Abstract
We have used a genetic approach to generate eight different mutant human cell lines in which NF-κB is constitutively activated. These independent clones have different phenotypes and belong to several different genetic complementation groups. In one clone inhibitor of κB(IκB) kinase is constitutively active, but in the seven others it is not, despite the fact that IκB is degraded in all eight clones. Thus, IκB kinase-independent mechanisms of IκB degradation and NF-κB activation are predominant in these mutants. Biochemical analyses of the mutants revealed that they fall into at least five different categories, differing in the sets of upstream kinases that are activated, confirming multiple mechanisms of NF-κB activation. By introducing a retroviral cDNA library into the Ras C6 cell line, with constitutively active NF-κB, followed by selection for functional complementation, we isolated a cDNA encoding a C-terminal fragment of enolase 1 and identified it as negative regulator of NF-κB.
Keywords: forward genetics, complementation groups, functional complementation, enolase 1, ras
The term NF-κB refers to a family of dimeric proteins with related promoter binding and transactivation properties. The prototypic NF-κB complex is a p65-p50 heterodimer (1). The subunits p65/RelA, RelB, and c-Rel stimulate transcription whereas the subunits p50 and p52 serve primarily to bind to DNA (2). NF-κB, present in all cell types, is sequestered as a latent complex in the cytoplasm through its interaction with inhibitor of κB (IκB). In most untransformed cell types, NF-κB is largely cytoplasmic and therefore remains transcriptionally inactive until a cell receives an appropriate stimulus. Exposure of cells to a variety of stimuli leads to the rapid phosphorylation of the IκB proteins on specific serine residues by the multisubunit IκB kinase (IKK) complex (3–5), followed by the polyubiquitination of IκB on specific lysine residues and its degradation by the 26S proteasome. These events release NF-κB, allowing it to translocate to the nucleus and regulate transcription (1, 6–8). The ability of p65/RelA, RelB, and c-Rel to stimulate transcription also depends on stimulus-induced phosphorylation of these specialized subunits (9–18). Activation of phosphatidylinositol 3-kinase (PI3-K)/Akt initiates a pathway, quite separate from the one leading to IκB degradation, leading to the phosphorylation and activation of p65/RelA, and this pathway is required for full activation of NF-κB (15, 19, 20).
NF-κB is activated in response to IL-1, tumor necrosis factor (TNF) and many other stimuli: UV or γ-irradiation, double-stranded RNA, phorbol esters, reactive oxygen species, lipopolysaccharide (ref. 21 and references therein). Although the mechanism of NF-κB activation described above applies to most of these activators, alternative or atypical mechanisms of activation have also been reported. Exposure of cells to UV leads to NF-κB activation by two entirely different mechanisms (22, 23). Early IκBα degradation (0.5–6 h) is not initiated by UV-induced DNA damage and does not require IKK. In the later response (15–20 h) IκB degradation and NF-κB liberation occur through the classical pathway as a result of proteolytic processing of IL-1 precursors and release of IL-1 (22). Reoxygenation of hypoxic Jurkat T cells results in the activation of NF-κB by yet another mechanism that involves the phosphorylation of IκB on tyrosine 42 (24). Tyrosine phosphorylation of IκB represents a proteolysis-independent activation mechanism. NF-κB is constitutively active in B lymphocytes. Doerre and Corley (25) have provided evidence that different B cell lines use different strategies to activate NF-κB, depending on which receptor subtype is expressed.
Several reports indicate that NF-κB can be activated by various viral oncoproteins and by oncogenic forms of Ras (26–30) and that NF-κB activity is required for these oncoproteins to induce cellular transformation. Inhibition of NF-κB blocks cellular transformation by oncogenic Ras (26, 31). Activation of the p65 NF-κB subunit by oncogenic Ras or IL-1 is mediated by the serine/threonine kinase Akt via the PI3-K pathway (15, 19).
Over the past few years several proteins have been shown to negatively regulate the activation of NF-κB. The 60-kDa protein SODD (silencer of death domains) is preassociated with the intracellular domain of TNFα receptor 1 (TNF-R1) and DR3. TNF-induced aggregation of TNF-R1 disrupts the TNF-R1–SODD complex, after which SODD rapidly dissociates from the receptor (32). SODD seems to regulate tightly the activation of NF-κB in the absence of TNF. Other proteins have been implicated in the down-regulation of stimulus-induced NF-κB activation: apoptosis signal-regulating kinase 1 (ASK1) (33), PKC-interacting cousin of thioredoxin (PICOT) (34, †), rafkinase inhibitor protein (RKIP) (35, 36), and CARD-inhibitor of NF-κB-activating ligands (CARDINAL) (37).
NF-κB is well established as a regulator of a large number of genes encoding cytokines, cytokine receptors, and cell adhesion molecules that drive immune and inflammatory responses (7, 38, 39). NF-κB activation has been connected with multiple aspects of oncogenesis, including the control of apoptosis, cell cycle differentiation, and migration (29, 40). Constitutively high levels of nuclear NF-κB have been observed in many tumors, but the basis of constitutive activation is largely unknown. Further insight into how this transcription factor, central for many stress responses, is constrained is important in elucidating its role in cancer.
