Abstract
Poly(ADP-ribosyl)ation has been suggested to be involved in regulation of DNA repair, transcription, centrosome duplication, and chromosome stability. However, the regulation of degradation of poly(ADP-ribose) and its significance are not well understood. Here we report a loss-of-function mutant Drosophila with regard to poly(ADP-ribose) glycohydrolase, a major hydrolyzing enzyme of poly(ADP-ribose). The mutant lacks the conserved catalytic domain of poly(ADP-ribose) glycohydrolase, and exhibits lethality in the larval stages at the normal development temperature of 25°C. However, one-fourth of the mutants progress to the adult stage at 29°C but showed progressive neurodegeneration with reduced locomotor activity and a short lifespan. In association with this, extensive accumulation of poly(ADP-ribose) could be detected in the central nervous system. These results suggest that poly(ADP-ribose) metabolism is required for maintenance of the normal function of neuronal cells. The phenotypes observed in the parg mutant might be useful to understand neurodegenerative conditions such as the Alzheimer's and Parkinson's diseases that are caused by abnormal accumulation of substances in nervous tissue.
Poly(ADP-ribosyl)ation process involves a posttranslational modification of target proteins catalyzed by the poly(ADP-ribose) polymerase (PARP) family of enzymes with NAD+ as the substrate, resulting in formation of long-branched polymers of ADP-ribose (1, 2). The covalently attached and negatively charged poly(ADP-ribose) units significantly affect several important biological functions, including DNA repair (3), transcription (4), regulation of telomere length, cell cycle progression, centrosome duplication (5, 6), and chromosome stability (7). One of the major members of the PARP family is PARP-1, which catalyzes poly(ADP-ribosyl)ation in response to DNA strand breaks. Recently, additional members of the PARP family of enzymes have been characterized; PARP-2, -3, tankyrase-1, -2, VPARP, and Ti-PARP (2). In Drosophila, only two PARP family members, corresponding to PARP-1 and tankyrase, have been reported (8, 9).
Poly(ADP-ribose) attached to acceptor proteins is hydrolyzed rapidly to produce free ADP-ribose residues by poly(ADP-ribose) glycohydrolase (PARG) (10–12). In contrast to PARPs, only one gene for PARG has been detected in mammals and insects (13). Thus, it is likely that the regulation of PARG activity is required to complete protein modification cycles initiated by different PARPs. In fact, PARG has been proposed to shuttle between the nucleus and the cytoplasm and becomes localized to the centrosomes during mitosis (14). There is evidence for an alternative form in the cytoplasm (M. K. Jacobson, personal communication), indicating that regulation of poly(ADP-ribosyl)ation by PARG is very dynamic.
Mutation of the parg gene, tej, in plants alters circadian rhythms (15), and increased sensitivity to DNA damage in parg knockout mouse embryonic stem cells has been reported (3). However, little is known about the effects of a loss-of-function mutations in animal models. To elucidate the role of poly(ADP-ribose) metabolism in vivo, we have generated such a parg mutant in Drosophila.
Materials and Methods
Flies and Mutagenesis. The Drosophila EP351 strain was obtained from the Bloomington Stock Center (http://fly.bio.indiana.edu), and EP351; +/TMS, Sb P[ry+ delta2–3]99B females were crossed with FM7/Y males. Offspring without P element, selected by eye color, were crossed with FM7/Y to produce parg27.1. Transgenic flies expressing PARG were made according to a standard protocol (16). pargEGFP consists of parg gene (nulceotides 33,667–37,056 of GenBank accession no. Z98254) fused to EGFP (Clontech) and a pCaSpeR-based vector. hs-parg is a parg cDNA driven by a heatinducible promoter of pCaSpeR-hs.
Genomic Southern Blotting. A cDNA fragment of parg exon 1 was labeled with digoxigenin–dUTP (Roche Diagnostics) by PCR amplification. HindIII-digested genomic DNA from adult flies was used for Southern blot analysis, as described (17), then detected with alkaline phosphatase-labeled anti-digoxigenin antibody by using nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate for color development.
