Abstract
The vacuole-type ATPases (V-ATPases) exist in various intracellular compartments of eukaryotic cells to regulate physiological processes by controlling the acidic environment. The crystal structure of the subunit C of Thermus thermophilus V-ATPase, homologous to eukaryotic subunit d of V-ATPases, has been determined at 1.95-Å resolution and located into the holoenzyme complex structure obtained by single particle analysis as suggested by the results of subunit cross-linking experiments. The result shows that V-ATPase is substantially longer than the related F-type ATPase, due to the insertion of subunit C between the V1 (soluble) and the Vo (membrane bound) domains. Subunit C, attached to the Vo domain, seems to have a socket like function in attaching the central-stalk subunits of the V1 domain. This architecture seems essential for the reversible association/dissociation of the V1 and the Vo domains, unique for V-ATPase activity regulation.
The vacuole-type ATPases (V-ATPases) are commonly found in many organisms involved in a variety of physiological processes (1). V-ATPases in eukaryotic cells (eukaryotic V-ATPases) translocate protons across the membrane consuming ATP. They reside within intracellular compartments, including endosomes, lysosomes, and secretory vesicles, and within plasma membranes of certain cells including renal intercalated cells, osteoclasts, and macrophages. Eukaryotic V-ATPases are responsible for various cell functions including the acidification of intracellular compartments, renal acidification, born resorption, and tumor metastasis (2).
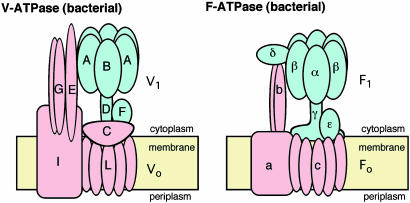
V-ATPase and the F-type ATP synthase (F-ATPase) are evolutionary related and share the rotary mechanism coupling ATP synthesis/hydrolysis and proton translocation across the membrane (2–4). However, these two types of ATPase show significant differences. Reversible association/dissociation of the V1 domain (soluble) and the Vo domain (membrane bound) is a unique activity regulation mechanism compared to F-ATPase (Fig. 1). For example, glucose deprivation has been shown to cause a rapid dissociation of the yeast V-ATPase into free V1 and Vo domains, which is reversible and independent of de novo protein synthesis (5, 6). Similar observations have been reported for Manduca sexta and mammalian complexes (7–9). Subunit composition and structure in the stalk region of V-ATPase, which connects the Vo and V1 domains, are suggested to be significantly different from those in F-ATPase (10) (Fig. 1). Thus, this region is possibly responsible for the association/dissociation of the complex.
Fig. 1.
Subunit composition of the V-ATPase from T. thermophilus and the F-ATPase from E. coli. The V1 and F1 domains are colored in blue, and the Fo and Vo domains are in pink.
V-ATPases are also found in archaea and some eubacteria (prokaryotic V-ATPases) (11). The V-ATPase from Thermus thermophilus is solely responsible for aerobic ATP synthesis in this bacteria, which lacks F-ATPase (12). The Thermus V-ATPase is composed of nine different subunits, which are arranged within the atp operon in the order of G–I–L–E–C–F–A–B–D, which encodes proteins with molecular sizes of 13, 71, 8, 20, 35, 12, 64, 54, and 25 kDa, respectively (10) (Fig. 1). This ATPase-active V1 domain is composed of four subunits with a stoichiometry of A3B3D1F1 (13), whereas the Vo domain, involved in proton translocation across the membrane, is an assembly of G, I, L, E, and C subunits. Each Thermus V-ATPase subunit shows a sequence similarity to its eukaryotic counterpart. An essential component, subunit C shows rather low (18%) but significant sequence similarity to subunit d of the yeast enzyme (10). This subunit is part of the central stalk of V-ATPase (10), and no counterpart exists in F-ATPase (Fig. 1).
Here, we report the crystal structure of the subunit C of the Thermus V-ATPase at 1.95-Å resolution. The structure was successfully placed into the low-resolution EM structure of the holo-V-ATPase according to the results of cross-linking experiments. The result strongly indicated that this subunit is, indeed, serving as a “socket” to attach V1 central stalk subunits onto the L subunit ring of Vo domain.
