Abstract
In vitro selection was used to investigate whether nucleic acid enzymes are capable of catalyzing photochemical reactions. The reaction chosen was photoreactivation of thymine cyclobutane dimers in DNA by using serotonin as cofactor and light of wavelengths longer than the absorption spectrum of DNA. Curiously, the dominant single-stranded DNA sequence selected, UV1A, was found to repair its internal thymine dimer substrate efficiently even in the absence of serotonin or any other cofactor. UV1C, a 42-nucleotide fragment of UV1A, repaired the thymine dimer substrate in trans (kcat/kuncat = 2.5 × 104), showing optimal activity with 305 nm light and thus resembling naturally occurring photolyase enzymes. Mechanistic investigation of UV1C indicated that its catalytic role likely exceeded the mere positioning of the substrate in a conformation favorable for photoreactivation. A higher-order structure, likely a quadruplex, formed by specific guanine bases within the deoxyribozyme, was implicated as serving as a light-harvesting antenna, with photoreactivation of the thymine dimer proceeding possibly via electron donation from an excited guanine base. In a primordial “RNA world,” self-replicating nucleic acid populations may have been vulnerable to deactivation via UV light-mediated pyrimidine dimer formation. Photolyase nucleic acid enzymes such as the one described here could thus have played a role in preserving the integrity of such an RNA world.
The RNA world hypothesis (1) postulates that RNA or RNA-like polymers, capable of genetic as well as catalytic function, may have constituted primitive “life” in the course of evolution. Currently, in vitro selection (2, 3) experiments from random sequence DNA and RNA libraries permit the identification of novel catalytic activities for nucleic acids, in support of the RNA world hypothesis. To date, such selections have indicated a substantially broader catalytic repertoire for RNA and DNA than found in naturally occurring ribozymes (4). We were interested in investigating whether reactions that use light energy could be catalyzed by nucleic acid enzymes.
Thymine (or pyrimidine) dimers are the major lesions formed in DNA as a result of exposure to UV light. Two major kinds of dimer are known, the cyclobutane and the (6—4) photoproduct (5). Different organisms use a variety of strategies to repair these lesions, among the more interesting of which is the use of light of substantially lower energy (longer wavelength) than the natural absorption of thymine dimers (>250 nm wavelength) to reactivate the dimers back to monomers. Such “photolyase” enzymes for the repair of both cyclobutane and (6—4) dimers have been studied extensively (6). The cyclobutane (CPD) photolyases harness a broad spectrum of light by using a number of chromophores such as methenyltetrahydrofolate (MTHF), flavin nucleotides, and tryptophan side chains (7). Photoexcitation culminates in electron donation from the excited-state flavin directly to the thymine dimer, leading to destabilization of the 5—5 and 6—6 bonds of the dimer, and thus, reversion to base monomers. Interestingly, studies have shown that a single strategically positioned tryptophan residue in the Escherichia coli enzyme's active site is able to provide a significant photoreactivation, even in the absence of the FADH and MTHF cofactors (8).
