Abstract
Smad proteins play pivotal roles in mediating the transforming growth factor β (TGF-β) transcriptional responses. We show in this report that PIAS3, a member of the protein inhibitor of activated STAT (PIAS) family, activates TGF-β/Smad transcriptional responses. PIAS3 interacts with Smad proteins, most strongly with Smad3. PIAS3 and Smad3 interact with each other at the endogenous protein level in mammalian cells and also in vitro, and the association occurs through the C-terminal domain of Smad3. We further show that PIAS3 can interact with the general coactivators p300/CBP, the first evidence that a PIAS protein can associate with p300/CBP. In contrast, PIASy, which inhibits Smad transcriptional activity and other transcriptional responses, is unable to interact with p300/CBP. The RING domain of PIAS3 is essential for interaction with p300/CBP, and a RING domain mutant PIAS3, which cannot bind p300/CBP, no longer activates TGF-β/Smad-dependent transcription. Furthermore, we show that PIAS3, Smad3, and p300 can form a ternary complex, which is markedly increased by TGF-β treatment. Taken together, our studies indicate that on TGF-β treatment, PIAS3 can form a complex with Smads and p300/CBP and activate Smad transcriptional activity.
Transforming growth factor β (TGF-β) regulates a wide variety of biological activities (1–5). Smad proteins can transduce the TGF-β signal at the cell surface into gene regulation in the nucleus (2–5). Smad2 and Smad3 are phosphorylated by the activated TGF-β receptor, form complexes with Smad4, and together accumulate in the nucleus to regulate transcription of target genes that play important roles in diverse cellular processes (2–5).
Smads can activate transcription by recruiting transcriptional coactivators (3–5). Transcriptional activation by Smad3 and Smad2 occurs, at least in part, by their ability to recruit general transcriptional coactivators p300/CBP (3–11). p300/CBP have intrinsic histone acetyltransferase (HAT) activity, which facilitates transcription by altering nucleosome structure through histone acetylation and thereby remodeling the chromatin template (12, 13). The C-terminal domains of Smad3 or Smad2 are necessary for the interaction with p300/CBP (6–11). In addition, P/CAF, another HAT-containing transcriptional coactivator, has been shown to associate with Smad3 on TGF-β receptor activation and to enhance TGF-β/Smad3 signaling (14). Smad4 plays an essential role in Smad-mediated transcriptional activation (2–5). This is partly due to the unique Smad activation domain (SAD), a 48-aa proline-rich regulatory element in the linker region of Smad4 (15). The SAD domain physically interacts with p300/CBP (15). In addition, the Smad4 interacting protein MSG1, which lacks intrinsic DNA binding, can recruit p300/CBP to Smad4 via SAD and function as a coactivator of Smad4 (16). The SAD domain also recruits SMIF, a Smad4 coactivator that is essential for Smad4 transcriptional activity (17). Another Smad coactivator is ARC105, which interacts with Smad2, Smad3, and Smad4 (18). In addition to activation of transcription, Smads can also repress transcription through recruitment of corepressors (4, 5).
PIAS family members were initially identified through interaction with STAT proteins (19), and they regulate the activities of many transcription factors through distinct mechanisms. PIAS1 and PIAS3 bind and inhibit STAT1 and STAT3 DNA-binding activities, respectively (19, 20). PIASxα and PIASxβ were identified through interactions with the androgen receptor and the homeodomain protein Msx2, respectively (21, 22). PIASxα and PIASxβ inhibit IL12-mediated and STAT4-dependent gene activation (23). PIAS1, PIAS3, PIASxα, and PIASxβ also regulate transcriptional activation by various steroid receptors (21, 24–26). PIASy has been shown to antagonize the activities of STAT1 (27), androgen receptor (28), p53 (29), LEF1 (30), Smads (31, 32), and Nurr1 (33). All PIAS family members possess E3 ligase activity for SUMO (small ubiquitin-related modifier), and the RING domain of the PIAS proteins is essential for this activity (30, 34–41).
Recently, in an effort to search for new regulators for TGF-β signaling pathways by a yeast three-hybrid screen, we identified PIASy (31). We have shown that PIASy can inhibit Smadmediated transcriptional responses by interacting with Smads and histone deacetylase (HDAC) (31). We continued in this study to test the effect of other PIAS family members on TGF-β/Smad signaling and found that PIAS3 can activate Smad-dependent transcription. We provide evidence that PIAS3 activates Smad transcriptional activity through its interaction with Smads and p300/CBP.
Materials and Methods
Plasmids. Flag-tagged various PIAS plasmids have been described (19, 20, 23, 27). Myc-tagged short and long forms of PIAS3 were constructed in the CS3+-6Myc vector. The Myc-tagged deletion mutants of PIAS3 were constructed by subcloning or PCR from deletion mutants of PIAS3 in a yeast two-hybrid system (41) into the CS3+-6Myc vector. CS2-PIAS3 was constructed by cloning the short form of PIAS3 cDNA in the CS2 vector. Flag- or Myc-tagged mouse PIAS3 RING domain mutants (C299S, H301A) were constructed by PCR and verified by sequencing. p300-HA and CBP-HA have been described (7).