Materials and Methods
Expression and Purification of Subunit C. The gene encoding subunit C was amplified by PCR using the following oligonucleotide primers: 5′-T T TGGGGCTAGACGCATATGGCGGACGAC-3′ (sense primer) and 5′-ATCGGCGATCACCGGGATCCTCACGG-3′ (antisense primer), possessing NdeI and BamHI sites, respectively (underlined). Amplified DNA was digested with NdeI and BamHI, followed by ligation between the NdeI and BamHI sites of pET-17b (Novagen) to obtain pET-C. Escherichia coli BL21-CodonPlus (DE3)-RP cells (Stratagene) harboring pET-C were grown in 2× YT medium (16 g of polypepton/10 g of yeast extract/5 g of NaCl, per 1 liter of medium) overnight at 37°C. The harvested cells, resuspended in 20 mM Hepes·NaOH (pH 7.0) and 1 mM EDTA, were sonicated, followed by incubation for 30 min at 65°C. Denatured E. coli proteins were removed by centrifugation at 19,000 × g for 60 min. The supernatant was applied to CM-Sepharose FF column (Amersham Pharmacia) equilibrated with 20 mM Hepes·NaOH (pH 7.0) and 1 mM EDTA. The protein was eluted with a linear gradient of sodium chloride, from 0 to 0.5 M. The fractions containing subunit C were collected and concentrated by ultrafiltration, then applied onto Superdex HR-200 (Amersham Pharmacia) equilibrated with 20 mM Mops·KOH (pH 7.0) and 0.1 M KCl. The fractions containing subunit C were collected and stored at 4°C until use.
Antibodies. The recombinant subunit C or F of T. thermophilus was injected into rabbits. The resultant antisera were affinity-purified with the subunit C or F immobilized onto Affi-Gel 10 (Bio-Rad). Preparation of recombinant F subunit is described elsewhere (unpublished data).
Isolation of Mutated V-ATPase. Mutations (A-His-8-tags/C-C267S/C-C322S, or/C-T105C or/C-A141C or/C-K220C or/C-Q278C or/C-Q315C) were introduced in atp operon in T. thermophilus chromosomal DNA by using the integration vector system (4, 5, 14). Chromosomal DNA was prepared from a transformed strain, and mutations were confirmed by sequencing with an ABI 310 sequencer. The mutated V-ATPases were solubilized from the membranes of recombinant T. thermophilus and purified as described (10).
Covalent Cross-Linking of the V-ATPases by Using 4-(N-maleimido)-benzophenone (MBP). Two endogenous cysteine residues, C267 and C322 in subunit C, were replaced by serine and an unique cysteine residue was introduced at T105, A141, L220, Q278, or Q315 by using the shuttle integration vector system (14–16). The mutated V-ATPases, dissolved at 0.7 mg/ml in 20 mM Tris·Cl (pH8.0), 0.1 mM EDTA, and 0.05% Triton X-100, were subjected to cross-linking experiments using MBP. MBP was dissolved in dimethylformamide at 100 mM, and 1 μl of the MBP solution was added to the protein solution, yielding a final concentration of 1 mM. For negative controls, 1 μl of dimethylformamide was added instead. After incubation in the dark for 30 min at 25°C, the unreacted MBP was quenched by addition of 10 mM DTT and the resultant enzymes were irradiated with a long-wavelength UV lamp for 5 min at 4°C. The cross-linked products were separated by SDS/15% PAGE and analyzed by the Western blot using anti-subunit C or F antibody. The anti-subunit C antibody also cross-reacts with subunit I; however, this did not cause any problem of assignment because no cross-linked product larger than subunit I was obtained.
Crystallization and Structure Determination of Subunit C. Hexagonal-column shaped crystals were obtained by the vapor diffusion method in a solution containing 0.1 M sodium citrate (pH 5.6), 22–26% 2-methyl-2,4-pentandiol, and 0.2 M ammonium acetate at 4°C. The crystals belong to a hexagonal space group P61 with the cell dimensions of a = b = 118.43 Å, c = 152.09 Å. The crystallographic asymmetric unit contains three subunit C molecules. The collected x-ray data sets were processed by using hkl program package (17) and ccp4 program suite (18). Initial phases were determined by using mlphare (19) using two Os derivatives with different soaking times, followed by density modification and noncrystallographic symmetry (NCS) averaging with dm (20). The initial model was built by arp/warp (21, 22) followed by manual remodeling using o (23). The final structure was refined by refmac 5 (24). A modest NCS restraint was applied in early stage of the refinement; however, the restrain was removed in the final stage of the refinement to yield the lowest free-R factor. The rms differences for the three molecules in the asymmetric unit are 0.64 Å (molecules A and B), 0.51 Å (molecules B and C), and 0.37 Å (molecules A and C), using 320 Cα atoms. Data collection and phasing statistics are summarized in Table 1. The refinement statistics and the model stereochemistry, which is excellent for a 1.95-Å resolution structure, are summarized in Table 2. The slightly high free-R factor could be due to the observed diffuse scattering, indicating that a small proportion of the crystals are rotationally disordered around the C axis. An example of the density is shown in Fig. 2.