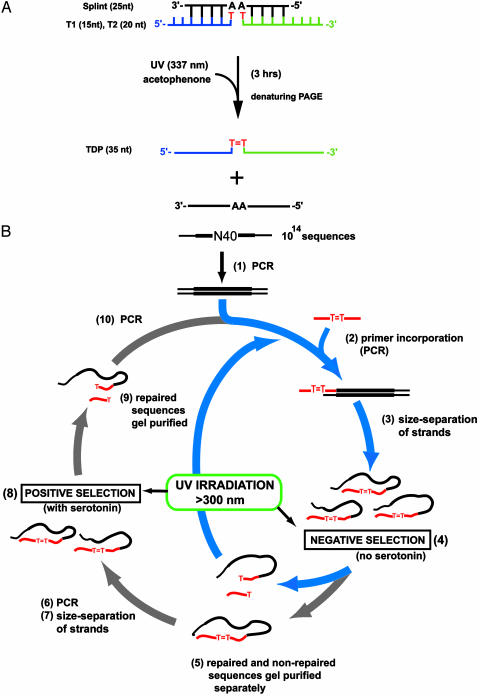
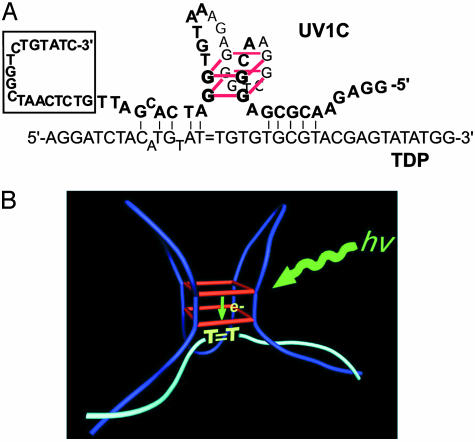
To investigate whether a photolyase nucleic acid enzyme could indeed exist, in vitro selection was carried out from a random-sequence DNA library. Our library (1014 different sequences, each containing a 40-nt random sequence flanked by 35- and 20-nt primer-binding sequences) incorporated the following design feature (Fig. 1A): the 35-nt 5′ element (“TDP”) consisted of 20- and 15-mer oligonucleotides (T1 and T2, respectively) covalently linked through a thymine dimer, but lacking a connecting phosphodiester linkage. This TDP substrate was incorporated by PCR into the random-sequence DNA library, providing a means for selecting out catalytically active molecules from the pool, because DNA molecules capable of efficient self-repair decreased in length from 95 to 80 nt, making size-based purification possible. Because tryptophan has been shown to be an adequate sensitizer for photoreactivation, the indole-containing compound serotonin (whose absorbance spectrum is red-shifted relative to that of tryptophan), was chosen as cofactor for our selection.
Fig. 1.
Substrate design and the in vitro selection cycle. (A) Synthesis of a thymine dimer-containing DNA oligonucleotide (TDP) lacking a key internal phosphodiester. A 25-nt complementary “splint” was used to align a 5′ 32P-labeled 15-nt oligonucleotide (T1) and a 20-nt oligonucleotide (T2), lacking 3′ and 5′ terminal phosphate groups, respectively. (B) The in vitro selection cycle.
Materials and Methods
DNA Oligonucleotides. DNA oligomers were obtained from University of British Columbia Nucleic Acid Protein Service (NAPS) Unit and size-purified on denaturing polyacrylamide gels. The starting library of random DNA sequences was obtained from University of Calgary Core DNA Services, and had the sequence 5′-AGGATCTACATGTATTGTGTGCGTACGAGTATATGGN40GTCTCAATCGGTCTGTATC-3′.
Synthesis of Thymine Dimer-Containing Oligonucleotide. For TDP formation, unlabeled T2 and 5′ 32P-labeled T1 oligonucleotides were annealed at 400 μM concentrations to a complementary splint oligonucleotide in buffered 40 mM MgCl2. The sequence of T1 was 5′-AGGATCTACATGTAT-3′, and that of T2 was 5′-TGTGTGCGTACGAGTATATG-3′. Solutions were degassed by freezing and thawing under vacuum three times. The DNA was irradiated in the presence of a 5 mM concentration of the triplet sensitizer acetophenone (shown specifically to favor the formation of cyclobutane thymine dimers; ref. 9) and 5% acetone for 3 h in a quartz cuvette (with a Photon Research Associates LN-1000 nitrogen laser with spectral output at 337 nm and a pulse rate of 7 Hz). The TDP construct (5′-AGGATCTACATGTAT = TGTGTGCGTACGAGTATATG-3′) was size-purified on 12% denaturing polyacrylamide gels and tested for the presence of the cyclobutane [rather than the (6—4)] thymine dimer by direct photoreversal experiments at 254 nm. At this wavelength, almost quantitative yield of the regenerated T1 and T2 strands was obtained.
