Our own ITC experiments using CHIP and full length Hsp70 confirm the dissociation constant to be ~ 1 µM (data not shown). Measured binding affinities and stoichiometries allowed us to design the next set of experiments using appropriate conditions.
CHIP ubiquitinates Hsp70 and Hsp90
It has been proposed that CHIP can ubiquitinate Hsp70 and Hsp90 (18, 29). Our first experiments investigated the details of ubiquitination of Hsp70 and Hsp90 by CHIP, by reconstituting ubiquitination in vitro using purified components. In these assays we used UbcH5c as the ubiquitin-conjugating enzyme E2, because it has been shown to function with CHIP (16). Because we had determined the dissociation constants for CHIP-Hsp70 and CHIP-Hsp90 complex formation, we were able to adjust the conditions of the ubiquitination assays so that most of the CHIP and chaperone molecules are bound to each other (concentration of each component is at least four times Kd).
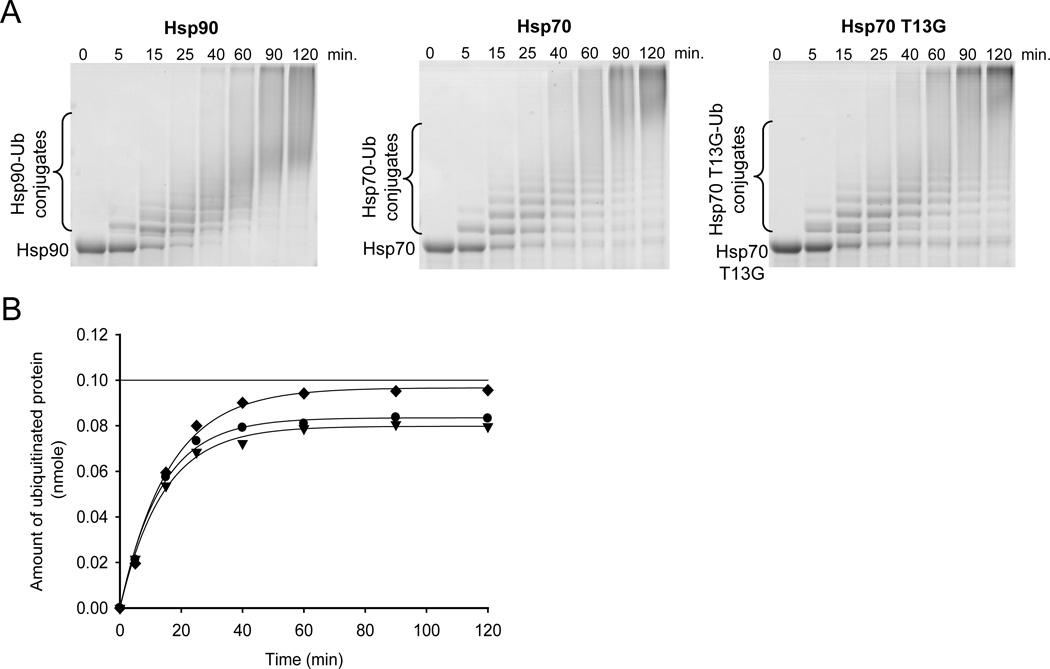
Given that ubiquitination is an ATP dependent reaction, it was necessary to have ATP (10 mM) in the reaction mixture. Under the conditions of the ubiquitination assay, therefore, Hsp70 is predominantly in the ATP bound, or ‘open’ conformation. To test whether the rate of Hsp70 ubiquitination is dependent upon the ‘open’ or ‘closed’ states of Hsp70 we created and tested the Hsp70 T13G mutant. It has been previously shown that Hsp70 with the T13G mutation binds ATP, but it is unable to undergo the ATP-binding induced conformational change (4, 30). The Hsp70 mutant is trapped in the ‘closed’ conformation. Figure 3A shows ubiquitination time-courses of Hsp90, Hsp70 (wt), and Hsp70 T13G. Comparable rates of ubiquitination of either Hsp70 or Hsp90 by CHIP were observed (Figure 3B). The rate of ubiquitination of Hsp70 (wt) and T13G are also very similar, indicating that the conformation of Hsp70 has no effect on how fast or the extent to which the chaperone is ubiquitinated.
FIGURE 3.
Ubiquitination of various substrates by CHIP. (A) In vitro ubiquitination of Hsp90, Hsp70, and Hsp70 T13G by CHIP. Coomassie-stained SDS-polyacrylamide gel analysis of the reaction at various times after initiation. Protein bands corresponding to unmodified protein and its ubiquitin conjugates are indicated. (B) Time course of ubiquitination of Hsp90 (diamonds), Hsp70 (circles), and Hsp70 T13G (triangles). Ubiquitination was quantified by determining the amount of protein in the unmodified band as a function of time. Data were fit to first-order rate equation. CHIP ubiquitinates Hsp90, Hsp70, and Hsp70 T13G at the rates of 0.063, 0.074, and 0.071 nmole of ubiquitinated protein/minute, respectively. Linear line at 0.1 nmole of ubiquitinated protein indicates 100% substrate ubiquitination.