Table 1. Summary of data collection and phase determination.
| Native
|
K2 OsCl6
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Refinement | Phasing | 2 mM, 2 days | 10 mM, 1.5 days | ||||||
| Resolution, Å | 1.95 (2.02-1.95) | 2.0 (2.07-2.00) | 2.5 (2.59-2.50) | 2.0 (2.07-2.00) | |||||
| Number of reflections | |||||||||
| Overall | 410,472 | 345,506 | 189,506 | 366,352 | |||||
| Unique* | 86,730 (8,633) | 80,380 (8,081) | 41,158 | 80,919 | |||||
| Completeness,* % | 99.3 (99.0) | 98.9 (98.5) | 99.7 (99.6) | 99.7 (99.8) | |||||
| I/σ* | 13.8 (4.3) | 12.4 (3.7) | 12.8 (7.8) | 12.9 (4.4) | |||||
| Rmerge,*† % | 6.8 (32.8) | 5.9 (29.0) | 6.0 (16.5) | 6.6 (28.1) | |||||
| Rderiv,‡ % | 8.3 | 9.4 | |||||||
| Heavy atom sites | 3 | 3 | |||||||
| Rcullis§ | 0.62 | 0.60 | |||||||
| X-ray source | X06SA/SLS | X06SA/SLS | ID13/ESRF | X06SA/SLS | |||||
| Phase determination | |||||||||
| Resolution, Å | 13.91 | 8.42 | 6.04 | 4.71 | 3.86 | 3.27 | 2.83 | 2.50 | Total |
| Reflections | 219 | 793 | 1,873 | 3,290 | 5,124 | 7,314 | 9,907 | 12,900 | 41,420 |
| Figure of merit | 0.30 | 0.35 | 0.48 | 0.50 | 0.43 | 0.43 | 0.45 | 0.39 | 0.43 |
Values for the highest resolution shell are given in parentheses.
Rmerge = ΣhΣf Ii(h) - <I(h)>|/ΣhΣi(h), where Ii(h) is the ith measurement.
Rderiv = Σ∥FPH| -|FP∥/Σ|FP|, where FPH and FP are protein and heavy atom derivative structure factors, respectively.
Rcullis = Σ∥FPH-FP| - FH(calc)∥/Σ|FPH-FP|. FPH and FP are defined above, and FH(calc) is the calculated heavy atom structure factor. Summation was done by using centric reflections only.
Table 2. Refinement statistics.
| Number of nonhydrogen atoms | 8,179 |
| Number of water molecules | 637 |
| Average temperature factor, Å2 | 35.4 |
| Resolution range,* Å | 40.0-1.95 (2.00-1.95) |
| R factor,*† % | 19.1 (23.7) |
| Number of reflections* | 78,514 (6,082) |
| Rfree,*‡ % | 24.9 (27.2) |
| Number of reflections* | 4,353 (315) |
| Estimated overall coordinate error based on Rfree, Å | 0.156 |
| rms deviations from ideal values | |
| Bond length, Å | 0.025 |
| Bond angles, ° | 1,936 |
| Ramachandran plot (non-Gly, non-Pro residues), % | |
| Residues in most favored regions | 96.1 |
| Residues in additionally allowed regions | 3.9 |
| Others | 0.0 |
Values for the highest resolution shell are given in parentheses.
R factor = ∑∥Fobs| - |Fcalc∥/∑|Fobs|.
Rfree was calculated for 5% of reflections randomly excluded from the refinement.
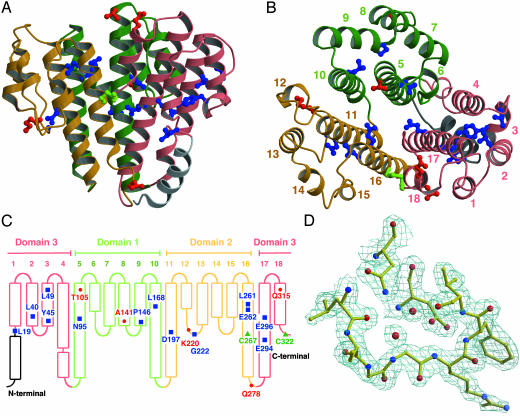
Fig. 2.