Immunoprecipitation, Immunoblot, and Detection of Ternary Complex. COS or 293T cells were transfected by Lipofectamine plus reagent. For interaction analysis between Smads and PIAS3, cells were lysed in TNE buffer (10 mM Tris·HCl, pH 7.8/150 mM NaCl/1 mM EDTA/1.0% Nonidet P-40). To detect interaction between PIAS3 and p300/CBP, cells were lysed in TNM buffer (20 mM Tris·HCl, pH 8.0/150 mM NaCl/3 mM MgCl2/0.5% Nonidet P-40), and the immunoprecipitates were washed five times each with 1 ml of buffer for 10–15 min. To detect PIAS3–Smad3–p300 ternary complex, transfected 293T cells were lysed in the TNMG buffer (50 mM Tris·HCl, pH 8.0/50 mM NaCl/5 mM MgCl2/10% glycerol/0.5% Nonidet P-40). All other procedures for immunoprecipitation and immunoblot were performed as described (42).
GST Pull-Down Assay. 35S-labeled PIAS3 was synthesized by in vitro translation from the CS2-PIAS3 by using the SP6 TNT coupled transcription/translation system (Promega). Other procedures were performed as described (31).
Luciferase Reporter Gene Assay. HaCaT cells in 60-mm dishes were transfected by DEAE-dextran and treated with or without 200 pM TGF-β for 18–24 h as described (43). Luciferase activities were normalized with cotransfected Renilla luciferase control. Results represent at least three independent transfections.
Results
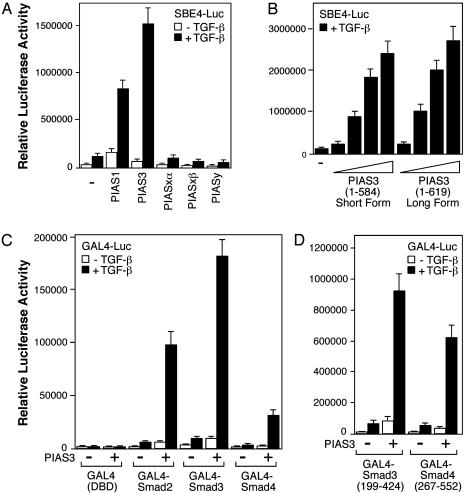
PIAS3 Can Activate Smad Transcriptional Activity. To analyze the effects of various PIAS proteins on TGF-β/Smad-dependent transcription, HaCaT keratinocytes were cotransfected with a PIAS expression plasmid along with the SBE4-Luc, a TGF-β responsive reporter gene that contains four copies of the Smadbinding element (SBE) (44). As shown in Fig. 1A, PIAS3, and to a lesser extent PIAS1, activated the TGF-β induction of the reporter gene. PIASxα had little effect, whereas PIASxβ and PIASy inhibited the reporter gene to different extents (Fig. 1 A). Thus, different PIAS family members have distinct effects on TGF-β/Smad mediated transcription.
Fig. 1.
PIAS3 can activate Smad transcriptional activity. (A) Different PIAS proteins have distinct effects on the SBE4-Luc reporter gene activity. HaCaT cells were cotransfected with the SBE4-Luc reporter gene and a Flag-PIAS plasmid as indicated. Cells were treated with or without TGF-β and analyzed for luciferase activity. (B) The short and long forms of PIAS3 have comparable activity to stimulate the SBE4-Luc reporter gene. (C) PIAS3 can activate the transcriptional activity of GAL4-Smad fusion proteins. HaCaT cells were cotransfected with the GAL4-Luc reporter gene along with the GAL4 DNA-binding domain (DBD), GAL4-Smad2, GAL4-Smad3, or GAL4-Smad4 in the absence or presence of Flag-PIAS3 plasmid, treated with or without TGF-β, and analyzed for luciferase activity. (D) The stimulatory effect of PIAS3 occurs through the transcriptional activation domains of Smad proteins.
There are two forms of PIAS3 proteins. The short form is 584 aa long, and the long form contains 619 aa. These two PIAS3 proteins differ by an internal 35 aa, which are present only in the long form of PIAS3. As shown in Fig. 1B, the two PIAS3 proteins have comparable capacity to activate the SBE4-Luc reporter gene.
To determine whether PIAS3 can directly activate Smad transcriptional activity, we used a GAL4 fusion assay by fusing Smad2, Smad3, and Smad4 to the GAL4 DNA-binding domain. As shown in Fig. 1C, PIAS3 greatly elevated the TGF-β-induced transcriptional activity of GAL4-Smad fusion proteins. Thus, PIAS3 activation of Smad-mediated transcriptional responses can occur directly by stimulation of Smad transcriptional activity.
PIAS family proteins possess SUMO E3 ligase activities, and Smad4 has been shown to be sumoylated at two lysines: K159 in the linker region and K113 the N-terminal domain (refs. 40, 45, and 46 and our unpublished results). To analyze whether sumoylation of Smad4 is involved in PIAS3-mediated activation, we used GAL4-Smad4 (267–552), which contains the SAD domain and the C-terminal domain but has no sumoylation sites of Smad4. As shown in Fig. 1D, PIAS3 can still markedly increase the activity of GAL4-Smad4 (267–552). Apparently, sumoylation of Smad4 is not required in PIAS3-mediated activation. Moreover, GAL4-Smad3 (199–424) and GAL4-Smad2 (231–424), both of which contain the C-terminal domain and part of the linker region, are also activated by PIAS3 (Fig. 1D and data not shown). Taken together, these observations indicate that the transcriptional activation domains of Smad proteins are necessary for the stimulatory effect of PIAS3.