RT-PCR. Total RNA was isolated with ISOGEN (Wako Pure Chemical, Osaka) from adult flies cultured at 29°C, then treated with RNase free DNase (Roche Diagnostics). RT-PCR was performed with a RT-PCR core kit following the manufacturer's protocol (Applied Biosystems) with primers for parg (forward, 5′-GGATCCACCGGTATGCAAGAATT-3′; reverse, 5′-TCGCAGACACTCCGTAAAGA-3′) and rp49 (forward, 5′-GAAGAAGCGCACCAAGGACT-3′; reverse, 5′-TTGAATCCGGTGGGCAGCAT-3′) (18).
Locomotor Activity and Survival. The parg27.1/FM7 genotype was cultured at 29°C with normal media. Eclosed parg27.1/Y and parg27.1/FM7 genotypes were kept at 29°C in a vial with blotting paper soaked with grape juice to prevent death of the mutants, and they were used for scoring locomotor activity. For this purpose, flies were put in a clear polystyrene vial with measures. Flies were shaken down to the bottom, and their position was scored after 20 sec. A perfect score was given if they climbed >10 cm within 20 sec. The average score of 10 experiments and the survival rate of the flies used for scoring were determined.
ELISA. Total nucleic acid from adult flies eclosed at 29°C was spotted on a nylon membrane, and the concentration was adjusted to 10 A260 then diluted 5- and 25-fold. After blocking with 5% skimmed milk/TBS-T (Tris-buffered saline plus 0.1% Tween 20), the membranes were incubated with anti-poly(ADP-ribose) antibody (10H) (19) and with alkaline phosphatase labeled second antibody (Sigma). Detecton of binding was carried out with nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate.
Immunostaining. Adult flies were frozen in carboxymethyl cellulose, then sectioned in a cryostat by the film method (20). These sections were fixed with formalin, and stained with antipoly(ADP-ribose) antibody (10H) and tetramethylrhodamine B isothiocyanate (TRITC)-labeled second antibody. Nuclei were stained with Hoechst dye 33342. For axon staining, formalinfixed, paraffin-embedded brain sections were immunostained by the avidin–biotin–peroxidase complex method with an antibody against phosphorylated neurofilaments (SMI 31, 1:1,000, Sternberger Monoclonal, Lutherville, MD). Diaminobenzidine was used as the chromogen, and sections were then counterstained with hematoxylin.
Electron Microscopy. Adult males of the mutant and wild-type OregonR, eclosed at 29°C and kept at 25°C, were used for electron-microscopic analysis according to a standard protocol.
Results
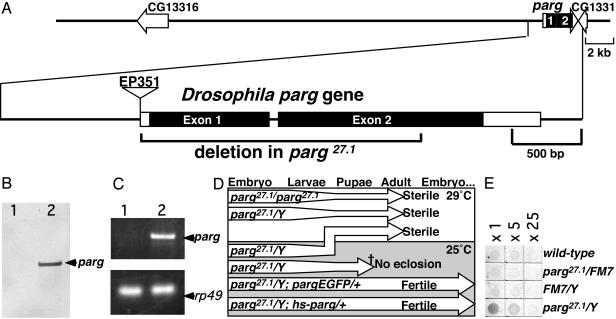
Isolation of a parg Loss-of-Function Drosophila melanogaster Mutant. The Drosophila parg gene (Fig. 1A) has been mapped to the X chromosome by the European Drosophila Genome Sequencing Consortium (GenBank accession no. Z98254), and a Drosophila parg cDNA has been submitted by Ame and Jacobson (GenBank accession no. AF079556). The ORF of parg is indicated to start from 68th adenine of AF079556. However, according to the annotation of the Drosophila Genome Project, the start is from the 203rd adenine. We found a polymorphic allele with a 10-bp deletion (169–178delACTCGGCAAG of AF079556) in some strains, in which the ORF from the 68th adenine was disrupted. Therefore, the regulation of translation of Drosophila parg might be variable. A mutant with a P element in 5′UTR of the parg gene, EP351 (Fig. 1 A) was produced by the Berkeley Drosophila Genome Project (GenBank accession no. AQ025499). EP351 proved to be viable in the homozygous state, expressing parg transcripts (data not shown). To make a loss-of-function mutant, we generated deletion mutants by imprecise excision of the P element. One of the mutants, parg27.1, lacked two-thirds of the parg ORF (nucleotides 34,622–36,079, GenBank accession no. Z98254), including the conserved catalytic domain (Fig. 1A). The allele, parg27.1, was stocked with a balancer X chromosome, either FM7 or FM7GFP. With the parg27.1/Y genotype, Southern blotting showed no band corresponding to the first exon of the parg gene (Fig. 1B) and RT-PCR did not detect any expression of parg-message (Fig. 1C). Almost all parg27.1/Y embryos laid by parg27.1/FM7GFP females crossed with FM7GFP/Y hatched, and two-thirds of the larvae developed to the pupal stage, but they showed lethality before eclosion at 25°C (Fig. 1D). When parg27.1/Y was cultured at 29°C continuously, approximately one-fourth of the parg27.1/Y embryo developed into morphologically normal adult flies (Fig. 1D). For this to occur, it was necessary to elevate the culture temperature from 25°C to 29°C before or just after pupation (Fig. 1D).