In the present study, we have used a genetic approach to identify negative regulators of NF-κB. We have previously reported the use of the HEK293 cell line stably expressing E-selectin-TK and E-selectin-zeocin constructs to isolate mutants defective in the IL-1-signaling pathway (41). Mutations were introduced into these cells by using ICR191, a frame-shift mutagen. Selection of mutagenized pools of cells in gancyclovir (GCV) for IL-1-unresponsive cells led to the isolation of mutant I1A, which has lost the expression of IL-1 receptor-associated kinase 1 (IRAK1) (41). Using the same mutagenized pools of 293 cells, we have now isolated mutant cell lines in which NF-κB is constitutively activated. These cells have several different phenotypes and belong to several different complementation groups. In a parallel study, parental HEK293 cells were stably transfected with oncogenic K-Ras, to generate a cell line (RasC6) that shows constitutive activation of NF-κB. Complementation of RasC6 cells has yielded a cDNA encoding a variant form of enolase 1, lacking ≈100 N-terminal residues.
Materials and Methods
Reagents and Cell Culture. Recombinant human IL-1β was provided by the National Cancer Institute. Recombinant human TNF was from Becton Dickinson. Polyclonal anti-IKKα, anti-IKKβ, anti-IKKγ, anti-p65/RelA, anti-p50, anti-IκBα, and anti-IκBβ were from Santa Cruz Biotechnology. Polyclonal anti-phospho-Akt, anti-phospho-p44/42 MAPK (mitogen-activated protein kinase) [extracellular signal-regulated kinase (ERK) 1/2], anti-Erk1/2, and anti-p90Rsk (Rsk, ribosomal S6 kinase) were from Cell Signaling Technology (Beverly, MA). Protein A-Sepharose and glutathione-agarose beads were from Pharmacia. Zeocin from Invitrogen was used at 50 μg/ml. GCV was obtained from the Cleveland Clinic Pharmacy. Cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FCS, penicillin G (100 μg/ml), and streptomycin (100 μg/ml).
Cell Line for Mutagenesis and Generation of Constitutive Mutants. The cell lines and protocols for mutagenesis have been described (41, 42). C6, C8, and C18 cells were subjected to five rounds of mutagenesis, to ≈50–70% lethality each time, with ICR191, an intercalating agent that induces frame-shift mutations (41). Several independent pools of mutagenized cells were generated. To obtain constitutive mutants, mutagenized pools were seeded onto several 150-mm plates. Each pool was subjected to selection in zeocin (50 μg/ml). Clones were picked and grown in nonselective medium. Each clone was expanded and tested for NF-κB activation by electrophoretic mobility-shift assay (EMSA).
Transfection and Reporter Assay. The NF-κB-dependent pE-selectin-luciferase reporter plasmid was a kind gift from P. DiCorleto (Cleveland Clinic Foundation). For the reporter gene assays, 2 × 106 cells, seeded on a 10-cm plate, were transiently cotransfected, by using 10 μg of pE-selectin-luciferase reporter plasmid and 1.0 μg of pSV2-βgal by the calcium phosphate method (43). After 24 h, the cells were harvested. The cells were either left untreated or stimulated with IL-1β or TNF for 4 h. Luciferase activities were determined with the Luciferase Assay System (Promega). The luciferase activity was normalized to the β-galactosidase activity to control for transfection efficiency.
Northern Transfers. Total RNA was isolated by using TRIzol reagent (GIBCO/BRL). RNA was fractionated by electrophoresis in a formaldehyde gel and transferred to Hybond-N, a positively charged nylon membrane according to the procedures provided by Amersham Pharmacia. Probes from IL-8 and GAPDH cDNAs were made by using the random priming kit from Amersham Pharmacia. Probe hybridization and washing were performed according to procedures provided by Amersham Pharmacia, and signals were visualized by autoradiography.
Gel Mobility and Supershift Assays. For EMSAs, where indicated, the cells were stimulated with IL-1β or TNF for 20 min. The NF-κB binding site (5′-GAGCAGAGGGAAATTCCGTAACTT-3′) from the IFN-inducible protein 10 (IP-10) gene was used as a probe (44). Complementary oligonucleotides, end-labeled with polynucleotide kinase and γ-32P-labeled ATP, were annealed by slow cooling. Approximately 20,000 cpm of probe were used per reaction mixture. Cytoplasmic extracts were prepared in binding reaction buffer as described (45, 46). The binding reaction was carried out at room temperature for 30 min in a total volume of 20 μl. The DNA–NF-κB complexes were separated on 5% polyacrylamide gels by electrophoresis in low ionic strength Tris-borate-EDTA buffer. For supershifts, ≈1 μg of polyclonal antibody against the NF-κB subunits p65/RelA, p50, or c-Rel was added to the binding reactions after 15 min, and the incubations were continued for 15 min more.
Immunoblotting. For immunoblotting, cells were washed once with PBS and lysed for 30 min at 4°C in RIPA buffer (1× PBS/1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS). Cellular debris was removed by centrifugation at 16,000 × g for 5 min. Cell extracts were fractionated directly by SDS/PAGE and transferred to poly(vinylidene difluoride) (PVDF) membranes. Immunoblot analysis was performed with the indicated primary antibodies, which were visualized with horseradish peroxidase-coupled goat anti-rabbit or anti-mouse immunoglobulins, by using the Enhanced Chemiluminescence (ECL) Western Blotting Detection System (Amersham Pharmacia).
Analysis of IKK Activity. IκBα expression plasmid (residues 1–54) was kindly provided by J. DiDonato (Cleveland Clinic Foundation). IκBα (amino acids 1–54) was expressed as a GST fusion protein in bacteria and purified after sonication at 4°C in 0.5% Nonidet P-40 lysis buffer as described (47). IKK assay was performed on whole cell lysates as described (5).