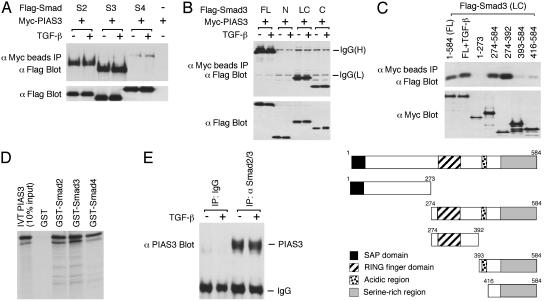
PIAS3 Interacts with Smad3 and Other Smads in Vivo and in Vitro. To examine whether PIAS3 can interact with Smads, Flag epitope-tagged Smad2, Smad3, or Smad4 and Myc-tagged mouse PIAS3 were cotransfected into COS cells. Cells were then subjected to immunoprecipitation with Myc antibody conjugated agarose beads followed by immunoblot with Flag antibody. As shown in Fig. 2A, PIAS3 interacted strongly with Smad3 and to a lesser extent with Smad2. PIAS3 also weakly associated with Smad4. TGF-β treatment had little effect on these interactions. Our subsequent analysis was mostly focused on Smad3–PIAS3 interaction. As shown in Fig. 2B, their interaction is mediated by the C-terminal domain of Smad3, which is necessary for transcriptional activation (2–5).
Fig. 2.
Interaction of PIAS3 with Smad3 and other Smad proteins. (A) Analysis of PIAS3 interaction with Smad2, Smad3, and Smad4 by immunoprecipitation (IP)-immunoblot assay in COS cells. TβRI (T204D) was cotransfected for TGF-β stimulation. Lysate controls are shown for expression levels of Smads. (B) PIAS3 interacts with the C-terminal domain of Smad3. FL, full length; N, N-terminal domain; LC, linker and C-terminal domain; C, C-terminal domain. (C) The RING domain of PIAS3 is involved in the interaction with Smad3. (Upper) Different domains of PIAS3 were cotransfected with Smad3 (199–424) into COS cells and analyzed as indicated. (Lower) The structures of the various PIAS3 deletion mutants. (D) 35S-labeled PIAS3 synthesized by in vitro translation (IVT) binds to GST-Smad proteins. (E) Endogenous Smad3 and Smad2 interact with PIAS3. HaCaT cells were treated with or without 200 pM TGF-β for 1 h. Cell lysates were immunoprecipitated with a goat antibody that specifically recognizes Smad3 and Smad2, followed by immunoblot with a rabbit PIAS3 antibody. The immunoblot was exposed for half an hour in the enhanced chemiluminescence (ECL, Amersham Biosciences) detection assay to obtain a strong signal for the interaction.
We also determined which domain of PIAS3 interacted with Smad3. PIAS3 has four structural features. The N-terminal SAP (SAF-A/B, Acinus, and PIAS) domain can mediate binding to DNA sequences that are present in nuclear matrix attachment regions (30, 47). The central RING domain is necessary for its SUMO E3 ligase activity. The acidic region in the C-terminal part is essential for interaction with certain proteins including the IFN regulatory factor 1 (IRF-1) (41) and TIF-2, a coactivator for nuclear receptors (48). The function of the C-terminal serine-rich region is unknown. To identify the Smad3-interacting domain in PIAS3, Myc-tagged deletion mutants of PIAS3 were cotransfected into COS cells along with Flag-tagged Smad3 (199–424), which contains the C-terminal domain and part of the linker region. The cell lysates were immunoprecipitated with Myc antibody-conjugated beads, followed by immunoblot with Flag antibody. As shown in Fig. 2C, full-length PIAS3, and two deletion mutants 274–584 and 274–392 that contain the RING domain, associated with Smad3. TGF-β treatment had almost no effect on the interactions with all these deletion mutants (data not shown). The N-terminal SAP domain and the C-terminal acidic region and serine-rich region had modest or no activity in binding to Smad3. Thus, the RING domain of PIAS3 is important for interaction with Smad3.
To determine whether PIAS3 can bind to Smad proteins in vitro, 35S-labeled PIAS3 synthesized by in vitro translation was incubated with GST or GST-Smad proteins, including Smad2, Smad3, and Smad4. As shown in Fig. 2D, PIAS3 binds to all three GST-Smad proteins, and the strongest interaction is with GST-Smad3. Interestingly, PIAS3 also binds well to GST-Smad4 in this assay, in contrast to the weak binding to Smad4 in vivo. It is possible that certain inhibitory factors or modifications may prevent Smad4 from binding to PIAS3 in vivo. It is also possible that the detected weak interaction between Smad4 and PIAS3 in vivo is mediated through Smad3 and Smad2.