Fig. 1.
Characterization of the Drosophila parg mutant. (A) The upper line denotes genes in the parg locus predicted by the European Drosophila Genome Sequencing Consortium (GenBank accession no. Z98254). The structure of the Drosophila parg gene is shown in the lower line, and the parg27.1 mutant deletion is indicated by the bracket. Boxes denote exons, and filled boxes denote coding regions. The position of a P element insertion is shown. (B) Genomic Southern blotting of the parg gene. Lane 1, parg27.1/Y; lane 2, wild type. (C) parg transcripts examined by RT-PCR. Lane 1, parg27.1/Y; lane 2, wild-type. rp49 was used as a control. (D) Effects of temperature on development of parg mutants. (E) Relative content of poly(ADP-ribose) examined by ELISA using an anti-poly(ADP-ribose) antibody.
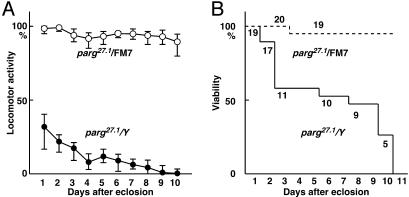
Neurological Disorders in the Loss-of-Function Mutant Raised at Permissive Temperature. Interestingly, the surviving parg27.1/Y adults showed neurological abnormalities, and reduced locomotor activity, which became progressively more severe (Fig. 2A), and most mutants died within 10 days after eclosion (Fig. 2B). parg27.1/parg27.1 homogygous females also developed to morphologically normal adults at 29°C and showed the same defects in locomotor activity and viability as parg27.1/Y males. In contrast, most sibling females with the parg27.1/FM7 genotype showed high locomotor activity and were viable at the 10th day (Fig. 2). The maximum life span of parg27.1/Y adults eclosed at 29°C and kept at 25°C was 16 days, whereas most parg27.1/FM7 adults survived for 1 month after eclosion. Generally, most wild-type adult flies live beyond 1 month.
Fig. 2.
Locomotor activity and survival of parg27.1/Y and parg27.1/FM7 genotypes. (A) Locomotor activity was measured as described in Materials and Methods. (B) Viability of the same group.
Most of parg27.1/Y adults dragged their wings and could not fly, and three-fourths of them developed a black spot(s) in one or both of the base joints of the second limb (data not shown). The limb became disabled with the appearance of the spot. Both parg27.1/Y and parg27.1/parg27.1 genotypes were sterile. The neurological disorders and sterility of the mutants were rescued by transgenes containing the parg ORF with 1 kb of its upstream sequence (Fig. 1D, pargEGFP), or parg cDNA with a heatinducible promoter (Fig. 1D, hs-parg).