Model client protein binds to Hsp70 and CHIP
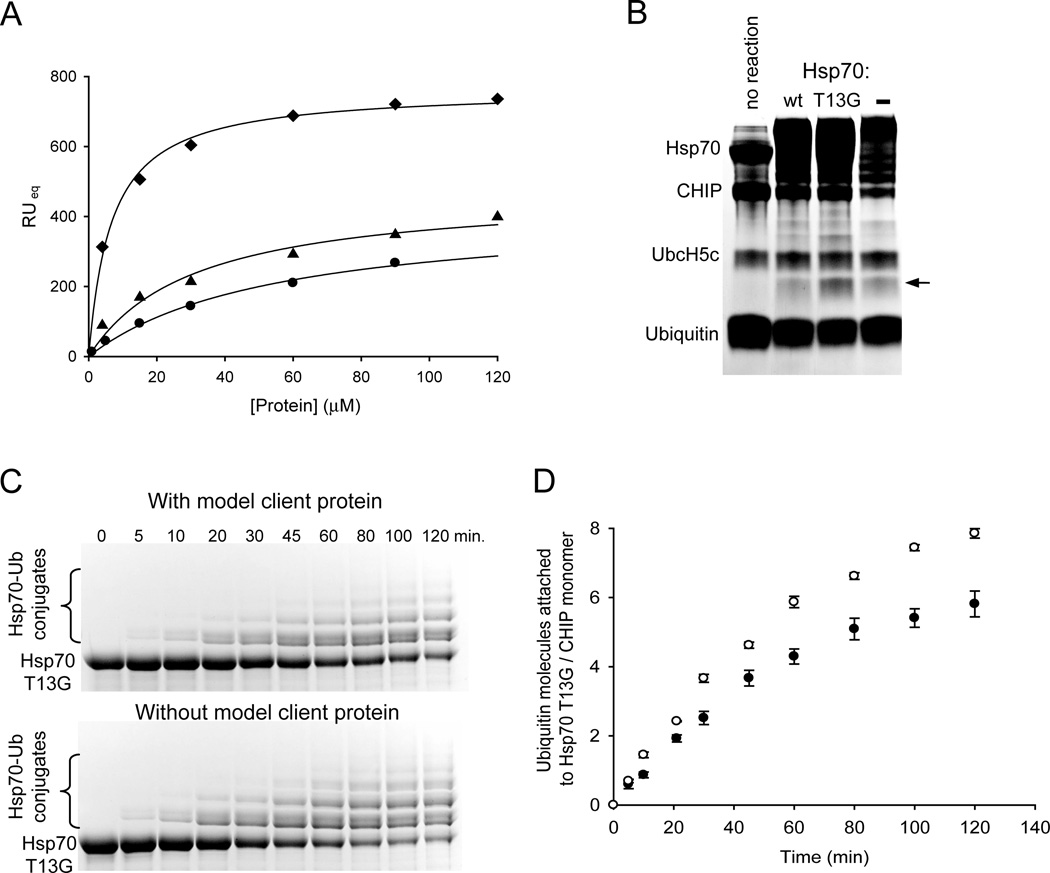
As the next step in elucidating the function of CHIP, we investigated the ubiquitination of an Hsp70 client protein and the effect of the client protein on Hsp70 ubiquitination rate. It was essential to first determine the Kd between the client protein and Hsp70. To ensure a homogeneous system, we based the design for the client on a previously characterized Hsp70 client peptide FYQLALT (3). Because a sample of denatured protein is composed of various species, it was not a good choice as a client protein for our experiments. We designed a biotinylated model client protein (sequence: FYQLALTGGGGKGKGKGKGKGKGKK-biotin) which includes an Hsp70 binding site (underlined) (3) plus lysine residues for ubiquitination separated by a short glycine linker. SPR experiments indicate that immobilized client protein interacts with Hsp70 and Hsp70 T13G with dissociation constants of ~ 50 µM and ~30 µM respectively (Figure 4A). The observation that Hsp70 T13G has a higher affinity for the client protein than wt Hsp70 is consistent with studies showing this mutant to be locked in the ‘closed’ conformation (4, 30). Interestingly, CHIP was also found to interact directly with the client peptide (Kd ~ 6.7 µM, Figure 4A). It has been previously suggested that CHIP may be able to bind unfolded proteins directly (31), but this possible interaction has not been well studied and its biological relevance is unclear. Although the model client protein binds to CHIP directly, we chose not to try a different client protein in subsequent ubiquitination assays. The designed client protein is well characterized, we know its dissociation constant with Hsp70, and thus is still a good choice for our experiments.
FIGURE 4.
Model client protein binds Hsp70 and CHIP, is ubiquitinated by CHIP, and affects the rate of Hsp70 ubiquitination. (A) Interactions of the model client protein with Hsp70, Hsp70 T13G, and CHIP monitored by SPR. Plot of the average response at equilibrium versus concentrations of proteins. Binding of immobilized client protein to Hsp70 (circles), Hsp70 T13G (triangles), and CHIP (diamonds) is depicted. The solid lines show fits to a one-site binding equation with dissociation constants of ~ 50 µM for Hsp70, ~ 30 µM for Hsp70 T13G, and ~ 6.7 µM for CHIP. (B) In vitro ubiquitination of the model client protein by CHIP. Coomassie-stained SDS-polyacrylamide gel analysis of 2 hour ubiquitination reactions in the presence of Hsp70, Hsp70 T13G, or in the absence of Hsp70. Protein band corresponding to the client protein conjugated to one ubiquitin is indicated with an arrow. Other components of the reaction are labeled. (C) In vitro ubiquitination of Hsp70 T13G in the presence and absence of the model client protein. Coomassie-stained SDS-polyacrylamide gel analysis of the reactions at various times after initiation. Time points are the same for both conditions. Unmodified Hsp70 T13G as well as its ubiquitin conjugate species are indicated. (D) Time course of Hsp70 T13G ubiquitination in the presence (solid circles) and absence (open circles) of the model client protein. Ubiquitination was quantified from 3 independent experiments as in (C) by determining the amount of ubiquitin attached to Hsp70 T13G as a function of time. The initial phase of the curve was fit to a linear equation and the obtained rates for Hsp70 T13G ubiquitination are ~ 0.07 and ~ 0.1 ubiquitin molecules attached to Hsp70 T13G/CHIP monomer/minute in the presence and absence of client protein, respectively.