Generation of Retroviral Library, Infection, and RT-PCR Analysis of the Complemented Clones. A retroviral cDNA library from HaCaT cells was a kind gift from H. Lodish, Massachusetts Institute of Technology. The library was amplified by using Epicurion coli (SURE2) supercompetent cells from Stratagene. High titers of ecotropic virus (5 × 105/ml) were generated after the transient transfection of the BOSC 23 packaging cell line (48). After transfection with the cDNA library, the supernatant media containing the retrovirus were collected. Supernatant suspensions containing the recombinant retroviruses were incubated with the Ras C6 cells stably expressing the ecotropic receptor for 6 h in normal medium containing 4 μg/ml Polybrene (Sigma). Six hours after infection, the supernatant was removed, and the cells were cultured for 48 h more in normal medium. The infected cells were then selected in GCV (5 μg/ml). Selection medium was replaced at least every 5 days, and clones were isolated after ≈20 days.
Results
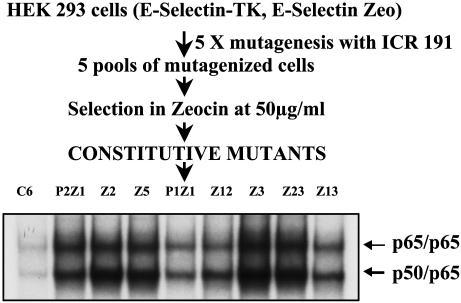
Isolation and Properties of Constitutive Mutants. The E-selectin promoter has a low basal activity and a high inducible activity. Five mutagenized pools of 293 cells stably expressing the E-selectin-zeocin and E-selectin-TK constructs were selected in zeocin (Fig. 1). Constitutive activation of NF-κB renders the cells resistant to zeocin and sensitive to GCV. Several zeocin-resistant clones were selected from each pool (Table 1). Because clones obtained from the same mutagenized pools could be siblings, only one or two clones from each pool were selected for further analysis. When two clones from the same pool were analyzed, their phenotypes made it clear that they were distinct. Each mutant clone was expanded and analyzed by drug selection for constitutive activation of NF-κB. As expected, all of the mutants were sensitive to GCV at a concentration between 1 μg/ml and 5 μg/ml and resistant to zeocin at a concentration of 50 μg/ml.
Fig. 1.
Scheme for generating constitutive mutants in human embryonic kidney (HEK) cells and EMSA of parental and eight mutant clones. Cell extracts were made from parental C6 and mutant cells. Extracts were analyzed by EMSA for the ability of NF-κB to bind to the NF-κB-consensus sequence of the IP-10 gene. The faster moving band is the p50/p65 heterodimer, and the slower moving band is the p65 homodimer.
Table 1. Constitutive mutants.
| Mutagenized pool | No. of constitutive mutants | Mutants selected for further analysis |
|---|---|---|
| C6P1 | 12 | C6P1Z1 and Z12 |
| C6P2 | 2 | C6P2Z1 and Z2 |
| C8P1 | 5 | Z3 and Z23 |
| C8P4 | 5 | Z5 |
| C18P3 | 1 | Z13 |
C6, C8, and C18 are three different initial clones. P1—P4 are independently mutagenized pools. Z1—Z23 are individual zeocin-resistant clones.
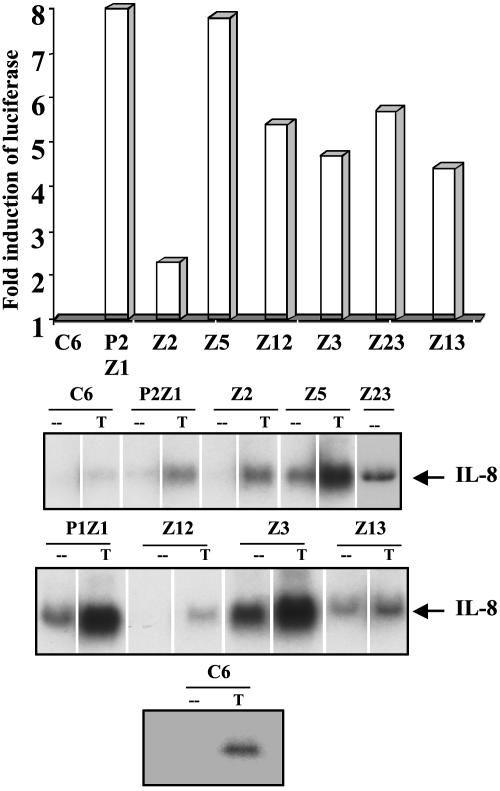
To confirm that NF-κB is activated in the mutant clones, the DNA binding activity of NF-κB in each mutant was tested by EMSA, using an oligonucleotide from the IP-10 promoter (44), which contains κB sites. All of the mutants show NF-κB–DNA binding (Fig. 1). A supershift assay using anti-p50 and anti-p65 antibodies confirmed that the faster migrating band is the p50-p65 heterodimer whereas the slower migrating band is the p65 homodimer (data not shown). An antibody to C-Rel did not cause any supershift (data not shown). To test whether the constitutively active NF-κB is capable of transactivation, the mutant cells were transfected transiently with an E-selectinluciferase construct, and the luciferase activity was monitored. The reporter is activated in all of the constitutive mutants (Fig. 2A) (data for P1Z1 not shown). Northern transfers of the constitutive mutants were assayed for IL-8 mRNA, which is up-regulated in response to cytokines such as IL-1 and TNF. As shown in Fig. 2B, the IL-8 gene is not up-regulated in untreated parental cells but is on treatment with TNF. The IL-8 message is elevated constitutively in all of the mutants except Z12, demonstrating that an endogenous NF-κB-dependent gene is up-regulated in the constitutive mutants. Furthermore, TNF-mediated IL-8 expression is elevated in some of the mutants (Fig. 2B).