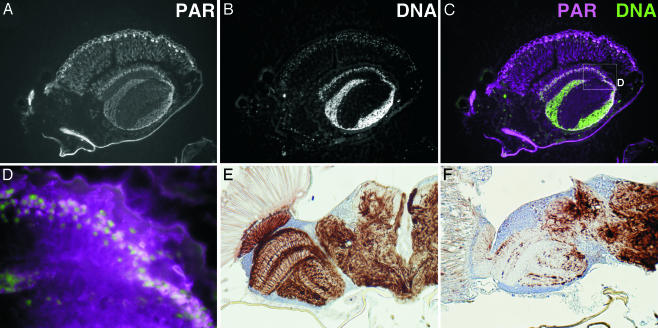
Accumulation of Poly(ADP-Ribose). The poly(ADP-ribose) content of adult flies was examined by ELISA using an anti-poly(ADP-ribose) antibody, 10H (19). The parg27.1/Y genotype showed strong staining, whereas the wild-type, parg27.1/FM7 and FM7/Y genotypes had only very faint signals (Fig. 1E). Incubation of poly(ADP-ribose) with extracts of the CNS of wild-type wandering larva eliminated the poly(ADP-ribose) signal, but there was no effect with extracts from parg27.1/Y larval CNS (data not shown). When adult wild-type flies were separated into head and body, both demonstrated significant capacity for poly(ADP-ribose) degradation. On the other hand, the bodies of parg mutant flies had activity that could be suppressed by NaF, an inhibitor of phosphodiesterase, whereas the heads showed only barely detectable activity (data not shown). These results indicate that the PARG protein is expressed in the CNS and that PARG must be the major or the only enzyme in this site that can degrade poly(ADP-ribose). To examine the location of poly(ADP-ribose) accumulation in the parg27.1/Y genotype, sections through adult flies were stained with anti-poly(ADP-ribose) antibody. Poly(ADP-ribose) was widely detected in the parg27.1/Y case, being particularly prominent in the CNS including the eye (Fig. 3 A–D) and the thoracic ganglion regions (data not shown), whereas poly(ADP-ribose) was not detectable in wild-type animals (data not shown). In the absence of the primary antibody, only autofluorescence of surface cuticle was detected (data not shown). With the parg27.1/Y genotype, strong signals were detected at the surface of brain, where nuclei of neuronal cells are clustered. There was some overlap of poly(ADP-ribose) and nuclear staining (Fig. 3 C and D). Poly(ADP-ribose) is known to be mainly metabolized in nuclei, but our findings indicated wide accumulation of poly(ADP-ribose) throughout the cell. No immunoreactivity of ubiquitinated proteins or polyglutamine repeat proteins was detected in the parg27.1/Y or wild-type genotypes (data not shown).
Fig. 3.
Accumulation of poly(ADP-ribose) and disruption of axon structures in the parg mutant. (A) Poly(ADP-ribose) in a parg27.1/Y adult as detected with anti-poly(ADP-ribose) antibody. (B) Nuclei are stained with Hoechst dye 33342. A part of the sagittal section of the head is shown. (C) Merging of poly(ADP-ribose) (magenta) and DNA (green). The box in C is shown in D at higher magnification. Axon staining of the horizontal section of adult head at 10 days after eclosion is shown in E for the wild type and in F for parg27.1/Y mutant.
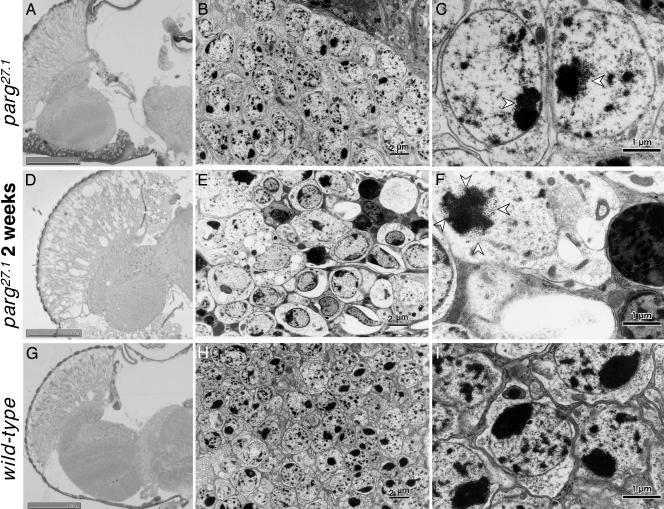
Neurodegenerative Changes. The normal structure of axons in the wild-type optic lobe (Fig. 3E), examined by immunostaining of phosphorylated neurofilament, was completely absent in the mutant brains (Fig. 3F). Although there were no gross microscopic changes in morphology a few days after eclosion (Fig. 4A), degeneration was remarkable 2 weeks thereafter (Fig. 4D). Electron microscopic analysis showed no typical degenerative change in neurons compared with wild-type (Fig. 4 B and H) at low magnification, but at high magnification aggregate(s) of uniform particles adjacent to nucleolus in the parg27.1/Y genotype were observed a few days after eclosion (Fig. 4C, arrowhead). These aggregates were more abundant in dying mutants 2 weeks later (Fig. 4F). Our findings are consistent with a recent report that poly(ADP-ribose) is abundant in chromocenters (21), which correspond to clustered heterochromatin. It is under investigation whether these particles are poly(ADP-ribose) or proteins modified by poly(ADP-ribosyl)ation. Most of the cells in the mutants were swollen, and the organelles and nuclei were no longer clearly visible (Fig. 4 E and F). Also, there were many condensed bodies indicative of cell death (Fig. 4 E and F). These features were not found in wild-type adults (Fig. 4 H and I).