Client protein is ubiquitinated by CHIP and slows down the rate of Hsp70 ubiquitination
We employed in vitro assay to study ubiquitination of the client protein by CHIP. As Figure 4B indicates, the client protein is ubiquitinated by CHIP in the presence of either Hsp70 or Hsp70 T13G and this ubiquitination was confirmed with anti-biotin antibody Western blot analysis (data not shown). The amount of client protein ubiquitination is, however, considerably higher in the presence of Hsp70 T13G than wt Hsp70. This difference is most likely due to the decrease in Kd and increase in half-life of Hsp70-client protein complex for mutant versus wild-type Hsp70 (3). The model client protein is also ubiquitinated in the absence of Hsp70 (Figure 4B). We presume this ubiquitination is due to the direct interaction of CHIP with the model client protein. The amount of client protein ubiquitination is, however, higher with the addition of Hsp70 T13G than in the absence of Hsp70. This observation indicates that Hsp70 T13G mediates ubiquitination of the client protein. Because client protein is evidently ubiquitinated through two pathways in the presence of Hsp70, ubiquitination mediated by Hsp70 and via the direct interaction with CHIP, we were unable to compare the rate of Hsp70 ubiquitination with that of the client protein.
By increasing the concentration of the client protein so that both Hsp70 and CHIP are saturated with the client protein, we were, however, able to compare the rates of Hsp70 T13G ubiquitination with or without bound client protein. As the time-course data (Figure 4C) and its quantification (Figure 4D) of Hsp70 T13G ubiquitination +/− client protein show, Hsp70 T13G is ubiquitinated robustly in both conditions, albeit somewhat slower in the presence of the client protein: ~ 0.07 and ~ 0.1 ubiquitin molecules attached/CHIP monomer/minute in the presence and absence of the client protein respectively. CHIP is mediating ubiquitination of both the client protein and Hsp70. The difference in the rates is, therefore, most likely due to the competition between Hsp70 and the client protein for the ubiquitin molecules. It has previously been proposed that a client protein bound to Hsp70 undergoes CHIP mediated degradation before Hsp70 itself (18). Our data show that even when a client protein is bound to Hsp70, the chaperone is ubiquitinated robustly.
Direct competition between protein folding and degradation machineries
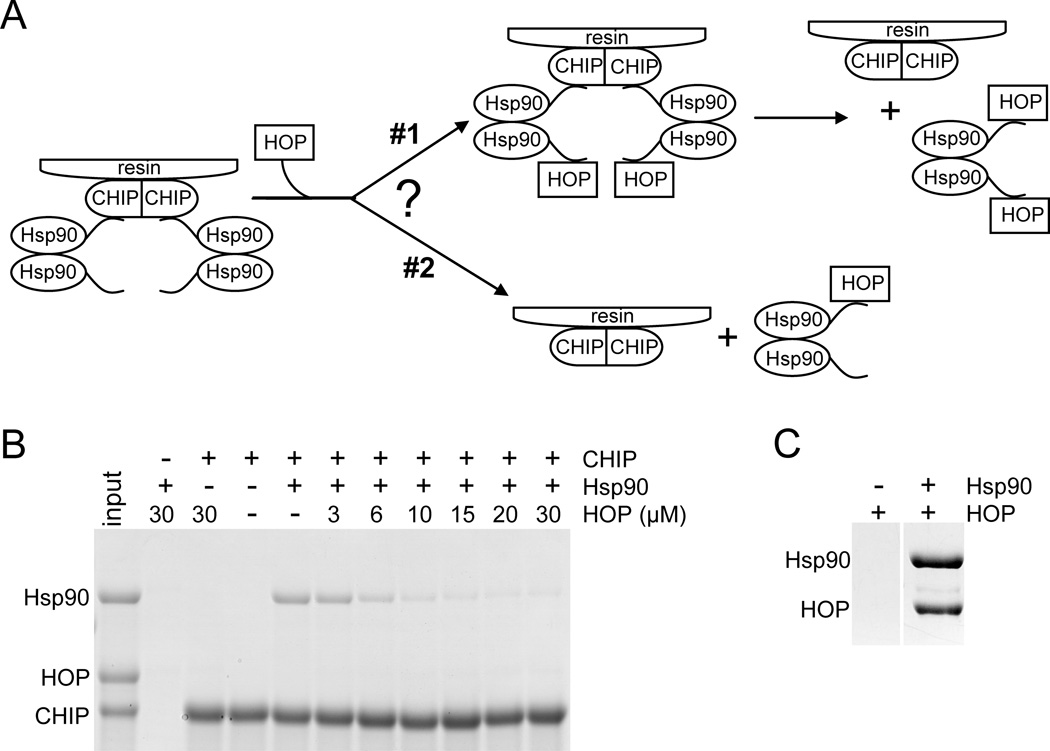
We next addressed the question whether the protein folding and degradation machineries are in direct competition or whether a complex exists which includes a component of each of the two opposing pathways. In other words, can either Hsp70 or Hsp90 bind to both HOP and CHIP at the same time? Because HOP and CHIP bind to Hsp70 and Hsp90 at the same site (C-terminus), only oligomerization of Hsp70 or Hsp90 could facilitate the formation of a complex which would include both HOP and CHIP. Because Hsp70 is a monomer (see experimental procedures and Figure S1), a HOP-Hsp70-CHIP complex cannot form. Hsp90 dimer, on the other hand, could theoretically form HOP-Hsp90 dimer-CHIP complex. We performed competition pull down experiments to search for the existence of such a complex. These experiments are described in a schematic in Figure 5A with results shown in Figure 5B. It is evident that CHIP does not pull-down HOP through Hsp90, but that HOP and CHIP compete for binding to Hsp90 (scenario #2 in Figure 5A). Control experiment shows that the HOP and Hsp90 interaction is stable and we can observe it in a pull-down experiment (Figure 5C). We conclude that the examined folding and degradation machineries are mutually exclusive and that as Hsp70, Hsp90 is in complex with either HOP or CHIP.