Fig. 2.
The E-selectin-luciferase reporter gene and the endogenous IL-8 gene are constitutively activated in the mutant cells. (A) Reporter assay. Cells were transfected transiently with the NF-κB-dependent reporter plasmid pE-selectin-luciferase (10 μg) and pSV2-βgal (1 μg) to normalize for transfection efficiencies. The cells were lysed 48 h after transfection and assayed for luciferase activity. Luciferase activity was normalized to β-galactosidase. The fold increase relative to C6 parental cells is shown. (B) IL-8 mRNA expression is constitutively activated in some of the mutant cells. Cells were either left untreated (-) or treated with TNF (T) at 20 ng/ml for 4 h. The cells were washed and lysed, and an equal amount of total RNA was assayed for the expression of IL-8 mRNA by the Northern method. A different analysis of the C6 cell line (Bottom) confirms that there is no basal expression of IL-8 in these cells.
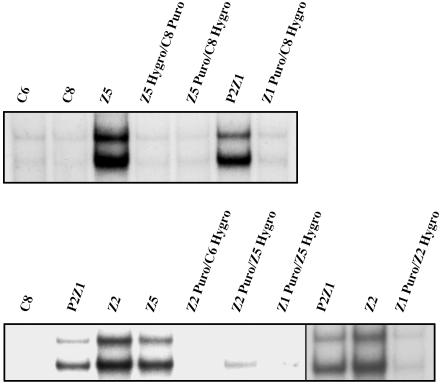
Dominance and Complementation. To assign each mutant to a complementation group, it is necessary to know whether the mutation is dominant or recessive. Parental cells containing either the puromycin or hygromycin drug selection markers were fused by using polyethylene glycol (PEG) to mutant cells carrying the reciprocal drug resistance marker. After fusion, the cells were selected in both the drugs to isolate heterokaryons, which were further selected in GCV and tested for NF-κB activation by EMSA (Fig. 3). Whereas mutants Z12 and Z23 show a partial dominance (data not shown), the rest of the mutants are recessive. Pairwise complementation analysis between mutants P2Z1, Z2, and Z5 was done by fusing a puromycin-resistant population of one mutant with a hygromycin-resistant population of the other. The heterokaryons were tested for constitutive NF-κB activation by EMSA (Fig. 3). Constitutive activation of NF-κB is greatly reduced or abolished in the heterokaryons, indicating that mutants C6P2Z1, Z2, and Z5 are in different complementation groups. The heterokaryons did not survive in zeocin, further supporting the EMSA results. The rest of the mutants (P1Z1, Z3, and Z13) have not been assigned to complementation groups.
Fig. 3.
Dominance and complementation. Each of the constitutive mutants tested was cotransfected independently with pBABE-Hygro and pBABE-Puro. The mutants P2Z1, Z2, and Z5 were fused either to parental C6 or C8 cells (dominance test) or to each other by using polyethylene glycol. The heterokaryons were selected in the presence of both hygromycin and puromycin. Pools of the clones resulting from each fusion were analyzed by EMSA by using an IP-10 probe.
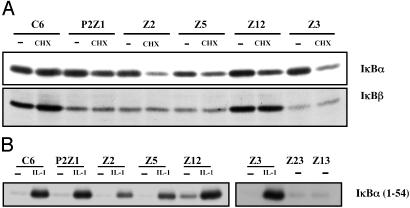
Biochemical Analysis of the Constitutive Mutants. To define the status of IκB proteins in the constitutive mutants, extracts of parental and mutant clones were assayed with IκBα,β antibodies. Because NF-κB up-regulates IκBα levels, the cells were treated with cycloheximide for 3 h before lysing them. There is constitutive degradation of IκBα in all of the mutants, and IκBβ is constitutively degraded in all except Z12 (Fig. 4A). Data for mutants P1Z1, Z13, and Z23 are not shown.
Fig. 4.
IκBα and IκBβ are constitutively degraded in most mutant cell lines whereas IKK is activated above basal levels only in mutant Z12. (A) Parental C6 and mutant cells were either untreated (-) or treated with cycloheximide (CHX) for 3 h. Cells were washed and lysed, and whole cell extracts were prepared. The amounts of protein in the extracts were quantitated, and equal amounts of the protein were analyzed by SDS/PAGE by using antibodies against IκBα. The same blot was probed with anti-IκBβ. The proteins were visualized by enhanced chemiluminescence. (B) IKK is activated above basal levels only in mutant Z12. Parental C6 and mutant cells were either untreated or treated with IL-1 for 15 min. The cells were washed and lysed, and the extracts were incubated with anti-IKKα antibody overnight at 4°C. The IKK complex was pulled down by using Protein A beads. The beads were washed, and a kinase assay was performed for 1 h in the presence of γ-labeled ATP and GST-tagged IκBα (residues 1–54) as a substrate. The beads were boiled in loading buffer, the proteins were separated by SDS/PAGE, and labeled IκBα was visualized by autoradiography.