Fig. 4.
Microscopic and electron-microscopic analyses of adult brain. Horizontal section through adult heads. (A–C) parg27.1/Y at a few days after eclosion. (D–F) parg27.1/Y at 2 weeks after eclosion. (G–I) Wild type. A, D, and G show light microphotographs of tissue stained with toluidine blue; B, E, and H show electron microphotographs; C, F, and I show higher magnification of parts of B, E, and H, respectively. Open arrowheads indicate granular structures typical in the parg27.1/Y genotype. (Scale bars, 2 μm in B, E, and H and 1 μm in C, F, and I.)
Discussion
The parg mutant isolated in this study lacks the entire region of the catalytic domain, and the parg27.1/Y genotype is clearly associated with neurodegeneration and accumulation of poly(ADP-ribose) in the CNS. The preferential accumulation of poly(ADP-ribose) or poly(ADP-ribosyl)ated proteins in the CNS has several possible explanations. First, the neuronal cells in CNS do not divide so that the accumulated poly(ADP-ribose) could not be diluted. However almost all cells in adult flies are nondividing, so that this cannot be a major factor. Second, the accumulation may reflect the relative amount of PARP activity in different tissues, because poly(ADP-ribose) would be expected to accumulate with a lack of degrading enzyme activity and is known that the brain has marked PARP activity in rats (22) and in mice (23). It is interesting that membrane depolarization induces poly(ADP-ribosyl)ation of nuclear proteins in brain cortical neurons (24), suggesting an importance of active poly(ADP-ribose) metabolism in neuronal function. Third, other enzyme(s) could be involved in the degradation of poly(ADP-ribose) in organs other than the CNS. For example, a phosphodiesterase that can degrade poly(ADP-ribose) has been described (25–27) and the brain might have lower phosphodiesterase activity as compared with other organs of Drosophila.
The fact that the parg mutants could not develop into adult flies at the normal temperature for development, 25°C, indicates that PARG is essential for the survival of Drosophila. Although PARG is clealy not required for morphogenesis of adult Drosophila, parg27.1/Y flies under permissive conditions, at 29°C, the observed neurodegeneration indicates an indispensable role in the maintenance of neuronal function and cell survival. It is known that some neurological disorders can be ameliorated by expression of heat shock proteins (28). Although the molecular mechanism underlying the neuronal cell death in our flies is not clear, it is possible that target proteins were irreversibly modified by poly(ADP-ribose) so that cellular events requiring its removal are disturbed. For example in mammals, PARG may regulate PARP-1 activity by removing poly(ADP-ribose) from heavily automodified PARP-1, because inactivation of PARP-1 by an inhibitor of PARG has been reported (29). Alternatively, because decondensation of chromatin structures by poly(ADP-ribose) synthesis has been reported (30), extensive accumulation of poly(ADP-ribose) in the mutant could lead to abnormal chromatin remodeling and disordered transcription, which might also explain the observed neurodegeneration. Consistent with this notion, the period length of the circadian oscillator of Arabidopsis was recently reported to be distorted by mutation of the tej gene, identified as a PARG orthologue in plants (15), presumably because of disordered regulation of transcription by persistent poly(ADP-ribosyl)ation of transcriptional factors.
A number of neurodegenerative disorders, including Alzheimer's disease, Parkinson's disease, prion disease, polyglutamine disease, the Tauopathies, and familial amyotrophic lateral sclerosis, are characterized by damage to neurons that can be caused by abnormal accumulation of protein aggregates seen as inclusion bodies (28, 31). These disorders could be caused by mutations of genes leading to abnormal aggregation of proteins normally hydrolyzed by proteases. At present, it is speculative whether abnormal accumulation of poly(ADP-ribose) might also be found in humans. However, progressive neurological deterioration and renal failure due to storage of glutamyl ribose-5-phosphate, which might be derived from the linkage portion between poly(ADP-ribose) and acceptor proteins, has been reported (32).