FIGURE 5.
HOP and CHIP binding to Hsp90 is mutually exclusive. (A) Schematic illustration of the experiment. An initial complex of two Hsp90 dimers bound to a CHIP dimer is attached to the glutathione resin via GST-tag on CHIP. As the concentration of HOP is increased, it competes with CHIP for binding to Hsp90. Two different scenarios are depicted. In the first, an Hsp90 dimer can bind to both CHIP and HOP simultaneously. In the second, an Hsp90 dimer can bind to either CHIP or HOP, but cannot bind to both simultaneously. Eventually, in both scenarios, at high concentrations of HOP, all the Hsp90 will be bound to HOP, and none will be bound to CHIP and the resin. (B) Coomassie-stained SDS-polyacrylamide gel of the resin-bound proteins as the concentration of HOP competitor is increased. In each experiment +/− 10 µM GST-CHIP and +/− 10 µM Hsp90 were incubated with indicated concentrations of HOP. Protein bands corresponding to Hsp90, HOP, and CHIP are indicated. The input lane contains 20% of CHIP and Hsp90 at the above concentrations and 10 µM HOP. Control experiments in lanes 2 and 3 show that neither Hsp90 nor HOP non-specifically bind to the resin and that HOP does not bind to CHIP. The decrease in the amount of Hsp90 bound to CHIP as HOP concentration increases indicates that HOP is competing with CHIP for binding to Hsp90. Because HOP is never pulled-down by CHIP through the Hsp90 dimer, it is evident that HOP competes with CHIP for binding to Hsp90 via scenario #2 as depicted in (A). (C) Hsp90 pulls-down HOP in a similar experiment as in (B). Coomassie-stained SDS-polyacrylamide gel of Ni-NTA resin elusions of protein mixtures: +/− 20 µM His6-Hsp90 and 20 µM HOP with its His6-tag cleaved off by TEV protease. Protein bands corresponding to Hsp90 and HOP are indicated. HOP does not bind non-specifically to the resin but binds to Hsp90. Because the shown lanes are not adjacent on the gel, other lanes between them were cut out.
Proposed model for the balance between protein folding and degradation
Having characterized the function of CHIP in vitro, we next studied the effects of CHIP on the folding/degradation balance in vivo. A major question remains: Does CHIP selectively ubiquitinate only the client proteins unable to fold correctly, but not those undergoing their normal folding process and if so, what is the mechanism of the selection? For this study we employed the Hsp90 inhibitor 17-AAG, which binds to the ATP binding pocket and renders the chaperone unable to facilitate client protein folding process (32). It has been demonstrated that upon 17-AAG treatment of cells, client proteins such as the oncoprotein HER2/neu undergo rapid CHIP-dependent degradation (17). Inhibition of Hsp90 with 17-AAG, thus, causes cells to shift from client protein folding to ‘degradation mode’. We first hypothesized that the switch between the two opposing pathways may lie in changes in the cellular concentration of CHIP. We therefore used quantitative Western blot analysis to determine the total cellular concentrations of Hsp70, HOP, Hsp90, and CHIP in BT474 breast cancer cells under normal conditions and upon 17-AAG treatment (Figure 6A). Before considering how cellular concentrations of the chaperones and co-chaperones change with 17-AAG treatment, we will first focus on protein concentrations at normal conditions and how they influence the folding/degradation balance.
FIGURE 6.