A crucial step in NF-κB activation is the phosphorylation of IκB on serines 32 and 36. Stimulus-induced protein kinase activity specific to the N-terminal regulatory serines of IκBα and IκBβ was identified by several groups (3–5). The IKK complex comprises the IKKα and IKKβ catalytic subunits and the regulatory subunit IKKγ. An immune-complex kinase assay was done to test whether the IKK complex is activated in the constitutive mutants. With the exception of mutant Z12, IKK is not activated above the background level in parental cells in any of the constitutive mutants (Fig. 4B) (data for P1Z1 not shown).
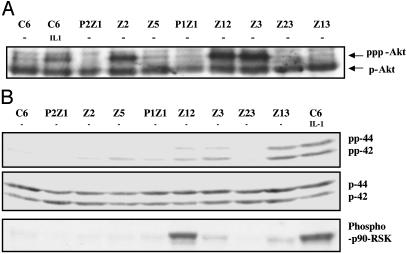
To test for the constitutive activation of other kinases that have been implicated in NF-κB-dependent signaling, Western transfers of cell extracts were probed with several phospho-specific antibodies. Akt, a serine-threonine kinase, is a downstream target for PI3-K and helps to activate NF-κB in response to stimuli such as TNF and IL-1 (15, 28). Parental C6 cells contain high levels of endogenous Akt (Fig. 5A). However, an additional, more slowly migrating band, probably a superphosphorylated or acetylated form of Akt that appears in IL-1-treated cells, was also seen in mutants Z2, Z12, and Z3, implying that the PI3-K/Akt pathway is constitutively activated in these three mutants (Fig. 5A).
Fig. 5.
Akt and p90rsk1 are activated in some constitutive mutants. (A) Akt is constitutively phosphorylated in mutants Z2, Z12, and Z3. Extracts of untreated and IL-1-treated (15 min) C6 cells and untreated mutant cells were analyzed by SDS/PAGE by using anti-phospho-Akt antibody. The faster migrating band corresponds to phosphorylated Akt (pAkt). The slower moving band in Z2, Z3, and Z12 lanes may correspond to a hyperphosphorylated or acetylated form of Akt. (B) The same extracts were analyzed by SDS/PAGE by using anti-phospho-p44/42 (Top), anti-p44/42 (Middle), and anti-phosphop90rsk1 (Bottom). The proteins were visualized by enhanced chemiluminescence. p90rsk1 and p44/42 are constitutively activated in mutants Z12, Z3, and Z13.
Mitogen-activated 90-kDa ribosomal S6 kinase (p90rsk1), a serine-threonine kinase, is involved in transducing signals induced by stimuli that activate Ras-MAPK cascades. It is phosphorylated and subsequently activated by the extracellularly regulated kinases p44erk1 and p42erk2. Schouten et al. (49) have reported that p90rsk1 can phosphorylate IκB on serine 32, both in vitro and in vivo. They showed that p90rsk1 is required for phorbol 12-myristate 13-acetate (PMA)-induced phosphorylation and subsequent degradation of IκBα. To determine whether p90rsk1 and the ERKs are activated in the constitutive mutants, cell extracts were separated by SDS/PAGE, and the Western transfers were probed with anti-p90rsk1 and anti-pErk (p42/44). Constitutive activation of these two kinases was seen in mutants Z12, Z3, and Z13 but not in parental C6 controls (Fig. 5B). The Western analyses for activated kinases, summarized in Table 2, indicate that the mutants can be divided into at least five different categories, differing in the sets of kinases that are activated (data for JNK not shown).
Table 2. Constitutive activation of kinases in different constitutive mutant clones.
| Mutant cell line | IKK | JNK | Akt | p90rsk1 | ERK |
|---|---|---|---|---|---|
| Z12 | ↑ | ↑ | ↑ | ↑ | ↑ |
| Z2 | — | — | ↑ | — | — |
| Z3 | — | — | ↑ | ↑ | ↑ |
| Z13 | — | — | — | ↑ | ↑ |
| P2Z1, P1Z1, Z5, Z23 | — | — | — | — | — |
A dash indicates that the kinase was not constitutively active. JNK, c-Jun N-terminal kinase.