DNA Library Preparation and Selection Procedure. The TDP primer was incorporated into the 5′-end of the random sequence library by using PCR. Owing to the inability of Taq polymerase to extend past the thymine dimer on the complementary strand during extension (10), the two resulting strands had different sizes and could be cleanly separated by using denaturing polyacrylamide gel electrophoresis. For the “negative” selection step, 1 μM of purified DNA was folded in 20 mM sodium phosphate, pH 7.0, and 40 mM NaCl, and irradiated on a Fotodyne transilluminator (with a 300-nm output) with an intervening polystyrene filter, such that the light transmitted had wavelengths of >300 nm. The irradiated pool was then gel purified to retrieve unmodified (95 nt) single strands, which were subjected to “positive” selection by irradiation in the presence of 10 μM serotonin (in the first five rounds of selection, a PCR amplification step was carried out between the negative and positive selection steps). Stringency was enhanced in the course of the selection by decreasing the time of irradiation, from 2 h in round 1 down to 10 s by round 25. After round 25, the pools were cloned by using standard protocols.
Kinetic and Spectrophotometric Analysis. The kinetic properties of UV1C were measured in 20 mM sodium phosphate, pH 7.0, 240 mM NaCl, at room temperature. Solutions (100 μl) containing 5′ 32P-labeled DNA were irradiated in a quartz cuvette and cell holder by using an Ushio Xenon short arc lamp, with wavelengths isolated with a SLM MC200 model monochromator, and bandwidth set at 4 nm. Time points were taken, and the DNA, after separation on 12% denaturing gels, was quantified by using Molecular Dynamics imagequant software. Single turnover reactions were carried out with 2.1 μM UV1C and 20 nM TDP substrate in 20 mM sodium phosphate, pH 7.0, with 240 mM NaCl. Samples were irradiated in a quartz cuvette with a 1-cm path-length at different wavelengths by using a 4-nm bandwidth. The photon flux of the lamp source at given wavelengths was determined by standard ferrioxalate actinometry (11), and used for light intensity corrections and quantum yield calculations. Quantum yields of thymine dimer repair were calculated by using the slope obtained from a least squares fit of the initial rates of reaction obtained from quantitation of gel data. The difference absorption spectra were recorded in a Cary dual beam UV spectrophotometer on DNA folded in 50 mM Tris·Cl, pH 8.0, supplemented with either 200 mM NaCl or 200 mM LiCl. The absorbance spectrum of DNA in lithium buffer was subtracted from the spectrum of DNA in sodium buffer, and signal was averaged over 10 scans to obtain the final difference spectrum. For the rate enhancement data, reaction mixtures contained 4 μM UV1C and 40 nM TDP substrate in 20 mM sodium phosphate, pH 7.0, plus 240 mM NaCl. Initial rates of reaction were divided by the background rates (measured on 40 nM TDP in the presence of 4 μM of an unrelated DNA oligomer).
Results and Discussion
Fig. 1B summarizes the in vitro selection cycle. In a negative selection step, the TDP-containing DNA pool was irradiated at >300 nm, in the absence of serotonin (step 4); DNA molecules surviving this step intact were subjected to positive selection (step 8) in the presence of 10 μM serotonin. Curiously, after 5 rounds of selection, a pool of DNA molecules emerged that robustly self-repaired in the negative selection step; this was surprising because neither DNA nor thymine dimers absorb significantly at >300 nm. The serotonin-dependent and -independent pools were now separately taken through 20 additional selection rounds (the serotonin-independent pool was subjected only to the negative selection step, whereas the serotonin-dependent pool underwent both positive and negative selection, as defined in Fig. 1B). Each pool was then cloned and sequenced. The serotonin-independent pool, the subject of this article, yielded only one dominant sequence, UV1A, whose sequence was distinct from two major sequences obtained from the serotonin-dependent pool (data not shown).