Approximate cellular concentrations of Hsp70, Hsp90, HOP, and CHIP, the various complexes they form, and their effects on the folding/degradation balance. (A) Determination of the cellular concentrations of Hsp70, Hsp90, HOP, and CHIP under normal conditions and upon 17-AAG treatment using quantitative Western blot analysis. BT474 breast cancer cells were left untreated or treated with 0.178 µM 17-AAG for 7 or 14 hours. Total cell lysates from a known number of cells were loaded in each lane and blotted for various proteins. Increasing amount of purified recombinant Hsp70 (0.02, 0.06, 0.1, 0.3, 0.5 µg), Hsp90 (0.02, 0.06, 0.1, 0.14, 0.18 µg), HOP (0.005, 0.025, 0.045, 0.065, 0.085 µg), and CHIP (0.0003, 0.001, 0.002, 0.003 µg) were run along side for concentration calibration. Dashed line separates the endogenous from purified proteins. Cell lysates were also blotted for qualitative comparison of HER2 and GAPDH (loading control) levels. (B) A schematic depicting the approximate total cellular concentrations of Hsp70, Hsp90 dimer, HOP, and CHIP under normal conditions and the dissociation constants between interacting partners. Kd values for the interactions of CHIP with Hsp70 and Hsp90 are as determined here and those for the interactions of HOP with Hsp70 and Hsp90 are as reported (5). (C) Calculated approximate concentrations of the various protein complexes formed by Hsp70, Hsp90, HOP, and CHIP as well as free protein components (not bound to a co-chaperone). Concentrations at normal conditions were calculated based on the total protein concentrations and dissociation constants in (B). Concentrations after 17-AAG treatment were calculated the same way; only the total Hsp70 concentration was increased from 10 µM to 39 µM. (D) A schematic depiction of the folding/degradation balance under normal conditions based on the concentrations shown in (C). In the cell, 10 µM Hsp70 is a component of various complexes depicted in the schematic along with their approximate concentrations: Hsp70 can be free of a co-chaperone, bound only to HOP, bound to HOP-Hsp90, or bound to CHIP. The folding process of a client protein bound to Hsp70, which is free of a co-chaperone or only bound to HOP (~ 9.48 µM), cannot be completed. Client protein bound to Hsp70 within the Hsp70/HOP/Hsp90 folding complex (~ 0.43 µM) will be passed from Hsp70 to Hsp90 for final steps of maturation. Client protein bound to Hsp70 which is associated with CHIP (~ 0.083 µM) will be ubiquitinated. Thus, a client protein is ~ 5 times more likely to be folded (outcome highlighted in bold) than ubiquitinated. There is, however, a low level of client protein ubiquitination. (E) A schematic depicting the shift to client protein degradation upon 17-AAG treatment based on the concentrations shown in (C). When Hsp90 is inhibited, the amount of Hsp70 which is a component of the Hsp70/HOP/Hsp90 folding complex (~ 0.83 µM) and Hsp70 which is part of the Hsp70/CHIP degradation complex (~ 0.095 µM) does not differ much from the amounts at normal conditions in (D). The client protein thus has a similar probability of encountering the Hsp70/HOP/Hsp90 folding complex or the Hsp70/CHIP ubiquitination complex under both conditions. Because Hsp90 is inhibited upon 17-AAG treatment, however, even if the client protein is bound to Hsp70/HOP/Hsp90 complex, its folding process will be initiated, but not completed. Although the probability of the client proteins to encounter the degradation complex is low, all proteins will ultimately be degraded, because there is no alternative pathway.
As depicted in Figure 6B, the approximate total cellular concentrations under normal conditions are ~ 10 µM for Hsp70 monomer, ~ 3 µM for HOP monomer, ~ 5 µM for Hsp90 dimer, and strikingly only ~ 0.1 µM for CHIP monomer. Two independent experiments confirmed the low concentration of CHIP. Based on these concentrations and the Kd values for the different pairs of proteins, we calculated the approximate in vivo concentrations of the various complexes these proteins can form (Figure 6C). We assume an even distribution of components throughout the cell. The first important conclusion from these calculations is that there is ~ 10 fold more CHIP bound to Hsp70 than to Hsp90. It is therefore evident that the decision whether or not to ubiquitinate a client protein will be made while the client is bound to Hsp70 and that client protein ubiquitination via Hsp90 is a minor pathway. In the cell, Hsp70 is in equilibrium as a component of various complexes: it can be free of a co-chaperone (not bound to HOP or CHIP), bound only to HOP, in complex with HOP-Hsp90, or associated with CHIP. We presume that a client protein bound to Hsp70 has the same probability as Hsp70 of participating in the above listed complexes, although all Hsp70 proteins may not have a client protein bound to them. Our data indicate that under normal conditions (Figure 6D) ~ 94.87 % (9.48 µM out of the total 10 µM) of Hsp70 is free or interacting with non-Hsp90 bound HOP. ~ 4.3 % (0.43 µM out of the total 10 µM) of Hsp70 is bound to HOP-Hsp90, thus forming the productive Hsp70/HOP/Hsp90 folding complex. A client protein bound to Hsp70, in this case, will be passed to Hsp90 for folding completion. Lastly, only ~ 0.83 % (0.083 µM out of the total 10 µM) of Hsp70 is in complex with CHIP indicating that Hsp70 bound client proteins have a very small chance of encountering CHIP and being ubiquitinated. Under normal conditions, therefore, a client protein has a higher probability of being folded than degraded, but it is evident that there is a constant low level of client protein degradation. To our knowledge, the association of only one other co-chaperone, Tpr2, with the C-terminus of Hsp70 has been well characterized (33). Presence of Tpr2 or any other co-chaperone which interacts with the C-terminus of Hsp70 or Hsp90, however, does not change the ratio of the folding and degradation complexes in a cell.
Upon 17-AAG treatment we observe a decrease in HER2 levels, as expected (Figure 6A). We also see an increase in the total Hsp70 cellular concentration from ~ 10 µM to ~ 39 µM. Such an increase in Hsp70 levels has consistently been observed by others, and is believed to be due to heat shock factor 1 (HSF1) dependent activation of transcription (34). Hsp90 and HOP levels remain the same at ~ 5 µM and ~ 3 µM respectively. An increase in CHIP levels upon Hsp90 inhibition is not the explanation for client protein degradation. The concentration of CHIP remained low and constant ~ 0.1 µM. It is evident that in this degradation mode, the amounts of Hsp70 in the Hsp70/HOP/Hsp90 folding complex and in complex with CHIP are very similar to those when Hsp90 is not inhibited: ~ 0.43 µM and ~ 0.83 µM Hsp70 is a component of the folding pathway under normal conditions and upon 17-AAG treatment respectively, and ~ 0.083 µM and ~ 0.095 µM Hsp70 is a component of the degradation pathway under normal conditions and upon 17-AAG treatment respectively (Figure 6E). The chances, therefore, of an Hsp70-bound client protein to encounter the folding or the degradation pathways in the degradation mode are very similar to those when Hsp90 is not inhibited. In this case, however, even if a client protein is in the Hsp70-HOP-Hsp90 complex, it will not be folded because Hsp90 is inactive. Unable to fold, the client protein will continue to interact with Hsp70, which in turn will increase its chances of encountering CHIP and ultimately being ubiquitinated and degraded.