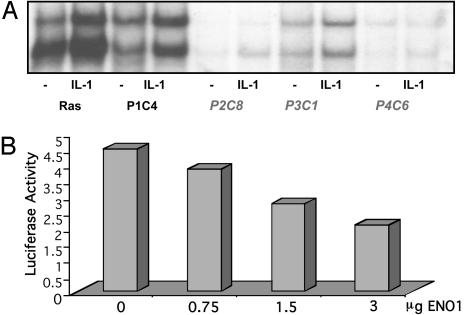
Phenotype and Complementation of Ras C6 Cells. C6 parental cells were stably transfected with pCGN-Kras12V, a plasmid encoding hygromycin resistance and hemagglutinin (HA)-tagged oncogenic K-Ras 12V, driven by the CMV promoter, and a new stable cell line (RasC6) was obtained by selection with hygromycin and zeocin. RasC6 cells express HA-tagged K-Ras12V, as confirmed by Western analysis (data not shown). They have constitutively activated NF-κB and therefore survive in zeocin and die in GCV. The constitutive activation of NF-κB in the RasC6 cell line was shown by EMSA and by Northern analysis of the expression of the endogenous NF-κB-dependent gene IL-8 (data not shown). RasC6 cells were infected with the HaCaT retroviral library and selected in GCV. Several clones were GCV-resistant and constitutive NF-κB activation was also repressed (Fig. 6A). The RT-PCR method was used to isolate complementing cDNAs, and three independent clones (P2C8, P3C1, and P4C6) were found to have been complemented with the same cDNA, which encodes a variant form of enolase 1, a truncated protein that lacks the amino terminal 100 aa. Truncated enolase 1 is already known to be a transcriptional repressor. It maps to a chromosomal breakpoint frequent in human cancers (50). Truncated enolase 1 shares 97% similarity with Myc-promoter binding protein-1 (MBP-1), which binds to and negatively regulates the c-myc P2 promoter. The complementing cDNA we have isolated codes for a truncated protein identical in sequence and length to the alternatively translated form of enolase 1 reported by Feo et al. (50). Structural analysis of α-enolase, the protein product of the enolase 1 gene, by Subramanian and Miller (51) has revealed that the N-terminal region of this protein is necessary for its function as a glycolytic enzyme but not for its ability to repress the c-myc P2 promoter. Truncated enolase 1 cDNA was subcloned into pCR 3.1 under control of the CMV promoter, and increasing amounts were transiently cotransfected with an NF-κB-dependent luciferase reporter into the RasC6 cell line. IL-1-induced activation was inhibited by increasing amounts of truncated enolase 1 (Fig. 6B). In a parallel control experiment, there was no effect of enolase 1 on the ability of IL-1 to activate an AP1-driven construct (data not shown).
Fig. 6.
Phenotype of complemented clones of RasC6 cells. (A) EMSA for NF-κB in RasC6 cells and enolase 1-complemented clones (P2C8, P3C1, and P4C6; no cDNA was recovered from P1C4), untreated or treated with IL-1 for 20 min. (B) Enolase 1 inhibits IL-1-stimulated NF-κB promoter activity in the RasC6 cell line. Cells were cotransfected transiently with increasing amounts of pCR 3.1 enolase 1 and with 1 μg of the NF-κB-dependent reporter E-selectinluciferase and pSV2-β-gal plasmid. Forty-eight hours posttransfection, cells were incubated with IL-1 for 4 h. Luciferase activity was normalized to β-galactosidase to correct for transfection efficiency.
Discussion
Using a dual drug selection strategy, we have isolated eight mutant cell lines in which NF-κB is constitutively active from pools of mutagenized HEK293 cells. As expected, each mutant is sensitive to GCV and resistant to puromycin. EMSAs revealed the presence of NF-κB p65-p65 homodimers and the p50-p65 heterodimers in each mutant. The ability of the mutants to transactivate the E-selectin-luciferase reporter indicates that the constitutively active NF-κB is transcriptionally competent. Furthermore, in all of the mutants except Z12, the endogenous IL-8 gene is up-regulated, indicating that the constitutively active NF-κB is capable of regulating its normal targets in the mutant cells. The level of IL-8 expression in the mutants is further induced on treatment with TNF, presumably due to a synergistic effect resulting from activation TNF-mediated NF-κB in addition to the basal activity of NF-κB. In mutant Z12, the IL-8 gene is not activated. The IL-8 promoter may require activation of transcription factors in addition to NF-κB and these factors may not be constitutively activated in mutant Z12. The resistance of Z12 cells to puromycin as well as the activation of the E-selectin promoter shows that NF-κB is functional in this cell line. There is no clear correlation between the extent of reporter activation and the level of IL-8 message in the mutant cells, probably because the activation of additional factors is required for IL-8 induction.
The mechanism of liberation of NF-κB in all of the mutants seems to be through the degradation of IκB. However, IκB may not always be degraded through the canonical pathway that is mediated by the IKK complex. As revealed by IKK immune-complex kinase assay, except for mutant Z12, NF-κB activation does not seem to be mediated by the IKKs. Although it is possible that a low level of IKK activation, not detected in the in vitro kinase assay, is enough to drive the pathway, it is more likely that IKK-independent mechanisms trigger the degradation of IκB and the release of NF-κB in seven of the eight mutants. There are several reports (22–24, 52) of alternative mechanisms or atypical pathways of NF-κB activation. In most of our mutants, IκBα and IκBβ are constitutively degraded without the apparent activation of IKK. This result is similar to the observation of Li and Karin (23) in cells exposed to short-wavelength UV radiation, which induced IκBα degradation via the 26S proteasome by a process that does not depend on the phosphorylation of Ser-32, Ser-36, or Tyr-42. The seven “atypical” constitutive mutants can be divided into four categories, based on the patterns of activation of upstream kinases (Table 2). All of these kinases have been shown to be involved in the stimulus-induced activation of NF-κB. However, at this point, we do not know whether the up-regulation of these kinases is a cause or effect of NF-κB activation in the constitutive mutants.
The constitutive activation of NF-κB in the mutant cell lines may be due to deregulated expression of activating cytokines or growth factors. Several studies in recent years have shown that autocrine production of cytokines confers constitutive NF-κB activity in colon, prostate, pancreatic, and other types of cancer (53). Our recent results reveal that the constitutive activation of NF-κB in most of the mutants is caused by deregulated expression of secretion of cytokines and other ligands, and some of these have been identified (54). Although the loss of cytoplasmic negative regulators such as SODD, CARD-inhibitor of NF-κB-activating ligands, apoptosis signal-regulating kinase 1, rafkinase inhibitor protein, PKC-interacting cousin of thioredoxin (PICOT), and phosphatase and tensin homolog (PTEN) might also result in the constitutive activation of NF-κB, we find that SODD, PICOT, and PTEN are not missing in our constitutive mutants.