Preliminary characterization of UV1A (sequence: 5′-AGGATCTACATGTAT = TGTGTGCGTACGAGTATATGGAGA ACGCGAGGCA AGGCTGGGAGA A ATGTGGATCACGATTGTCTCAATCGGTCTGTATC-3′) showed that it did indeed self-repair at an accelerated rate, relative to controls, with light of wavelengths >300 nm. To test whether a “substrate” component within the UV1A sequence could be separated from a “catalytic” component, the 5′ constant (TDP) region was detached from UV1A, leaving an oligonucleotide UV1B. UV1B was then further shortened by deletion of the 3′-most 20 nucleotides, to 42-nt UV1C (the underlined sequence within UV1A, above). Both UV1B and UV1C were found to be fully catalytic with respect to the substrate oligonucleotide TDP. All subsequent analysis was thus carried out with the 42-nt UV1C deoxyribozyme. Multiple turnover kinetic analysis (data not shown) yielded a kcat value of 4.5 min–1 and KM of 0.58 ± 0.14 μM at 3.4 × 10–9 einsteins·min–1 illumination (the uncatalyzed photoreactivation rate of TDP was 0.00018 min–1, giving kcat/kuncat = 25,000). Both the measured kcat and KM compared favorably with those of a catalytic antibody for thymine dimer photoreactivation (1.2 min–1 and 6.5 μM at 1.26 × 10–7 einsteins·min–1) (12, 13); although poorer than E. coli photolyase itself (14). The recognition between the TDP substrate and the UV1C deoxyribozyme appeared to involve a significant degree of tertiary interaction, because only modest stretches of Watson–Crick complementarity was found between them.
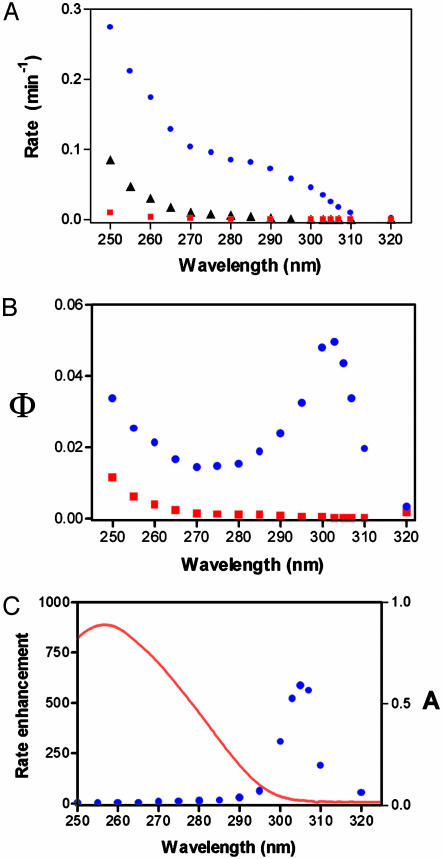
Fig. 2A shows the action spectrum of UV1C under single turnover conditions, within the 250–320 nm spectral range (normalized for constant light intensity). TDP (0.02 μM) was irradiated in the presence of an excess (2.1 μM) of (i) UV1C; (ii) the 25-nt splint (Fig. 1 A) that renders TDP double-stranded; and (iii) an unrelated, 42-nt, single-stranded DNA. Although the splint contributed to somewhat enhanced TDP repair, especially between 250 and 270 nm, UV1C caused significantly higher reactivation rates across the spectrum, with a notable “shoulder” at >275 nm. Fig. 2B shows Φ, or quantum yield, for TDP reactivation in the presence of excess UV1C and of excess complement (splint) DNA. The UV1C quantum yield of 0.05, although low compared to those of CPD photolyases (≈0.7) (14), was of the order of that for (6—4) photolyases (≈0.1) (15). The heightened efficiency of UV1C-dependent reactivation between 280 and 320 nm was particularly evident. Fig. 2C plots the ratio of the quantum yields reported in Fig. 2B (equivalent, also, to kUV1C-catalyzed/kuncatalyzed, under single-turnover conditions). Notably, the peak of UV1C catalytic activity, at 305 nm, was a significantly longer wavelength than the absorbance maximum of folded DNA (shown in blue) and also far from the absorption (<250 nm) of thymine dimers themselves. Such a property, of utilization of near UV light by UV1C, was reminiscent of protein photolyase enzymes. That UVIC was in fact catalyzing photo-reactivation (and not a nuclease or glycosidase activity) was verified by the successful reutilization of the UV1C-cleaved (using 303–307 nm light) T1 product to regenerate the active substrate TDP by using the protocol shown in Fig. 1 A. (Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 2.