Amelioration of hypoxia reperfusion injury of neuronal cells with inhibitors of PARP was recently reported (33–35). It is possible that ischemia reperfusion injury of the brain and also neurological diseases of unknown etiology may result from an imbalance of poly(ADP-ribose) metabolism. The neurotoxicity found in parg knockout flies could, for example, be related to the PARP-1-dependent cell death described by Yu et al. (36) by altering the ratio of PARP-1 to PARG. Understanding of metabolic disturbances and associations with regulation of target proteins by poly(ADP-ribosyl)ation may lead to the development of new strategies for treatment and prevention of the above diseases.
Acknowledgments
We thank Ms. N. Sugae for electron microscopic analysis and Ms. K. Izumi for making frozen sections (Finetec, Tokyo). We also are grateful to Drs. Y. Hiromi, M. Okabe (National Institute of Genetics, Mishima, Japan), K. Furukubo-Tokunaga, and M. Kurusu (Institute of Biological Sciences, University of Tsukuba) for comments and discussion. This work was supported by Grants-in-Aid from the Ministry of Health, Labour and Welfare, and the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Abbreviations: PARG, poly(ADP-ribose) glycohydrolase; PARP, poly(ADP-ribose) polymerase.
References
- 1.de Murcia, G. & Shall, S. (2000) PolyADP-Ribosylation Reactions (Oxford Univ. Press, Oxford).
- 2.Bouchard, V. J., Rouleau, M. & Poirier, G. G. (2003) Exp. Hematol. 31, 446–454. [DOI] [PubMed] [Google Scholar]
- 3.Masutani, M., Nakagama, H. & Sugimura, T. (2003) Genes Chromosomes Cancer 38, 339–348. [DOI] [PubMed] [Google Scholar]
- 4.Kraus, W. L. & Lis, J. T. (2003) Cell 113, 677–683. [DOI] [PubMed] [Google Scholar]
- 5.Kanai, M., Tong, W. M., Sugihara, E., Wang, Z. Q., Fukasawa, K. & Miwa, M. (2003) Mol. Cell. Biol. 23, 2451–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Augustin, A., Spenlehauer, C., Dumond, H., Menissier-De Murcia, J., Piel, M., Schmit, A. C., Apiou, F., Vonesch, J. L., Kock, M., Bornens, M. & De Murcia, G. (2003) J. Cell Sci. 116, 1551–1562. [DOI] [PubMed] [Google Scholar]
- 7.Menissier de Murcia, J., Ricoul, M., Tartier, L., Niedergang, C., Huber, A., Dantzer, F., Schreiber, V., Ame, J. C., Dierich, A., LeMeur, M., et al. (2003) EMBO J. 22, 2255–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uchida, K., Hanai, S., Ishikawa, K., Ozawa, Y., Uchida, M., Sugimura, T. & Miwa, M. (1993) Proc. Natl. Acad. Sci. USA 90, 3481–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith, S., Giriat, I., Schmitt, A. & de Lange, T. (1998) Science 282, 1484–1487. [DOI] [PubMed] [Google Scholar]
- 10.Miwa, M. & Sugimura, T. (1971) J. Biol. Chem. 246, 6362–6364. [PubMed] [Google Scholar]
- 11.Juarez-Salinas, H., Sims, J. L. & Jacobson, M. K. (1979) Nature 282, 740–741. [DOI] [PubMed] [Google Scholar]
- 12.Davidovic, L., Vodenicharov, M., Affar, E. B. & Poirier, G. G. (2001) Exp. Cell Res. 268, 7–13. [DOI] [PubMed] [Google Scholar]
- 13.Lin, W., Ame, J. C., Aboul Ela, N., Jacobson, E. L. & Jacobson, M. K. (1997) J. Biol. Chem. 272, 11895–11901. [DOI] [PubMed] [Google Scholar]
- 14.Ohashi, S., Kanai, M., Hanai, S., Uchiumi, F., Maruta, H., Tanuma, S. & Miwa, M. (2003) Biochem. Biophys. Res. Commun. 307, 915–921. [DOI] [PubMed] [Google Scholar]