Structure of subunit C. (A) Side view. (B) Top view from the cytoplasmic side. (C) Schematic representation. The two cysteine residues are marked in green, conserved residues are marked in blue, and residues that were mutated in the cross-linking experiments are marked in red. (D) 2Fo – Fc electron density map of a loop around G222 at 1.5σ. All figures with molecules and densities were prepared with molscript (43), bobscript (44), and raster3d (45).
Electron Microscopy and Single Particle Analysis. Protein sample was diluted to 50 μg/ml and by contrasting with 2% uranyl acetate as negative stain. Images were recorded with a Philips-CM100 transmission electron microscope at 80 kV, ×51,500 magnification. The micrographs were digitized by using a Leafscan 45 densitometer at a step-size of 10 μm (1.94 Å per pixel on the specimen scale). All image processing was performed with imagic-5 (25, 26). A data set of 14,173 single particle images was manually selected. Images of the particles were coarsened to 3.88 Å per pixel. The data sets were analyzed by the reference-free alignment-by-classification procedure (27) followed by the class averages (28) to determine relative orientations, resulting in initial 3D reconstructions gained from implementation of the exact back projection technique (29). Reprojections were taken from each 3D model and used to identify additional atypical views and to further refine the class averages within each subpopulation data set. The resolution was estimated as 30 Å by the Fourier shell correlation method between two independent 3D reconstructions (30).
Modeling of the L Subunit Ring. To study the assembly of subunits C and L, the L subunit ring was modeled with 12 molecules of subunit c (homologous to the V-ATPase subunit L) structure of E. coli F-ATPase determined by NMR (PDB entry 1C17). For F-ATPase, the number of subunits c per enzyme varies depending on the organism. So far, examples with 10-, 11-, and 14-c-subunit rings have been reported (31–33). There is no solid evidence how many L subunits are involved in the Thermus V-ATPase rotor ring. Therefore, we have used an average size, 12-L-subunit ring for the following discussion. However, the discussion is valid for 10- to 14-L-subunit ring forms of the enzyme.
Results and Discussion
Subunit C Structure. Subunit C is composed of 323 amino acid residues. The molecule has a funnel shape with dimensions of 45 × 50 × 50 Å (Fig. 2 A and B). There is a polar cavity inside (8-Å diameter, 25-Å depth), open to only one side. The surface of the subunit is mainly polar, and no possible membrane anchor region was observed. Although the subunit C forms a trimer in the crystals, the small trimer interface (17.5 Å2 per section) suggests that the physiological status of the protein is a monomer. Subunit C has two cysteine residues (C267 and C322), which form a disulfide bridge in the crystal structure. Subunit C is located in the reducing environment of the cytoplasm (10) and this disulfide bridge is likely to be an artifact formed during sample preparation under aerobic conditions. Disulfide-bridge formation plays an important role in regulation of chloroplast ATPases (34); however, this seems not to be the case here, because the cysteine residues are not conserved. This is also supported by the fact that the V-ATPase with the mutant subunit C, with cysteine residues replaced by serine residues, is properly assembled and fully functional (data not shown).
Subunit C is a highly helical molecule, composed of six central helices and 12 peripheral helices (Fig. 2 A and B). The six central helices, as long as 50 Å in length, surround the polar cavity. The peripheral helices form a rim of ≈25 Å height around the central helices. Helix 1, which is longer than other peripheral helices, in composed of two segments connected by a short loop.
The structure consists of three domains related by a distorted threefold symmetry; each of domain is composed of two central and four peripheral helices (Fig. 2B). The first and second domains contains the residues 78–183 and 184–281, respectively, and the third domain contains residues 282–323 and 18-77; the third domain is composed of both the N- and C-terminal sequences (Fig. 2 B and C). These domains can be superimposed with an rms difference of 2.0 Å (domains 1 and 2, 64 Cα atoms), 2.1 Å (domains 2 and 3, 80 Cα atoms), and 2.2 Å (domains 1 and 3, 54 Cα atoms). This result suggests that the molecule is a product of gene triplication, although no obvious sequence similarity is observed.