Wavelength dependence for the catalytic activity of the UV1C deoxyribozyme. (A) Action spectrum for photoreactivation of 20 nM TDP substrate by excess (2.1 μM) UV1C (blue circles); 2.1 μM of the 25-nt complementary splint oligomer (black triangles); and 2.0 μM of an unrelated 42-nt single-stranded DNA (red squares). All measurements were made under single turnover conditions, and all rates are shown corrected for variations in light intensity at the different wavelengths. (B) Quantum yield (Φ) of photoreactivation by UV1C (blue circles) and by the complementary splint oligomer (red squares). (C) The photoreactivation rate enhancement (blue circles) by UV1C over that by an unrelated, control DNA oligomer. The absorption spectrum of the DNAzyme–substrate complex (shown as a red line) helps to highlight the red shift in the spectral region for optimal rate enhancement relative to the absorption maximum of DNA.
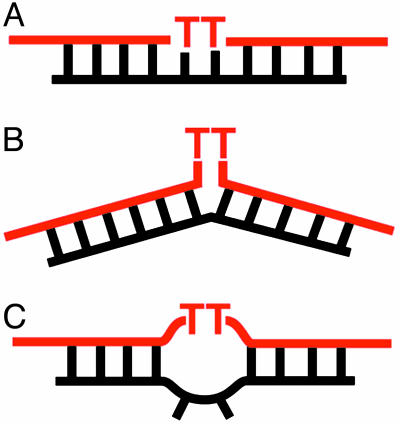
To explore whether the catalytic role of UV1C was primarily to orient the TDP thymine dimer to a conformation more amenable to photoreactivation, a number of model oligonucleotide constructs were synthesized (Fig. 3), and their reactivation rates were measured in the absence of UV1C. TDP was base-paired to (i) a perfectly complementary splint; (ii) a complementary splint as in i, but lacking the two adenines expected to base pair to the thymine dimer; and iii a complementary splint, containing two cytosines (or thymines) to replace the above-mentioned adenines. In case ii, it was anticipated that the thymine dimer might bulge out of the double helix in a manner preferred by various DNA repair enzymes (16, 17). Interestingly, all three constructs, in the absence of UV1C, underwent photoreactivation at or close to the background, uncatalyzed rate. Therefore, it was concluded that the catalytic role of UV1C was not solely to orient the thymine dimer in any one of the above conformations.
Fig. 3.
Oligonucleotide constructs used to test whether UV1C induced specific conformations in the TDP substrate. TDP hybridized to a perfectly complementary, 25-nt splint (A), a “flipped” splint in which the two adenines complementary to the TDP thymine dimer have been deleted (B), and a “mismatch” splint wherein the above-mentioned adenines have been replaced by either two thymines or two cytosines, creating mismatches with the thymine dimer (C).