We also analyzed whether endogenous Smad3 and Smad2 can interact with PIAS3. HaCaT cells were treated with or without TGF-β. Cell lysates were immunoprecipitated with a goat antibody that specifically recognizes Smad3 and Smad2 (Santa Cruz Biotechnology), followed by immunoblot with a rabbit highaffinity PIAS3 antibody (Santa Cruz Biotechnology). Two major bands were detected. One band migrated at the expected PIAS3 size. The other band was the IgG band. Because the rabbit PIAS3 antibody had only minimal cross-reactivity toward the goat IgG band, the IgG signal was not overwhelming even though the immunoblot was exposed for half an hour by chemiluminescence detection. As shown in Fig. 2E, endogenous PIAS3 and Smad3/2 interacted with each other at basal state, and TGF-β treatment had little effect on the interaction. The expression levels of Smad3, Smad2, and PIAS3 were not affected by TGF-β treatment (data not shown).
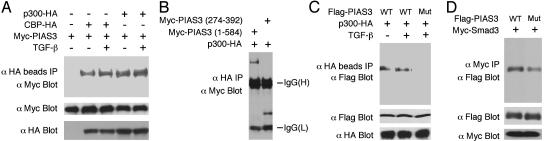
PIAS3 Can Interact with the General Coactivators p300/CBP. We then investigated the mechanism by which PIAS3 stimulates Smad transcriptional activity. To determine whether PIAS3 can activate Smad-dependent transcription by binding to general transcriptional coactivators p300/CBP, we first asked whether PIAS3 and p300/CBP can interact with each other. Myc-PIAS3 and p300-HA or CBP-HA were cotransfected into 293T cells and treated with or without TGF-β. As shown in Fig. 3A, PIAS3 can interact with p300 and CBP, and TGF-β treatment has little effect on their interactions. The PIAS3 deletion mutant 274–392, which contains the RING domain, can bind to p300 on its own (Fig. 3B).
Fig. 3.
The RING domain of PIAS3 is essential for interaction with p300/CBP. (A) PIAS3 can interact with p300/CBP. Myc-PIAS3 along with p300-HA or CBP-HA and TβRI (T204D) for TGF-β stimulation was cotransfected into 293T cells. (B) The RING domain of PIAS3 can interact with p300. (C) The RING domain mutant PIAS3 cannot bind to p300. COS cells were cotransfected with p300-HA along with Flag-tagged wild-type PIAS3 or RING mutant PIAS3 (C299S, H301A) in the absence or presence of TβRI (T204D) for TGF-β stimulation. (D) The RING domain mutant PIAS3 has a reduced ability to bind Smad3.
To further determine the role of the PIAS3 RING domain for interaction with p300, we generated a mutant PIAS3 that changed the cysteine 299 to serine and histidine 301 to alanine, which is predicted to disrupt the ability of PIAS3 to form a functional RING domain. The PIAS3 (C299S, H301A) mutant was then analyzed for its ability to interact with p300. The RING domain mutant PIAS3 expresses less protein than the wild-type PIAS3 when the same amount of DNA was transfected. However, by introduction of more DNA, the same or even higher amounts of RING domain mutant PIAS3 protein can be produced. As shown in Fig. 3C, mutation of the RING domain essentially eliminated the ability of PIAS3 to interact with p300, even though more RING domain mutant PIAS3 DNA was used to generate the same protein level as the wild-type PIAS3. Similar results were also obtained for CBP (data not shown). We also examined whether this RING domain mutant can still interact with Smad3. As shown in Fig. 3D, binding of Smad3 by the RING domain mutant was reduced but not abolished. Taken together, these observations suggest that the RING domain of PIAS3 is essential for interaction with p300/CBP.
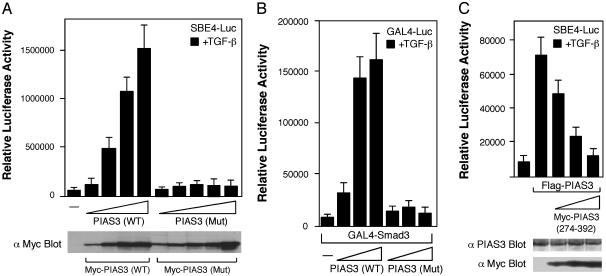
We then asked whether the RING domain mutant is able to activate Smad-dependent transcription. As shown in Fig. 4A, wild-type PIAS3 increased SBE4-luc transcription in a dose-dependent manner, whereas the RING domain mutant PIAS3 essentially lost the ability to activate SBE4-Luc transcription. The wild-type PIAS3 and the RING domain mutant PIAS3 were expressed at comparable levels in these experiments, as shown in a representative immunoblot in Fig. 4A Lower. In addition, the RING domain mutant PIAS3 also failed to activate the transcriptional activity of GAL4-Smad3 (Fig. 4B).
Fig. 4.
The RING domain mutant PIAS3 cannot activate Smad-dependent transcription. (A) RING domain mutant PIAS3 cannot activate SBE4-luc reporter gene. (Upper) HaCaT cells were cotransfected with the SBE4-Luc reporter gene and increasing amount of wild type or RING mutant (C299S, H301A) of Myc-PIAS3 as indicated. (Lower) A representative immunoblot with the Myc antibody using the transfected extracts from Upper. Higher amount of RING domain mutant PIAS3 plasmids were used to achieve comparable expression as the wild-type PIAS3. (B) RING domain mutant PIAS3 cannot activate GAL4-Smad3 transcriptional activity. HaCaT cells were cotransfected with the GAL4-Luc reporter gene, GAL4-Smad3, and increasing dose of wild type or RING mutant (C299S, H301A) of PIAS3 as indicated. Higher doses of RING domain mutant PIAS3 plasmids were used to obtain comparable expression levels as the wild-type PIAS3. (C) The PIAS3 RING-like domain (amino acids 274–392) functions as a dominant-negative mutant to inhibit full-length PIAS3. (Upper) HaCaT cells were cotransfected with the SBE4-Luc reporter gene and the full-length Flag-PIAS3 along with increasing amount of Myc-PIAS3 (274–392) as indicated. (Lower) Representative immunoblots with the PIAS3 and the Myc antibodies.