Thirteen highly conserved residues were identified by multiple alignment of subunit C and d sequences of V-ATPases (35). Five of them are polar residues; E262 (helix 16) and E294 (helix 17) exist within the internal polar cavity. N95 (helix 5), D197 (helix 11), and E296 (helix 17) are located on the central helices and form hydrogen bond/salt bridge networks with residues from peripheral helices. L19 (helix 1), L40 (helix 2), Y45 (helix 3), L49 (helix 3), L168 (helix 10), and L261 (helix 16) are part of the hydrophobic cores between the central and peripheral helices and, together with other hydrophobic residues, stabilizes the rim structure. P146 (loop 8–9) and G222 (loop 12–13) are at the lower end of the rim; these residues seem important to maintain turn structures in the loops. Eleven of the conserved residues seem to be involved in maintaining the rim structure (Fig. 2 A and B), suggesting the observed rim structure is conserved among C (bacterial) and d (eukaryotic) subunits of V-ATPases.
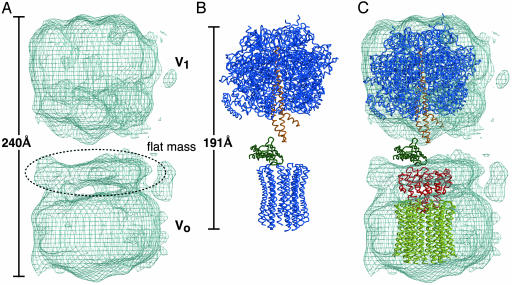
V-ATPase Structure by Electron Microscopy. Single particle analysis using electron microscopy showed that the Thermus V-ATPase has a dumbbell shape with dimensions of 130 × 120 × 240 Å (Fig. 3A). The V-ATPase structure is similar to those of other prokaryotic (36, 37) and eukaryotic (38, 39) V-ATPases. The peripheral stalks, which were observed for other V-ATPase structures, are not clearly visible for our reconstitution model, and this likely due to the disorder of the region and rotationally averaging effect. One domain, which shows approximate sixfold symmetry in the view parallel to the membrane normal, has been assigned as V1 domain. Consequently, the other domain has been assigned as Vo domain, which has an oval shape with a flat “lid” like mass attached on the cytoplasmic side. When the crystal structure of yeast F1c10 complex (PDB entry 1QO1) was compared to the EM structure, the density for the V1-ATPase was shown to accommodate the F1-ATPase model well. However, it is impossible to fit both F1 and Fo domains to the EM density because the V-ATPase is significantly longer (240 Å) than the F-ATPase (191 Å) (Fig. 3 A and B). This difference is due to the considerably longer stalk region of V-ATPase as reported by Chaban et al. (40). The observed “flat mass” attached to Vo domain (Fig. 3A) is the main contributor to this longer stalk.
Fig. 3.
The structure of the holo V-ATPase compared with the F-ATPase. (A) The V-ATPase structure by single particle analysis using electron microscopy. (B) X-ray structure of F1c10-ATPase from yeast (PDB ID 1QO1). Central stalk subunits γ and ε are highlighted in orange and green, respectively, whereas the other subunits (α3,β3,ε,c10) are colored in blue. (C) Superimposition of subunit C (red) with the modeled rotor ring (yellow). Yeast F1-ATPase (α3,β3,γ,ε) is also fitted to the Vo domain as a reference. The color scheme of the F1-ATPase is the same as in B.
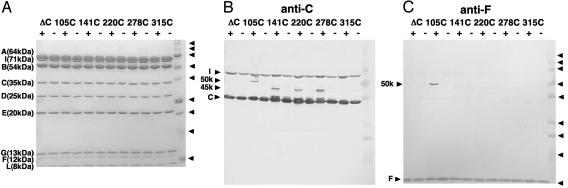
Covalent Cross-Linking of Subunit C by Using MBP. To identify the location of subunit C relative to the other subunits in the holoenzyme, we produced five site-directed cysteine mutants and performed cross-linking experiments using the photoactivatable cross-linker MBP. In the presence of MBP, these introduced cysteine residues are cross-linked to an adjacent residue by UV irradiation. A unique cysteine residue was introduced at position Thr-105, Ala-141, Lys-220, Gln-278, or Gln-315 (Fig. 2C). For four of the five mutants, we obtained cross-linked products, which were immunostained by an anti-subunit C antibody (Fig. 4B). Among them, the cross-linked product for Thr-105, with a molecular mass of ≈50 kDa, immunostained by the anti-subunit F antibody indicating Thr-105 is in the vicinity of subunit F (Fig. 4C). The cross-linked products for Ala-141, Lys-220 (both at the rim end), and Gln-278 (at the tip of the central helix bundle) show a molecular mass of ≈45 kDa indicating these are composed of subunit C (35 kDa) and L (8 kDa), because there is no other subunit that can form a product as small as 45 kDa with subunit C. The result clearly indicates that the lower surface of subunit C in Fig. 2 A, including the rim ends and the central helices, has a wide interface with the L subunit ring, whereas a part of the upper surface (Fig. 2) forms a contact with subunit F, which is a central stalk subunit of V1-ATPase.