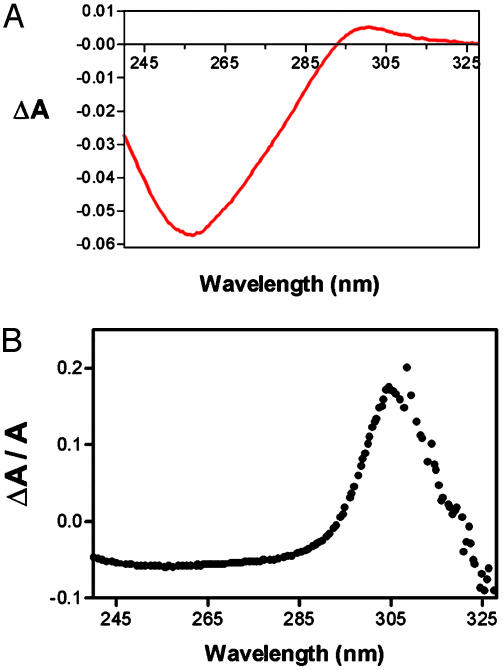
A clue to UV1C's mode of catalysis emerged from analysis of its ionic requirements. UV1C did not require magnesium but did require sodium or potassium (however, significantly, not lithium) ions for activity. Furthermore, methylation protection analysis of folded UV1A revealed the protection of blocks of guanines in sodium, but not lithium-containing buffers. These observations, taken together with the high guanine-content of UV1C and its parent, UV1A, suggested that they fold to form guanine-quadruplexes (which are stabilized specifically by Na and K, but not by Li or Mg ions; ref. 18). Recently, Mergny et al. (19) have shown, by using difference spectral analysis, that G-quadruplex absorption incorporates a modestly enhanced “tail” in the 290- to 305-nm region. Fig. 4A shows a difference spectrum, ΔA, recorded for UV1C (spectra recorded in buffered 200 mM LiCl subtracted from those recorded in NaCl). The shape of this difference spectrum is fully consistent with UV1C folding to a G-quadruplex in sodium but not in lithium solution. Fig. 4B, which plots ΔA/A (the difference spectrum standardized to absorbance), shows a dominant peak at ≈305 nm, a feature that strikingly resembles the peaks at 305 nm in both the quantum yield (Fig. 2B) and rate enhancement (Fig. 2C) plots. The markedly efficient reactivation by UV1C at 305 nm therefore appears to have a basis in the deoxyribozyme's spectral absorption. To test whether contaminating acetophenone from the synthesis of the thymine dimer in TDP played a part in the observed catalysis (although TDP had been extensively gel-purified following exposure to acetophenone), acetophenone was deliberately added to UV1C and substrate. However, no change in rate was observed (data not shown).
Fig. 4.
Absorbance difference spectra of the deoxyribozyme-substrate complex recorded in sodium-containing and lithium-containing buffers. (A) The difference spectrum was obtained by subtracting the lithium spectrum from the sodium spectrum. UV1C (2 μM) was folded in the presence of 2 μM of a pseudosubstrate (a 35-mer oligonucleotide of the sequence of TDP, but containing two undimerized thymines in place of the thymine dimer) in buffered 200 mM NaCl or 200 mM LiCl. Absorbance measurements were taken by using a dual beam UV spectrophotometer in a quartz cuvette with a 1-cm path length. (B) The difference spectrum shown in A, normalized with respect to overall absorbance.
Fig. 5A shows a proposed model for the folded structure of UV1C, based on the methylation protection data. Fig. 5B illustrates a model for the hypothesized mechanism of UV1C. The guanine quadruplex within the folded structure of UV1C acts as an antenna for “long wavelength” (near UV) light, whose energy is then used with relative efficiency for the repair of the thymine dimer substrate. The precise mechanism of repair remains unclear; it is conceivable, by analogy with photolyase protein enzymes, that the photoexcited G-quadruplex donates one or more electrons toward the reactivation of the thymine dimers (of the four natural bases in DNA, guanine is known to be the most readily oxidized; ref. 20).
Fig. 5.
(A) A model for the folded structure of the UV1C deoxyribozyme, with possible Watson–Crick base pairs formed between it and the substrate, TDP. UV1C itself does not incorporate the sequence shown within the boxed area (inclusion of this sequence, the original 3′ invariant region from the in vitro selection library, generates the deoxyribozyme UV1B). Correspondingly, joining the 3′ terminus of TDP (after exclusion of the 3′-most two guanines) to the 5′ terminus of UV1B defines the single-turnover deoxyribozyme UV1A. (B) A model for the mode of catalysis by the UV1C deoxyribozyme. A short guanine quadruplex within the folded deoxyribozyme is able to absorb near-UV light at 305 nm wavelength. The absorbed energy is then transmitted efficiently to the bound substrate, possibly in the form of one or more electrons donated from the guanines of the quadruplex to the thymine dimer, which may be stacked next to the quadruplex.