Our interest in understanding the basis of constitutive activation of NF-κB stems from the fact that deregulated expression of NF-κB is a hallmark of many tumors (29, 40, 55, 56). By complementing the mutants with a cDNA library, we may identify novel genes that can function as negative regulators of the pathway. We introduced the HaCaT cDNA library into Ras C6 cells and isolated clones with a reverted phenotype. The cDNA cloned as a novel repressor of signaling from mutant Ras to NF-κB encodes a variant form of enolase 1. The complementing cDNA is virtually identical with that encoding Myc-promoter binding protein-1 (MBP-1), which is identical in sequence and length to the alternatively translated form of enolase 1 reported by Feo et al. (50). This variant is a transcriptional repressor and has been mapped to a chromosomal breakpoint frequently found in human cancers (50). Structural analysis of enolase 1 has revealed that the N-terminal portion of this protein is necessary for its function as a glycolytic enzyme but not for its ability to repress the c-myc P2 promoter (51). Our complementing enolase 1 cDNA encodes only the portion of enolase that is responsible for its ability to repress the c-myc P2 promoter. Much further work will be required to determine how general is the ability of truncated enolase 1 to inhibit NF-κB activation in different circumstances and to define its mechanism of action.
Despite repeated attempts, we were not successful in complementing several of the mutant cell lines, by using the HaCat cDNA library and others. We do not know why these experiments failed. The next step is attempting to complement mutant cell lines with constitutively active NF-κB by using insertional mutagenesis to mark physically the affected gene.
Acknowledgments
This work was supported by National Institutes of Health Grant P01CA62220 (to G.R.S.).
Abbreviations: EMSA, electrophoretic mobility-shift assay; ERK, extracellular signal-regulated kinase; GCV, gancyclovir; IκB, inhibitor of κB; IKK, IκB kinase; PI3-K, phosphatidylinositol 3′-kinase; RSK, ribosomal S6 kinase; SODD, silencer of death domains; TNFα, tumor necrosis factor α; IP-10, IFN-inducible protein 10.
Footnotes
Babichev, Y., Tamir, A., Kalifa, R., Witte, S., Altman, A. & Isakov, N. (2001) Keystone Symposium Abstract Book A201, p. 57.
References
- 1.Verma, I. M., Stevenson, J. K., Schwarz, E. M., Van Antwerp, D. & Miyamoto, S. (1995) Genes Dev. 9, 2723-2735. [DOI] [PubMed] [Google Scholar]
- 2.Liou, H. C. & Baltimore, D. (1993) Curr. Opin. Cell Biol. 5, 477-487. [DOI] [PubMed] [Google Scholar]
- 3.Regnier, C. H., Song, H. Y., Gao, X., Goeddel, D. V., Cao, Z. & Rothe, M. (1997) Cell 90, 373-383. [DOI] [PubMed] [Google Scholar]
- 4.DiDonato, J. A., Hayakawa, M., Rothwarf, D. M., Zandi, E. & Karin, M. (1997) Nature 388, 548-554. [DOI] [PubMed] [Google Scholar]
- 5.Mercurio, F., Zhu, H., Murray, B. W., Shevchenko, A., Bennett, B. L., Li, J., Young, D. B., Barbosa, M., Mann, M., Manning, A., et al. (1997) Science 278, 860-866. [DOI] [PubMed] [Google Scholar]
- 6.Baeuerle, P. A. & Henkel, T. (1994) Annu. Rev. Immunol. 12, 141-179. [DOI] [PubMed] [Google Scholar]
- 7.Siebenlist, U., Franzoso, G. & Brown, K. (1994) Annu. Rev. Cell Biol. 10, 405-455. [DOI] [PubMed] [Google Scholar]
- 8.Thanos, D. & Maniatis, T. (1995) Cell 80, 529-532. [DOI] [PubMed] [Google Scholar]
- 9.Anrather, J., Csizmadia, V., Soares, M. P. & Winkler, H. (1999) J. Biol. Chem. 274, 13594-13603. [DOI] [PubMed] [Google Scholar]
- 10.Bird, T. A., Schooley, K., Dower, S. K., Hagen, H. & Virca, G. D. (1997) J. Biol. Chem. 272, 32606-32612. [DOI] [PubMed] [Google Scholar]
- 11.Diehl, J. A., Tong, W., Sun, G. & Hannink, M. (1995) J. Biol. Chem. 270, 2703-2707. [DOI] [PubMed] [Google Scholar]
- 12.Martin, A. G. & Fresno, M. (2000) J. Biol. Chem. 275, 24383-24391. [DOI] [PubMed] [Google Scholar]
- 13.Naumann, M. & Scheidereit, C. (1994) EMBO J. 13, 4597-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakurai, H., Chiba, H., Miyoshi, H., Sugita, T. & Toriumi, W. (1999) J. Biol. Chem. 274, 30353-30356. [DOI] [PubMed] [Google Scholar]