Fig. 4.
Cross-linking of subunit C and other subunit of V-ATPase by using the photoactivable sulfhydryl reagent MBP. (A) SDS/PAGE of V-ATPases containing a single cysteine mutant of subunit C stained by Coomassie brilliant blue (CBB). All mutants (mutations are indicated in the figure) are based on the cysteine-less mutant of the subunit (ΔC). Each of isolated mutant VoV1-ATPase (50 μg of protein) was incubated in the presence (+) or absence (–) of 1 mM MBP. Indicated to the right of the panel are the molecular masses of marker proteins (150, 100, 75, 50, 37, 25, 15, and 10 kDa). Because of the low yields, the cross-linked products are not visible by CBB staining but are visible only by immunostaining in B and C.(B) Western blot using an anti-C antibody. The lanes are the same as in A.(C) The same as B but immunostained by anti-F antibody.
Location of Subunit C in the Holoenzyme. For the Thermus V-ATPase, subunit C can form a stable subcomplex with the rotor L subunit ring and is classified as a Vo domain subunit (10). This is the same for subunit d of eukaryotic V-ATPases. However, for some other prokaryotic V-ATPases, such as Na+-pumping V-ATPases from Clostridium fervidus (40) and Enterococcus hirae (41), the subunit C is part of V1 domain. These data indicate that subunit C is at the Vo and V1 domain interface. In the C. fervidus V-ATPase structure, EM studies using various subcomplexes showed that the “flat mass” attached to Vo domain (Fig. 3A) contains subunit C in the center surrounded by the soluble domain of subunit I, part of the peripheral stalk(s).
Taking all of the results into account, we were able to fit the subunit C and L structures into the EM density of the holoenzyme as shown in Fig. 3C. For the fitting, we used the rotor ring containing 12 L subunits (see Materials and Methods for details). In Fig. 3C, V1 domain part is fitted with F1-ATPase (α3β3γε). Subunit C is fitted to the center of the observed “flat mass,” attached to the Vo domain. The thickness of the flat mass is equal to the height of the peripheral helix rim of subunit C (25 Å). The flat mass is wider than subunit C parallel to the membrane; the extra density most likely forms a part of the peripheral stalk(s) as the EM study showed (40). In the model, subunit C caps one end of the subunit-L ring and the internal cavity of subunit C is open toward the upper side. In the model, the rim end, which accommodates residues cross-linked to subunit C (Ala-141 and Lys-220), forms a contact with the flat upper surface of the subunit-L ring. The central helix bundle of subunit C is penetrating into the central cavity of the subunit-L ring by 12 Å in depth. This is consistent with the fact that Gln-278, at the tip of the central helix bundle, is also cross-linked to subunit L. The diameter of the central cavity of the subunit-L ring (30 Å) is large enough to accommodate the central helix bundle of subunit C (the largest diameter, 28 Å). Thr-105, which is on the upper surface of subunit C, is cross-linked to subunit F, which is a part of the central stalk (10). It is clear in Fig. 3C that subunit F should be located at the equivalent position to the ε-subunit of F-ATPase.
In the EM studies using the subcomplexes, the central stalk subunit E of the C. fervidus V1 domain (equivalent of F-ATPase γ-subunit, subunit D for the Thermus V-ATPase) was shown to form a contact with subunit C (40). This indicates not only subunit F but also subunit D of V1 domain forms contacts with subunit C. A cross-linking experiment (42) indicates the subunits D and F are forming “DF central shaft,” and subunit C likely recognizes this complex. Because the torque needs to be transmitted through the stalk between V1 and Vo domain, a tight coupling between subunit C and the DF central shaft is required. The central cavity of subunit C, which accommodates two conserved polar residues inside, could be involved in this coupling.
Subunit C could play an important role in the reversible association/dissociation of the V1 and Vo domains in vivo. In F-ATPase, both ε- and γ-subunits (equivalent for the V-ATPase subunit F and D, respectively) directly bind to the rotor ring (31). In contrast, at the V1 and Vo interface of V-ATPase, subunit C forms a socket like structure to accommodate the DF central shaft. Thus, the Vo–V1 interface is significantly different from its counterpart in F-ATPase and this could be relevant for the unique reversible association/dissociation of V-ATPase. Further biochemical characterization is essential to elucidate how the V1 and Vo dissociation/association is regulated by various signals such as pH and glucose concentration.