Because the PIAS3 (274–392) that contains the RING domain is sufficient to bind to both p300 and Smad3, we analyzed whether it can act as a dominant-negative mutant to inhibit the activity of the full-length PIAS3 in the reporter gene assay. As shown in Fig. 4C, introduction of PIAS3 (274–392) significantly inhibited the activation function of PIAS3 in a dose-dependent manner. In the control experiment, the expression level of PIAS3 was only modestly reduced in the presence of the cotransfected PIAS3 (274–392) (Fig. 4C). Thus, the PIAS3 (274–392) can function as a dominant-negative mutant.
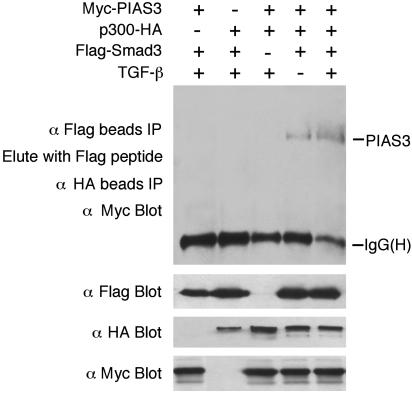
PIAS3, Smad3, and p300 Can Form a Ternary Complex, and TGF-β Treatment Increases This Complex Formation. The observations above prompted us to ask whether PIAS3, Smad3, and p300 can form a ternary complex. Myc-PIAS3, Flag-Smad3, and p300-HA were cotransfected into 293T cells and treated with or without TGF-β. Cell lysates were immunoprecipitated with Flag antibody coupled to agarose beads. Immuno complexes were eluted with the Flag peptide, and the eluate was used in a second immunoprecipitation with the HA antibody beads. Finally, the precipitate was analyzed by immunoblot with the Myc antibody. As shown in Fig. 5, only when the three components were coexpressed did we detect a specific ternary complex. Moreover, the ternary complex formation is significantly increased in the presence of TGF-β treatment, which may explain, at least in part, PIAS3 stimulation of Smad transcriptional activity in a TGF-β-dependent manner.
Fig. 5.
PIAS3, Smad3, and p300 can form a ternary complex. Myc-PIAS3, Flag-Smad3, p300-HA, and TβRI (T204D) for TGF-β stimulation were cotransfected into 293T cells. Ternary complex was detected by sequential immunoprecipitation with Flag and HA beads, followed by immunoblot with the Myc antibody.
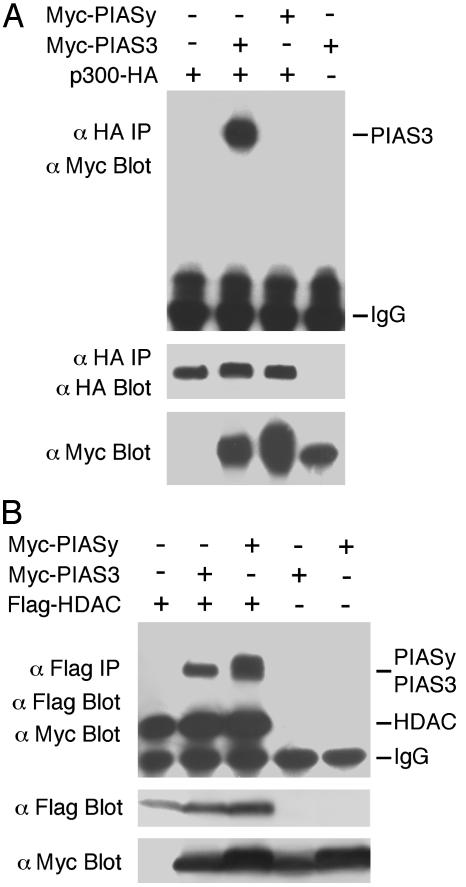
Comparison of the Abilities of PIAS3 and PIASy to Interact with p300 and HDAC1. The observations above suggest that PIAS3 interaction with p300 is important for its activation function. In a related study, we have shown that PIASy can inhibit Smad transcriptional activity through recruitment of HDAC1 (31). A natural question is whether PIAS3 only interacts with p300/CBP and PIASy only associates with HDAC. We therefore compared the abilities of PIAS3 and PIASy to interact with p300 and HDAC1 by using the coimmunoprecipitation-immunoblot assay. As shown in Fig. 6A, whereas PIAS3 and p300 interaction is readily detected, PIASy has no detectable binding to p300. In contrast, both PIAS3 and PIASy can bind to HDAC1 (Fig. 6B). The highly specific interaction of PIAS3 with p300 may correlate with its activation function as described in Discussion.
Fig. 6.