Based on these observations, we propose the following model for the balance between client protein folding and degradation. Under normal circumstances, Hsp70-bound client proteins are ubiquitinated by CHIP at a steady low rate, but they are more likely to be folded than ubiquitinated. When a client protein cannot fold properly or its kinetics of folding are greatly reduced due to Hsp90 inhibition, a mutation, or a cellular stressor, increased association with Hsp70 will greatly augment its chances of getting ubiquitinated and degraded. Our data showing that CHIP ubiquitinates Hsp90 and Hsp70 (even while ubiquitinating a client protein) also indicates a background level of chaperone degradation. Because only ~ 0.83 % of Hsp70 and ~ 0.16 % of Hsp90 is bound to CHIP, CHIP-mediated turnover of the chaperones will be low.
DISCUSSION
To ensure cellular health and survival, cells must produce folded, functional proteins. They must also have the capacity to respond rapidly to changing conditions, and to remove aberrant or damaged proteins. Various types of protein quality control regulators exist. Here we focus on the interplay between the folding and degradation pathways for Hsp90-dependent client proteins, in which the ubiquitin ligase, CHIP, is the key player.
To understand the underlying physical/chemical basis of the balance between folding and degradation, we first characterized and quantified the interaction of CHIP with the components of the folding complex in vitro. We determined that the C-terminal sequences of Hsp70 or Hsp90 are necessary and sufficient for CHIP binding and that CHIP ubiquitinates Hsp70 and Hsp90 at similar rates. The rate of ubiquitination of Hsp70 by CHIP is also independent of the conformational state of Hsp70, and CHIP ubiquitinates Hsp70 robustly even when a client protein is bound to the chaperone. By considering the binding affinities of Hsp70 and Hsp90 for CHIP, along with their intracellular concentrations, we conclude that ubiquitination of client proteins occurs predominantly in the client protein-Hsp70-CHIP complex. Ubiquitination in the client protein-Hsp90-CHIP complex represents a minor pathway, because the binding affinity of CHIP for Hsp70 is tighter than for Hsp90 and also the in vivo concentration of Hsp70 is higher than that of Hsp90. Thus CHIP is found predominantly in complex with Hsp70.
Our data also indicate that dimeric Hsp90 cannot bind to both HOP and CHIP simultaneously. Thus, the folding and degradation processes cannot co-exist in a single complex; rather, they are competing machineries. Under ‘normal’ conditions the probability that a client protein will be folded is much higher than the probability that it will be ubiquitinated and degraded, because the concentration of the Hsp70-HOP-Hsp90 complex is significantly higher than that of the Hsp70-CHIP complex. Nevertheless, even under ‘normal’ conditions there is a low background level of both client protein and chaperone ubiquitination and degradation.
When Hsp90 is inhibited, for example by treatment of cells with 17-AAG, Hsp70 levels increase, CHIP levels stay constant, and the distribution of Hsp70 in the folding versus degradation complexes also remains constant. However, because Hsp90 is inhibited, encounters of client proteins with the Hsp70-HOP-Hsp90 complex are non-productive. Incompletely folded client proteins are released, continue rebinding to Hsp70, and eventually are degraded in the Hsp70-CHIP-client protein complex. We propose that CHIP does not actively recognize a client protein that is incompletely or incorrectly folded, nor does the switch from folding to degradation require synthesis of more CHIP. Thus, the system can respond rapidly to changes in cellular conditions by channeling more Hsp70-bound client proteins to degradation rather than to folding.
In our analysis of the balance between protein folding and degradation we focused on Hsp90-dependent client proteins. Many client proteins, however, only require Hsp70 for proper folding. The proposed model also applies to these Hsp70-only dependent client proteins. A small percentage of these clients will constantly be turned-over by CHIP.
The kinetics of client protein transfer from Hsp70 to Hsp90 and the kinetics of client protein ubiquitination clearly also contribute to the overall probability of a client protein to be folded or degraded. Quantitative measurements for all the steps are not yet available. We do know, however, that the rates of ATP hydrolysis by Hsp70 and Hsp90 are similar to the rate of protein ubiquitination by CHIP. Human Hsp90 hydrolyses ATP with a kcat of ~ 0.09 min−1 (19) and the kcat for client peptide stimulated hydrolysis of ATP by Hsp70 is ~ 0.1–0.3 min−1 (35). We determined that at substrate concentrations much above KM, the rate of ubiquitination is ~ 0.1 ubiquitin molecules attached/CHIP monomer/minute. As a first approximation, we therefore assume that the rates of client protein folding and ubiquitination are similar, and that the fate of a client protein is determined by whether it is bound to an active Hsp70-HOP-Hsp90 complex or to an Hsp70-CHIP complex.
Predictions of our model
First, under ‘normal’ conditions, in the absence of 17-AAG, we predict a low but non-zero ubiquitination and degradation of HER2 and other client proteins. The half life of HER2 in vivo in normal and in CHIP −/− cells has previously been measured using cycloheximide to inhibit protein synthesis (17). It was observed that HER2 is degraded in a CHIP-dependent fashion, with a half life of less than 1 hour in ‘normal’ non-Hsp90 inhibited cells and more than 5 hours in CHIP −/− cells. This result supports our conclusion that even during normal conditions a significant proportion of newly synthesized HER2 is degraded by CHIP while undergoing the folding process, strongly suggesting the continuous ubiquitination of client proteins by CHIP.