Acknowledgments
We thank Drs. Clemens Schulze-Briese and Takashi Tomizaki (PX06SA/SLS, Paul Scherrer Institute, Villigen, Switzerland) and Dr. David Flot (ID13/European Synchrotron Radiation Facility) for help with data collections, Dr. Jon Nield for aiding in the single particle image processing, and Dr. Bernadette Byrne for critical reading of the manuscript. Part of this work was supported by the Leverhulme Trust (to E.S., B.H., and J.B.) and by the Biotechnology and Biological Sciences Research Council (to S.I.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: V-ATPase, vacuole-type ATPase; F-ATPase, F-type ATP synthase; MBP, 4-(N-maleimido)benzophenone.
Data deposition: The atomic coordinates and the structure factors are deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1R5Z).
References
- 1.Nishi, T. & Forgac, M. (2002) Nat. Rev. Mol. Cell Biol. 3, 94–103. [DOI] [PubMed] [Google Scholar]
- 2.Yoshida, M., Muneyuki, E. & Hisabori, T. (2002) Nat. Rev. Mol. Cell Biol. 2, 669–677. [DOI] [PubMed] [Google Scholar]
- 3.Imamura, H., Nakano, M., Noji, H., Muneyuki, E., Ohkuma, S., Yoshida, M. & Yokoyama, K. (2003) Proc. Natl. Acad. Sci. USA 100, 2312–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yokoyama, K., Nakano, M., Imamura, H., Yoshida, M. & Tamakoshi, M. (2003) J. Biol. Chem. 278, 24255–24258. [DOI] [PubMed] [Google Scholar]
- 5.Kane, P. M. (1995) J. Biol. Chem. 270, 17025–17032. [PubMed] [Google Scholar]
- 6.Parra, K. J. & Kane, P. M. (1998) Mol. Cell. Biol. 18, 7064–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sumner, J. P., Dow, J. A., Earley, F. G., Klein, U., Jager, D. & Wieczorek, H. (1995) J. Biol. Chem. 270, 5649–5653. [DOI] [PubMed] [Google Scholar]
- 8.Forgac, M. (2000) J. Exp. Biol. 203, 71–80. [DOI] [PubMed] [Google Scholar]
- 9.Trombetta, E. S., Ebersold, M., Garrett, W., Pypaert, M. & Mellman, I. (2003) Science 299, 1400–1403. [DOI] [PubMed] [Google Scholar]
- 10.Yokoyama, K., Nagata, K., Imamura, H., Ohkuma, S., Yoshida, M. & Tamakoshi, M. (2003) J. Biol. Chem. 278, 42686–42691. [DOI] [PubMed] [Google Scholar]
- 11.Yokoyama, K., Muneyuki, E., Amano, T., Mizutani, S., Yoshida, M., Ishida, M. & Ohkuma, S. (1998) J. Biol. Chem. 273, 20504–20510. [DOI] [PubMed] [Google Scholar]
- 12.Yokoyama, K., Ohkuma, S., Taguchi, H., Yasunaga, T., Wakabayashi, T. & Yoshida, M. (2000) J. Biol. Chem. 275, 13955–13961. [DOI] [PubMed] [Google Scholar]
- 13.Yokoyama, K., Oshima, T. & Yoshida, M. (1990) J. Biol. Chem. 265, 1946–1950. [PubMed] [Google Scholar]
- 14.Tamakoshi, M., Uchida, M., Tanabe, K., Fukuyama, S., Yamagishi, A. & Oshima, T. (1997) J. Bacteriol. 179, 4811–4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamakoshi, M., Yamagishi, A. & Oshima, T. (1998) Gene 222, 125–132. [DOI] [PubMed] [Google Scholar]
- 16.Tamakoshi, M., Yaoi, T., Oshima, T. & Yamagishi, A. (1999) FEMS Microbiol. Lett. 173, 431–437. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski, Z. (1993) in Data Collection and Processing, eds. Sawyer, L., Isaacs, N. & Bailey, S., (SERC Daresbury Laboratory, Warrington, U.K.), pp. 56–62.
- 18.Collaborative Computational Project No. 4 (1994) Acta. Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 19.Otwinowski, Z. (1991) in Daresbury Study Weekend Proceedings, eds. Wolf, W., Evans, P. R. & Leshe, A. G. W. (SERC Daresbury Laboratory, Warrington, U.K.), pp. 23–38.