Analysis of PIAS3 and PIASy for interactions with p300 and HDAC1. (A) PIAS3 but not PIASy can interact with p300. Myc-PIAS3 or Myc-PIASy along with p300-HA was cotransfected into 293T cells. Cell lysates were immunoprecipitated with the HA antibody and immunoblotted with the Myc antibody. The part of the membrane containing p300-HA was immunoblotted with the HA antibody to detect the immunoprecipitated p300-HA. (B) Both PIAS3 and PIASy can interact with HDAC1. Myc-PIAS3 or Myc-PIASy along with Flag-HDAC1 was cotransfected into 293T cells. Cell lysates were immunoprecipitated with the Flag antibody, followed by immunoblot with the Flag and the Myc antibodies.
Discussion
We have shown in this report that PIAS3 can activate Smad-dependent transcription. PIAS3 and Smad3 interact with each other, and this interaction can be detected at the level of the endogenous proteins and also in vitro. We have also shown that PIAS3 can bind to the general coactivators p300/CBP. Interestingly, the RING domain of PIAS3, which is essential for the SUMO E3 ligase activity, is also important for interaction with p300/CBP and activation of Smad transcriptional activity. We have further shown that PIAS3, Smad3, and p300 can form a ternary complex, which is significantly increased by TGF-β treatment. Taken together, these results suggest that PIAS3 stimulates Smad transcriptional activity through formation of a complex with Smad proteins and p300/CBP.
Although the PIAS3–Smad3 association and the PIAS3–p300 interaction are little affected by TGF-β (Figs. 2 and 3), PIAS3–Smad3–p300 ternary complex formation is significantly increased in the presence of TGF-β. This most likely results from TGF-β-inducible interaction between Smad3 and p300 (6–8, 10, 11). Because the formation of the PIAS3–Smad3–p300 ternary complex is significantly increased in the presence of TGF-β, it may explain, at least in part, the strong stimulation of Smad transcriptional activity by PIAS3 in the presence of TGF-β.
PIAS proteins have sumoylation ligase activities. One natural question is whether sumoylation of certain targets is necessary for the stimulatory effect of PIAS3 on Smads. Smad4 has been shown to be sumoylated at two lysines (K159 and K113) in the linker region and the N-terminal domain (refs. 40, 45, and 46 and our unpublished results). Because PIAS3 is still able to activate the transcriptional activity of GAL4-Smad4 (267–552), which does not bear the two sumoylation sites, sumoylation of Smad4 is apparently not involved in the PIAS3-mediated activation. Although the RING domain of PIAS3 is necessary for p300 interaction, p300 may not be a substrate for sumoylation by PIAS3 in our system, because a recent study has shown that sumoylation of p300 correlates with transcriptional repression (49). Whether sumoylation of Smad3 or other targets is necessary for the stimulatory effect of PIAS3 on Smad transcriptional activity remains to be determined.
PIAS3 was initially identified through its inhibition of STAT3 DNA-binding activity (19). PIAS3 can also inhibit the DNA-binding activity of microphthalmia transcription factor (MITF) (50). We also analyzed whether PIAS3 can affect Smad DNA-binding activity. In some experiments we observed a modest increase of Smad DNA-binding activity by PIAS3 (data not shown). This effect, however, was not reproducible in all of the DNA-binding experiments we carried out. Although the reasons for these differences are not clear, we can firmly conclude that PIAS3 does not significantly affect Smad DNA-binding activity.
PIAS3 can use multiple modes to regulate transcription. In addition to exerting an inhibitory effect on STAT3 and MITF DNA-binding activity, PIAS3 can induce SUMO-1 modification of IFN regulatory factor 1 (IRF-1) and repress its transcriptional activity (41). PIAS3 can also coactivate or inhibit transcriptional responses mediated by various steroid receptors (24, 25). In a related study, PIAS3 is shown to modulate the ability of TIF2, a coactivator for nuclear receptors, to mediate ligand-induced transcription either positively or negatively (48). The mechanisms involved are not clear. It will be interesting to determine whether recruitment of p300 by PIAS3 is involved in the stimulatory effects on nuclear receptors.
PIAS proteins have distinct roles in regulation of Smad transcriptional activity. We have recently shown that PIASy can inhibit Smad transcriptional activity through interaction with Smads and HDAC (31). In this study, we have identified PIAS3 as an activator for Smad transcriptional activity. We have shown that PIAS3 but not PIASy interacts with p300, whereas both can interact with HDAC. This is similar to the two ZEB proteins that are highly related but have opposite functions (51, 52). ZEB-1/δEF1 binds and synergizes with Smad proteins to activate transcription, whereas ZEB-2/SIP1 also binds Smads but acts as a repressor (51, 52). Both ZEB-1 and ZEB-2 interact with the C-terminal-binding protein (CtBP) corepressor (52). However, only ZEB-1 interacts with p300 and P/CAF. Recruitment of p300 or P/CAF leads to the displacement of CtBP from ZEB-1 and enables it to activate transcription, whereas ZEB-2 represses transcription by binding to CtBP (52). It remains to be determined whether the displacement model is also used by PIAS3 for its activation function. In any case, the specific interaction of PIAS3 with p300 is likely to play an important role in PIAS3-mediated transcriptional activation.