Second, over-expression of CHIP should increase client protein degradation. It has consistently been demonstrated that exogenous over-expression of CHIP results in increased degradation of Hsp90-dependent client proteins (17, 36). Our data clearly predict that an increase in CHIP concentration will increase the fraction of Hsp70 that is bound to CHIP, resulting in a shift from predominantly client protein folding to predominantly client protein degradation. It was estimated that in these experiments exogenous over-expression increased the cellular concentration of CHIP about 20-fold (36). With a new total concentration of CHIP of approximately 2 µM, we estimate that 0.38 µM Hsp70 would be in complex with HOP/Hsp90, where client proteins can fold, but 1.6 µM Hsp70 will be in complex with CHIP, where client proteins are ubiquitinated. In other words, upon CHIP over-expression the folding/degradation balance completely flips; approximately four times more Hsp70 is now present in the degradation rather than in the folding complex.
Third, in our model, CHIP does not distinguish between a client protein undergoing its normal folding process and a mutant protein that is incapable of folding correctly. Thus, a shift to the degradation mode should induce ubiquitination not only of mutant proteins, but also wild-type proteins. It has been shown that an increase in CHIP cellular concentration leads to degradation of a cystic fibrosis causing mutant of Hsp70/Hsp90 client protein CFTR, CFTRΔF508, which is defective in folding and trafficking (36). As predicted by the model, an increase in CHIP cellular concentration also leads to degradation of wild-type CFTR, which is capable of folding correctly.
Finally, our model predicts a constant low level of CHIP-mediated ubiquitination of Hsp70 and Hsp90 with consequent degradation in ‘normal’ cells. CHIP-mediated turnover of Hsp70 has been studied extensively (18). An overall increase in Hsp70 levels in the absence of CHIP was noted. A decrease in Hsp70 levels was also observed upon over-expression of CHIP. Furthermore, CHIP greatly decreased the half-life of both Hsp70 and Hsp90, with degradation of Hsp70 being more pronounced. Again, these results are consistent with the predictions of our model, both qualitatively and quantitatively.
Hsp90-dependent protein folding machinery is complex and involves various co-chaperones and regulators (6). Here, we study the balance between the Hsp90-mediated folding process and the degradation machinery. By focusing on the proteins which are directly involved in the protein folding/degradation decision process, we were able to quantitatively analyze the system and thus propose a testable model. It is possible that other co-chaperones may influence the folding/degradation balance. The presented model, however, provides a quantitative framework for this multi-protein system, which can in the future be built-upon or modified as new data emerges.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Elizabeth Rhoades for allowing us to use her cell culture facilities and Aitziber Lopez Cortajarena, Meredith Jackrel, Robielyn Ilagan, Clarence Chen, Tijana Grove, and Genaro Pimienta-Rosales for their helpful comments on the manuscript.
Footnotes
Abbreviations: HOP, Hsp70/Hsp90 organizing protein; TPR, tetratricopeptide repeat; CHIP, C-terminus of Hsc70 interacting protein; βME, β-mercaptoethanol; SPR, surface plasmon resonance; ITC, isothermal titration calorimetry.
SUPPORTING INFORMATION AVAILABLE
Oligomeric state of recombinant Hsp70 in solution. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Neckers L. Heat shock protein 90: the cancer chaperone. J. Biosci. 2007;32:517–530. doi: 10.1007/s12038-007-0051-y. [DOI] [PubMed] [Google Scholar]
- 2.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeda S, McKay DB. Kinetics of peptide binding to the bovine 70 kDa heat shock cognate protein, a molecular chaperone. Biochemistry. 1996;35:4636–4644. doi: 10.1021/bi952903o. [DOI] [PubMed] [Google Scholar]
- 4.Wei J, Gaut JR, Hendershot LM. In vitro dissociation of BiP-peptide complexes requires a conformational change in BiP after ATP binding but does not require ATP hydrolysis. J. Biol. Chem. 1995;270:26677–26682. doi: 10.1074/jbc.270.44.26677. [DOI] [PubMed] [Google Scholar]
- 5.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70–Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 6.Wandinger SK, Richter K, Buchner J. The Hsp90 chaperone machinery. J. Biol. Chem. 2008;283:18473–18477. doi: 10.1074/jbc.R800007200. [DOI] [PubMed] [Google Scholar]
- 7.Kosano H, Stensgard B, Cristine Charlesworth M, McMahon N, Toft D. The assembly of progesterone receptor-hsp90 complexes using purified proteins. J. Biol. Chem. 1998;273:32973–32979. doi: 10.1074/jbc.273.49.32973. [DOI] [PubMed] [Google Scholar]
- 8.Prodromou C, Siligardi G, O’Brien R, Woolfson DN, Regan L, Panaretou B, Ladbury JE, Piper PW, Pearl LH. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 1999;18:754–762. doi: 10.1093/emboj/18.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flom G, Behal RH, Rosen L, Cole DG, Johnson JL. Definition of the minimal fragments of Sti1 required for dimerization, interaction with Hsp70 and Hsp90 and in vivo functions. Biochem. J. 2007;404:159–167. doi: 10.1042/BJ20070084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onuoha SC, Coulstock ET, Grossmann JG, Jackson SE. Structural studies on the co-chaperone Hop and its complexes with Hsp90. J. Mol. Biol. 2008;379:732–744. doi: 10.1016/j.jmb.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Yi F, Doudevski I, Regan L. HOP is a monomer: investigation of the oligomeric state of the co-chaperone HOP. Protein Sci. 2010;19:19–25. doi: 10.1002/pro.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin L, Patterson C. Indentification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 1999;19:4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. Chaperoned ubiquitylation-crystal structures of the CHIP U Box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 14.Xu Z, Devlin KI, Ford MG, Nix JC, Qin J, Misra S. Structure and interactions of the helical and U-Box domains of CHIP, the C terminus of HSP70 interacting protein. Biochemistry. 2006;45:4749–4759. doi: 10.1021/bi0601508. [DOI] [PubMed] [Google Scholar]