- 20.Cowtan, K. (1994) Joint CCP4 and ESF-EACBM Newslett. Protein Crystallogr. 31, 34–38. [Google Scholar]
- 21.Lamzin, V. S. & Wilson, K. S. (1993) Acta Crystallogr. D 49, 129–149. [DOI] [PubMed] [Google Scholar]
- 22.Perrakis, A., Sixma, T. K., Wilson, K. S. & Lamzin, V. S. (1997) Acta Crystallogr. D 53, 448–455. [DOI] [PubMed] [Google Scholar]
- 23.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 24.Murshudov, G. N., Lebedev, A., Vagin, A. A., Wilson, K. S. & Dodson, E. J. (1999) Acta Crystallogr. D 55, 247–255. [DOI] [PubMed] [Google Scholar]
- 25.Orlova, E. V., Serysheva, I. I., van Heel, M., Hamilton, S. L. & Chiu, W. (1996) Nat. Struct. Biol. 3, 547–552. [DOI] [PubMed] [Google Scholar]
- 26.van Heel, M., Gowen, B., Matadeen, R., Orlova, E. V., Finn, R., Pape, T., Cohen, D., Stark, H., Schmidt, R., Schatz, M. & Patwardhan, A. (2000) Q. Rev. Biophys. 33, 307–369. [DOI] [PubMed] [Google Scholar]
- 27.Schatz, M. & van Heel, M. (1990) Ultramicroscopy 32, 255–264. [DOI] [PubMed] [Google Scholar]
- 28.van Heel, M. (1987) Ultramicroscopy 21, 111–124. [DOI] [PubMed] [Google Scholar]
- 29.Radermacher, M. (1988) J. Electron Microsc. Tech. 9, 359–394. [DOI] [PubMed] [Google Scholar]
- 30.Ruprecht, J. & Nield, J. (2001) Prog. Biophys. Mol. Biol. 75, 121–164. [DOI] [PubMed] [Google Scholar]
- 31.Stock, D., Leslie, A. G. & Walker, J. E. (1999) Science. 286, 1700–1705. [DOI] [PubMed] [Google Scholar]
- 32.Stahlberg, H., Müller, D. J., Suda, K., Fotiadis, D., Engel, A., Meier, T., Matthey, U. & Dimroth, P. (2001) EMBO Rep. 2, 229–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seelert, H., Poetsch, A., Dencher, N. A., Engel, A., Stahlberg, H. & Muller, D. J. (2000) Nature 405, 18–19. [DOI] [PubMed] [Google Scholar]
- 34.Tavakoli, N., Kluge, C., Golldack, D., Mimura, T. & Dietz, K. J. (2001) Plant J. 28, 51–59. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boekema, E. J., van Breemen, J. F., Brisson, A., Ubbink-Kok, T., Konings, W. N. & Lolkema, J. S. (1999) Nature 401, 37–38. [DOI] [PubMed] [Google Scholar]
- 37.Kakinuma, Y., Yamato, I. & Murata, T. (1999) J. Bioenerg. Biomembr. 31, 7–14. [DOI] [PubMed] [Google Scholar]
- 38.Domgall, I., Venzke, D., Luttge, U., Ratajczak, R. & Bottcher, B. (2002) J. Biol. Chem. 277, 13115–13121. [DOI] [PubMed] [Google Scholar]
- 39.Wilkens, S. & Forgac, M. (2001) J. Biol. Chem. 276, 44064–44068. [DOI] [PubMed] [Google Scholar]
- 40.Chaban, Y., Ubbink-Kok, T., Keegstra, W., Lolkema, J. S. & Boekema, E. J. (2002) EMBO Rep. 10, 982–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murata, T., Kawano, M., Igarashi, K., Yamato, I. & Kakinuma, Y. (2001) Biochim. Biophys. Acta 1505, 75–81. [DOI] [PubMed] [Google Scholar]
- 42.Arata, Y., Baleja, J., D. & Forgac, M. (2002) Biochemistry 41, 11301–11307. [DOI] [PubMed] [Google Scholar]
- 43.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24, 946–950. [Google Scholar]
- 44.Esnouf, R. M. (1997) J. Mol. Graphics 15, 133–138. [DOI] [PubMed] [Google Scholar]
- 45.Merritt, E. A. & Bacon, D. J. (1997) Methods Enzymol. 277, 505–524. [DOI] [PubMed] [Google Scholar]