Acknowledgments
We thank K. Shuai, H. Yokosawa, and R. Grosschedl for various PIAS constructs, X.-H. Feng and Y. Shi for p300 and CBP plasmids, R. Janknecht for Myc-tagged vector, J. Ding for assistance, and N. Denissova and many colleagues for discussions. This work was supported by the National Foundation for Cancer Research, the Emerald Foundation, a Burroughs Wellcome Fund New Investigator Award, a Kimmel Scholar Award from the Sidney Kimmel Foundation for Cancer Research, and the National Institutes of Health (all to F.L.).
Abbreviations: HDAC, histone deacetylase; PIAS, protein inhibitor of activated STAT; SBE, Smad-binding element; SUMO, small ubiquitin-related modifier; TGF-β, transforming growth factor β.
References
- 1.Roberts, A. B. & Sporn, M. B. (1990) in The Transforming Growth Factor-Betas, eds Sporn, M. B. & Roberts, A. B. (Springer, Heidelberg), pp. 419–472.
- 2.Heldin, C.-H., Miyazono, K. & ten Dijke, P. (1997) Nature 390, 465–471. [DOI] [PubMed] [Google Scholar]
- 3.Derynck, R., Zhang, Y. & Feng, X.-H. (1998) Cell 95, 737–740. [DOI] [PubMed] [Google Scholar]
- 4.Massagué, J. & Wotton, D. (2000) EMBO J. 19, 1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi, Y. & Massagué, J. (2003) Cell 113, 685–700. [DOI] [PubMed] [Google Scholar]
- 6.Janknecht, R., Wells, N. J. & Hunter, T. (1998) Genes Dev. 12, 2114–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng, X.-H., Zhang, Y., Wu, R. Y. & Derynck, R. (1998) Genes Dev. 12, 2153–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pouponnot, C., Jayaraman, L. & Massagué, J. (1998) J. Biol. Chem. 273, 22865–22868. [DOI] [PubMed] [Google Scholar]
- 9.Topper, J. N., DiChiara, M. R., Brown, J. D., Williams, A. J., Falb, D., Collins, T. & Gimbrone, M. A. J. (1998) Proc. Natl. Acad. Sci. USA 95, 9506–9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen, X., Hu, P. P., Liberati, N. T., Datto, M. B., Frederick, J. P. & Wang, X.-F. (1998) Mol. Biol. Cell 9, 3309–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishihara, A., Hanai, J. I., Okamoto, N., Yanagisawa, J., Kato, S., Miyazono, K. & Kawabata, M. (1998) Genes Cells 3, 613–623. [DOI] [PubMed] [Google Scholar]
- 12.Roth, S. Y., Denu, J. M. & Allis, C. D. (2001) Annu. Rev. Biochem. 70, 81–120. [DOI] [PubMed] [Google Scholar]
- 13.Vo, N. & Goodman, R. H. (2001) J. Biol. Chem. 276, 13505–13508. [DOI] [PubMed] [Google Scholar]
- 14.Itoh, S., Ericsson, J., Nishikawa, J.-I., Heldin, C.-H. & ten Dijke, P. (2000) Nucleic Acids Res. 28, 4291–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Caestecker, M. P., Yahata, T., Wang, D., Parks, W. T., Huang, S., Hill, C. S., Shioda, T., Roberts, A. B. & Lechleider, R. J. (2000) J. Biol. Chem. 275, 2115–2122. [DOI] [PubMed] [Google Scholar]
- 16.Yahata, T., de Caestecker, M. P., Lechleider, R. J., Andriole, S., Roberts, A. B., Isselbacher, K. J. & Shioda, T. (2000) J. Biol. Chem. 275, 8825–8834. [DOI] [PubMed] [Google Scholar]
- 17.Bai, R., Koester, C., Ouyang, T., Hahn, S. A., Hammerschmidt, M., Peschel, C. & Duyster, J. (2002) Nat. Cell Biol. 4, 181–190. [DOI] [PubMed] [Google Scholar]
- 18.Kato, Y., Habas, R., Katsuyama, Y., Naar, A. M. & He, X. (2002) Nature 418, 641–646. [DOI] [PubMed] [Google Scholar]
- 19.Chung, C. D., Liao, J., Liu, B., Rao, X., Jay, P., Berta, P. & Shuai, K. (1997) Science 278, 1803–1805. [DOI] [PubMed] [Google Scholar]
- 20.Liu, B., Liao, J., Rao, X., Kushner, S. A., Chung, C. D., Chang, D. D. & Shuai, K. (1998) Proc. Natl. Acad. Sci. USA 95, 10626–10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moilanen, A.-M., Karvonen, U., Poukka, H., Yan, W., Toppari, J., Janne, O. A. & Palvimo, J. J. (1999) J. Biol. Chem. 274, 3700–3704. [DOI] [PubMed] [Google Scholar]
- 22.Wu, L., Wu, H., Ma, L., Sangiorgi, F., Wu, N., Bell, J., Lyons, G. & Maxson, R. (1997) Mech. Dev. 65, 3–17. [DOI] [PubMed] [Google Scholar]
- 23.Arora, T., Liu, B., He, H., Kim, J., Murphy, T. L., Murphy, K. M., Modlin, R. L. & Shuai, K. (2003) J. Biol. Chem. 278, 21327–21330. [DOI] [PubMed] [Google Scholar]