In conclusion, unexpected similarities between the UV1C deoxyribozyme and photolyase protein enzymes (although UV1C utilizes a narrower spectral window than do protein photolyases), and the broader discovery that nucleic acids, even in the absence of extraneous sensitizing cofactors, are able to catalyze such photochemical transformations, suggest that photochemical reactions might well have constituted a significant part of the chemical/catalytic repertoire of a putative RNA world. A primordial role for the formation and breakage of intermolecular thymine dimers as a means of generating progressively larger nucleic acids has been hypothesized by Lewis and Hanawalt (21). The atmosphere of the early Earth was believed to have been subjected to high levels of UV radiation, owing to the absence of a stratospheric ozone layer. This would have posed a serious threat to a largely nucleic acid population of an RNA world. Evidence presented here that a string of nucleotides is able to repair itself from UV damage has implications for the survival of early life forms.
Supplementary Material
Acknowledgments
We thank Peter Unrau for comments on the manuscript and Gary Leach for use of the N2 laser. This work was supported by Natural Sciences and Engineering Research Council (Canada) and Canadian Institutes of Health Research. D.S. is a Senior Scholar of the Michael Smith Foundation for Health Research.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Gilbert, W. (1986) Nature 319, 618. [Google Scholar]
- 2.Ellington, A. D. & Szostak, J. W. (1990) Nature 346, 818–822. [DOI] [PubMed] [Google Scholar]
- 3.Tuerk, C. & Gold, L. (1990) Science 249, 505–510. [DOI] [PubMed] [Google Scholar]
- 4.Jaschke, A. (2001) Curr. Opin. Struct. Biol. 11, 321–326. [DOI] [PubMed] [Google Scholar]
- 5.Matsunaga, T. Hieda, K. & Nikaido, O. (1991) Photochem. Photobiol. 54, 403–410. [DOI] [PubMed] [Google Scholar]
- 6.Carell, T, Burgdorf, L. T., Kundu, L. M. & Cichon, M. (2001) Curr. Opin. Chem. Biol. 5, 491–498. [DOI] [PubMed] [Google Scholar]
- 7.Sancar, A. (2003) Chem. Rev. 103, 2203–2237. [DOI] [PubMed] [Google Scholar]
- 8.Kim, S. T., Li, Y. F. & Sancar, A. (1992) Proc. Natl. Acad. Sci. USA 89, 900–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang, S. Y., ed. (1976) Photochemistry & Photobiology of Nucleic Acids: Chemistry (Academic, New York), Vol. 1.
- 10.Wellinger, R.-E. & Thoma, F. (1996) Nucleic Acids Res. 24, 1578–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calvert, G. C. & Pitts, J. N., Jr. (1966) Photochemistry (Wiley, New York), pp. 783–786.
- 12.Cochran, A. G., Sugasawara, R. & Schultz, P. G. (1988) J. Am. Chem. Soc. 110, 7888–7890. [Google Scholar]
- 13.Jacobsen, J. R., Cochran, A. G., Stephans, J. C., King, D. S. & Schultz, P. G. (1995) J. Am. Chem. Soc. 117, 5453–5461. [Google Scholar]
- 14.Kim, S.-T. & Sancar, A. (1991) Biochemistry 30, 8623–8630. [DOI] [PubMed] [Google Scholar]
- 15.Hitomi, K., Kim, S. T., Iwai, S., Harima, N., Otoshi, E., Ikenaga, M. & Todo, T. (1997) J. Biol. Chem. 272, 32591–32598. [DOI] [PubMed] [Google Scholar]
- 16.Vande Berg, B. J. & Sancar, G. B. (1998) J. Biol. Chem. 273, 20276–20284. [DOI] [PubMed] [Google Scholar]
- 17.Bhattacharyya, A. & Lilley, D. M. (1989) Nucleic Acids Res. 17, 6821–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sen, D. & Gilbert, W. (1990) Nature 344, 410–414. [DOI] [PubMed] [Google Scholar]
- 19.Mergny, J. L., Phan, A. T. & Lacroix, L. (1998) FEBS Lett. 435, 74–78. [DOI] [PubMed] [Google Scholar]
- 20.Steenken, S. & Jovanovic, S. V. (1997) J. Am. Chem. Soc. 119, 617–618. [Google Scholar]
- 21.Lewis, R. J. & Hanawalt, P. C. (1982) Nature 298, 393–396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.