- 15.Panda, S., Poirier, G. G. & Kay, S. A. (2002) Dev. Cell 3, 51–61. [DOI] [PubMed] [Google Scholar]
- 16.Rubin, G. M. & Spradling, A. C. (1982) Science 218, 348–353. [DOI] [PubMed] [Google Scholar]
- 17.Hanai, S., Uchida, M., Kobayashi, S., Miwa, M. & Uchida, K. (1998) J. Biol. Chem. 273, 11881–11886. [DOI] [PubMed] [Google Scholar]
- 18.Endo, K., Akiyama, T., Kobayashi, S. & Okada, M. (1996) Mol. Gen. Genet. 253, 157–165. [DOI] [PubMed] [Google Scholar]
- 19.Kawamitsu, H., Hoshino, H., Okada, H., Miwa, M., Momoi, H. & Sugimura, T. (1984) Biochemistry 23, 3771–3777. [DOI] [PubMed] [Google Scholar]
- 20.Kawamoto, T. & Shimizu, M. (2000) Histochem. Cell Biol. 113, 331–339. [DOI] [PubMed] [Google Scholar]
- 21.Tulin, A., Stewart, D. & Spradling, A. C. (2002) Genes Dev. 16, 2108–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menegazzi, M., Grassi-Zucconi, G., Carcerero De Prati, A., Ogura, T., Poltronieri, P., Nyunoya, H., Shiratori-Nyunoya, Y., Miwa, M. & Suzuki, H. (1991) Exp. Cell Res. 197, 66–74. [DOI] [PubMed] [Google Scholar]
- 23.Ogura, T., Takenouchi, N., Yamaguchi, M., Matsukage, A., Sugimura, T. & Esumi, H. (1990) Biochem. Biophys. Res. Commun. 172, 377–384. [DOI] [PubMed] [Google Scholar]
- 24.Homburg, S., Visochek, L., Moran, N., Dantzer, F., Priel, E., Asculai, E., Schwartz, D., Rotter, V., Dekel, N. & Cohen-Armon, M. (2000) J. Cell Biol. 150, 293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Futai, M., Mizuno, D. & Sugimura, T. (1967) Biochem. Biophys. Res. Commun. 28, 395–399. [DOI] [PubMed] [Google Scholar]
- 26.Miwa, M., Tanaka, M., Shinshi, H., Takeuchi, M., Matsushima, T., Sugimura, T. & Shall, S. (1975) J. Biochem. (Tokyo) 77, 3p–4p. [PubMed] [Google Scholar]
- 27.Shinshi, H., Miwa, M., Kato, K., Noguchi, M., Matsushima, T. & Sugimura, T. (1976) Biochemistry 15, 2185–2190. [DOI] [PubMed] [Google Scholar]
- 28.Taylor, J. P., Hardy, J. & Fischbeck, K. H. (2002) Science 296, 1991–1995. [DOI] [PubMed] [Google Scholar]
- 29.Ying, W., Sevigny, M. B., Chen, Y. & Swanson, R. A. (2001) Proc. Natl. Acad. Sci. USA 98, 12227–12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tulin, A. & Spradling, A. (2003) Science 299, 560–562. [DOI] [PubMed] [Google Scholar]
- 31.Bahr, B. A. & Bendiske, J. (2002) J. Neurochem. 83, 481–489. [DOI] [PubMed] [Google Scholar]
- 32.Williams, J. C., Butler, I. J., Rosenberg, H. S., Verani, R., Scott, C. I. & Conley, S. B. (1984) N. Engl. J. Med. 311, 152–155. [DOI] [PubMed] [Google Scholar]
- 33.Endres, M., Wang, Z. Q., Namura, S., Waeber, C. & Moskowitz, M. A. (1997) J. Cereb. Blood Flow Metab. 17, 1143–1151. [DOI] [PubMed] [Google Scholar]
- 34.Eliasson, M. J., Sampei, K., Mandir, A. S., Hurn, P. D., Traystman, R. J., Bao, J., Pieper, A., Wang, Z. Q., Dawson, T. M., Snyder, S. H. & Dawson, V. L. (1997) Nat. Med. 3, 1089–1095. [DOI] [PubMed] [Google Scholar]
- 35.Virag, L. & Szabo, C. (2002) Pharmacol. Rev. 54, 375–429. [DOI] [PubMed] [Google Scholar]
- 36.Yu, S. W., Wang, H., Poitras, M. F., Coombs, C., Bowers, W. J., Federoff, H. J., Poirier, G. G., Dawson, T. M. & Dawson, V. L. (2002) Science 297, 259–263. [DOI] [PubMed] [Google Scholar]