- 24.Kotaja, N., Aittomaki, S., Silvennoinen, O., Palvimo, J. J. & Janne, O. A. (2000) Mol. Endocrinol. 14, 1986–2000. [DOI] [PubMed] [Google Scholar]
- 25.Junicho, A., Matsuda, T., Yamamoto, T., Kishi, H., Korkmaz, K., Saatcioglu, F., Fuse, H. & Muraguchi, A. (2000) Biochem. Biophys. Res. Commun. 278, 9–13. [DOI] [PubMed] [Google Scholar]
- 26.Nishida, T. & Yasuda, H. (2002) J. Biol. Chem. 277, 41311–41317. [DOI] [PubMed] [Google Scholar]
- 27.Liu, B., Gross, M., ten Hoeve, J. & Shuai, K. (2001) Proc. Natl. Acad. Sci. USA 98, 3203–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gross, M., Liu, B., Tan, J., French, F., Carey, M. & Shuai, K. (2001) Oncogene 20, 3880–3887. [DOI] [PubMed] [Google Scholar]
- 29.Nelson, V., Davis, G. E. & Maxwell, S. A. (2001) Apoptosis 6, 221–234. [DOI] [PubMed] [Google Scholar]
- 30.Sachdev, S., Bruhn, L., Sieber, H., Pichler, A., Melchior, F. & Grosschedl, R. (2001) Genes Dev. 15, 3088–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long, J., Matsuura, I., He, D., Wang, G., Shuai, K. & Liu, F. (2003) Proc. Natl. Acad. Sci. USA 100, 9791–9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imoto, S., Sugiyama, K., Sato, N., Yamamoto, T. & Matsuda, T. (2003) J. Biol. Chem. 278, 34253–34258. [DOI] [PubMed] [Google Scholar]
- 33.Galleguillos, D., Vecchiola, A., Fuentealba, J. A., Ojeda, V., Alvarez, K., Gomez, A. & Andres, M. E. (2003) J. Biol. Chem., in press. [DOI] [PubMed]
- 34.Jackson, P. K. (2001) Genes Dev. 15, 3053–3058. [DOI] [PubMed] [Google Scholar]
- 35.Johnson, E. S. & Gupta, A. A. (2001) Cell 106, 735–744. [DOI] [PubMed] [Google Scholar]
- 36.Kahyo, T., Nishida, T. & Yasuda, H. (2001) Mol. Cell 8, 713–718. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt, D. & Muller, S. (2002) Proc. Natl. Acad. Sci. USA 99, 2872–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kotaja, N., Karvonen, U., Janne, O. A. & Palvimo, J. J. (2002) Mol. Cell. Biol. 22, 5222–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin, X., Sun, B., Liang, M., Liang, Y. Y., Gast, A., Hildebrand, J., Brunicardi, F. C., Melchior, F. & Feng X.-H. (2003) Mol. Cell 11, 1389–1396. [DOI] [PubMed] [Google Scholar]
- 40.Lee, P. S., Chang, C., Liu, D. & Derynck, R. (2003) J. Biol. Chem. 278, 27853–27863. [DOI] [PubMed] [Google Scholar]
- 41.Nakagawa, K. & Yokosawa, H. (2002) FEBS Lett. 530, 204–208. [DOI] [PubMed] [Google Scholar]
- 42.Liu, F., Pouponnot, C. & Massagué, J. (1997) Genes Dev. 11, 3157–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Denissova, N. G., Pouponnot, C., Long, J., He, D. & Liu, F. (2000) Proc. Natl. Acad. Sci. USA 97, 6397–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zawel, L., Dai, J., Buckhaults, P., Zhou, S., Kinzler, K., Vogelstein, B. & Kern, S. (1998) Mol. Cell 1, 611–617. [DOI] [PubMed] [Google Scholar]
- 45.Lin, X., Liang, M., Liang, Y. Y., Brunicardi, F. C., Melchior, F. & Feng X.-H. (2003) J. Biol. Chem. 278, 18714–18719. [DOI] [PubMed] [Google Scholar]
- 46.Ohshima, T. & Shimotohno, K. (2003) J. Biol. Chem., in press. [DOI] [PubMed]
- 47.Kipp, M., Gohring, F., Ostendorp, T., van Drunen, C. M., van Driel, R., Przybylski, M. & Fackelmayer, F. O. (2000) Mol. Cell. Biol. 20, 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jimenez-Lara, A. M., Heine, M. J. & Gronemeyer, H. (2002) FEBS Lett. 526, 142–146. [DOI] [PubMed] [Google Scholar]
- 49.Girdwood, D., Bumpass, D., Vaughan, O. A., Thain, A., Anderson, L. A., Snowden, A. W., Garcia-Wilson, E., Perkins, N. D. & Hay, R. T. (2003) Mol. Cell 11, 1043–1054. [DOI] [PubMed] [Google Scholar]
- 50.Levy, C., Nechushtan, H. & Razin, E. (2002) J. Biol. Chem. 277, 1962–1966. [DOI] [PubMed] [Google Scholar]
- 51.Postigo, A. A. (2003) EMBO J. 22, 2443–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Postigo, A. A., Depp, J. L., Taylor, J. J. & Kroll, K. L. (2003) EMBO J. 22, 2453–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]