- 15.Connell P, Ballinger CA, Jiang J, Wu J, Thompson LJ, Höhfeld J, Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 16.Murata S, Minami Y, Minami M, Chiba T, Tanaka K. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO J. 2001;2:1133–1138. doi: 10.1093/embo-reports/kve246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu W, Marcu M, Yuan X, Mimnaugh E, Patterson C, Neckers L. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc. Natl. Acad. Sci. USA. 2002;99:12847–12852. doi: 10.1073/pnas.202365899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian S, McDonough H, Boellmann F, Cyr DM, Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006;440:551–555. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLaughlin SH, Smith HW, Jackson SE. Stimulation of the weak ATPase activity of human Hsp90 by a client protein. J. Mol. Biol. 2002;315:787–798. doi: 10.1006/jmbi.2001.5245. [DOI] [PubMed] [Google Scholar]
- 20.Liu F, Wu S, Hu S, Hsiao C, Wang C. Specific interaction of the 70-kDa heat shock cognate protein with the tetratricopeptide repeats. J. Biol. Chem. 1999;274:34425–34432. doi: 10.1074/jbc.274.48.34425. [DOI] [PubMed] [Google Scholar]
- 21.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol. Cell. 2006;21:873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Benaroudj N, Batelier G, Triniolles F, Ladjimi MM. Self-association of the molecular chaperone HSC70. Biochemistry. 1995;34:15282–15290. doi: 10.1021/bi00046a037. [DOI] [PubMed] [Google Scholar]
- 23.Gao B, Eisenberg E, Greene L. Effect of constitutive 70-kDa heat shock protein polymerization on its interaction with protein substrate. J. Biol. Chem. 1996;271:16792–12797. doi: 10.1074/jbc.271.28.16792. [DOI] [PubMed] [Google Scholar]
- 24.Murphy PJM, Kanelakis KC, Galigniana MD, Morishima Y, Pratt WB. Stoichiometry, abundance, and functional significance of the hsp90/hsp70-based multiprotein chaperone machinery in reticulocyte lysate. J. Biol. Chem. 2001;276:30092–30098. doi: 10.1074/jbc.M103773200. [DOI] [PubMed] [Google Scholar]
- 25.Benaroudj N, Fouchaq B, Ladjimi MM. The COOH-terminal peptide binding domain is essential for self-association of the molecular chaperone HSC70. J. Biol. Chem. 1997;272:8744–8751. doi: 10.1074/jbc.272.13.8744. [DOI] [PubMed] [Google Scholar]
- 26.Yi F, Regan L. A novel class of small molecule inhibitors of Hsp90. ACS Chem. Biol. 2008;3:645–654. doi: 10.1021/cb800162x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith DF, Sullivan WP, Marion TN, Zaitsu K, Madden B, McCormick DJ, Toft DO. Identification of a 60-kilodalton stress-related protein, p60, which interacts with hsp90 and hsp70. Mol. Cell. Biol. 1993;13:869–876. doi: 10.1128/mcb.13.2.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seidl J, Knuechel R, Kunz-Schughart LA. Evaluation of membrane physiology following fluorescence activated or magnetic cell separation. Cytometry A. 1999;36:102–111. doi: 10.1002/(sici)1097-0320(19990601)36:2<102::aid-cyto3>3.3.co;2-4. [DOI] [PubMed] [Google Scholar]
- 29.Morales JL, Perdew GH. Carboxyl terminus of hsc70-interacting protein (CHIP) can remodel mature aryl hydrocarbon receptor (AhR) complexes and mediate ubiquitination of both the AhR and the 90 kDa heat-shock protein (hsp90) in vitro. Biochemistry. 2007;46:610–621. doi: 10.1021/bi062165b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sousa MC, McKay DB. The hydroxyl of threonine 13 of the bovine 70-kDa heat shock cognate protein is essential for transducing the ATP-induced conformational change. Biochemistry. 1998;37:15392–15399. doi: 10.1021/bi981510x. [DOI] [PubMed] [Google Scholar]
- 31.Rosser MFN, Washburn E, Muchowski PJ, Patterson C, Cyr DM. Chaperone functions of the E3 ubiquitin ligase CHIP. J. Biol. Chem. 2007;282:22267–22277. doi: 10.1074/jbc.M700513200. [DOI] [PubMed] [Google Scholar]
- 32.Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc. Natl. Acad. Sci. USA. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brychzy A, Rein T, Winklhofer KF, Hartl FU, Young JC, Obermann WMJ. Cofactor Tpr2 combines two TPR domains and a J domain to regulate the Hsp70/Hsp90 chaperone system. EMBO J. 2003;22:3613–3623. doi: 10.1093/emboj/cdg362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bagatell R, Paine-Murrieta GD, Taylor CW, Pulcini EJ, Akinaga S, Benjamin IJ, Whitesell L. Induction of a heat shock factor 1-dependent stress response alters the cytotoxic activity of Hsp90-binding agents. Clin. Cancer Res. 2000;6:3312–3318. [PubMed] [Google Scholar]
- 35.Ha J-H, Johnson ER, McKay DB, Sousa MC, Takeda S, Wilbanks SM. Structure and mechanism of Hsp70 proteins. In: Bukau B, editor. Molecular Chaperones and Folding Catalysts: Regulation, Cellular Function and Mechanism. Amsterdam: Harwood Academic Publishers; 1999. pp. 573–607. [Google Scholar]
- 36.Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.