- 15.Sizemore, N., Leung, S. & Stark, G. R. (1999) Mol. Cell. Biol. 19, 4798-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, D. & Baldwin, A. S. (1998) J. Biol. Chem. 273, 29411-29416. [DOI] [PubMed] [Google Scholar]
- 17.Wang, D., Westerheide, S. D., Hanson, J. L. & Baldwin, A. S., Jr. (2000) J. Biol. Chem. 275, 32592-32597. [DOI] [PubMed] [Google Scholar]
- 18.Zhong, H., Voll, R. E. & Ghosh, S. (1998) Mol. Cell 1, 661-671. [DOI] [PubMed] [Google Scholar]
- 19.Madrid, L. V., Wang, C. Y., Guttridge, D. C., Schottelius, A. J., Baldwin, A. S., Jr., & Mayo, M. W. (2000) Mol. Cell. Biol. 20, 1626-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang, Y., Zhou, H., Chen, A., Pittman, R. N. & Field, J. (2000) J. Biol. Chem. 275, 9106-9109. [DOI] [PubMed] [Google Scholar]
- 21.Pahl, H. L. (1999) Oncogene 18, 6853-6866. [DOI] [PubMed] [Google Scholar]
- 22.Bender, K., Gottlicher, M., Whiteside, S., Rahmsdorf, H. J. & Herrlich, P. (1998) EMBO J. 17, 5170-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, N. & Karin, M. (1998) Proc. Natl. Acad. Sci. USA 95, 13012-13017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imbert, V., Rupec, R. A., Livolsi, A., Pahl, H. L., Traenckner, E. B., Mueller-Dieckmann, C., Farahifar, D., Rossi, B., Auberger, P., Baeuerle, P. A., et al. (1996) Cell 86, 787-798. [DOI] [PubMed] [Google Scholar]
- 25.Doerre, S. & Corley, R. B. (1999) J. Immunol. 163, 269-277. [PubMed] [Google Scholar]
- 26.Finco, T. S., Westwick, J. K., Norris, J. L., Beg, A. A., Der, C. J. & Baldwin, A. S., Jr. (1997) J. Biol. Chem. 272, 24113-24116. [DOI] [PubMed] [Google Scholar]
- 27.Cahir McFarland, E. D., Izumi, K. M. & Mosialos, G. (1999) Oncogene 18, 6959-6964. [DOI] [PubMed] [Google Scholar]
- 28.Norris, J. L. & Baldwin, A. S., Jr. (1999) J. Biol. Chem. 274, 13841-13846. [DOI] [PubMed] [Google Scholar]
- 29.Rayet, B. & Gelinas, C. (1999) Oncogene 18, 6938-6947. [DOI] [PubMed] [Google Scholar]
- 30.Sun, S. C. & Ballard, D. W. (1999) Oncogene 18, 6948-6958. [DOI] [PubMed] [Google Scholar]
- 31.Jo, H., Zhang, R., Zhang, H., McKinsey, T. A., Shao, J., Beauchamp, R. D., Ballard, D. W. & Liang, P. (2000) Oncogene 19, 841-849. [DOI] [PubMed] [Google Scholar]
- 32.Jiang, Y., Woronicz, J. D., Liu, W. & Goeddel, D. V. (1999) Science 283, 543-546. [DOI] [PubMed] [Google Scholar]
- 33.Mochida, Y., Takeda, K., Saitoh, M., Nishitoh, H., Amagasa, T., Ninomiya-Tsuji, J., Matsumoto, K. & Ichijo, H. (2000) J. Biol. Chem. 275, 32747-32752. [DOI] [PubMed] [Google Scholar]
- 34.Witte, S., Villalba, M., Bi, K., Liu, Y., Isakov, N. & Altman, A. (2000) J. Biol. Chem. 275, 1902-1909. [DOI] [PubMed] [Google Scholar]
- 35.Yeung, K. C., Rose, D. W., Dhillon, A. S., Yaros, D., Gustafsson, M., Chatterjee, D., McFerran, B., Wyche, J., Kolch, W. & Sedivy, J. M. (2001) Mol. Cell. Biol. 21, 7207-7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeung, K., Janosch, P., McFerran, B., Rose, D. W., Mischak, H., Sedivy, J. M. & Kolch, W. (2000) Mol. Cell. Biol. 20, 3079-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouchier-Hayes, L., Conroy, H., Egan, H., Adrain, C., Creagh, E. M., MacFarlane, M. & Martin, S. J. (2001) J. Biol. Chem. 276, 44069-44077. [DOI] [PubMed] [Google Scholar]
- 38.Baeuerle, P. A. & Baichwal, V. R. (1997) Adv. Immunol. 65, 111-137. [PubMed] [Google Scholar]
- 39.Barnes, P. J. & Karin, M. (1997) N. Engl. J. Med. 336, 1066-1071. [DOI] [PubMed] [Google Scholar]
- 40.Baldwin, A. S. (2001) J. Clin. Invest. 107, 241-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, X., Commane, M., Burns, C., Vithalani, K., Cao, Z. & Stark, G. R. (1999) Mol. Cell. Biol. 19, 4643-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pellegrini, S., John, J., Shearer, M., Kerr, I. M. & Stark, G. R. (1989) Mol. Cell. Biol. 9, 4605-4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook, J., Frisch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 44.Majumder, S., Zhou, L. Z. H., Chaturvedi, R., Babcock, G., Aras, S. & Ransohoff, R. M. (1998) J. Neurosci. Res. 54, 169-180. [DOI] [PubMed] [Google Scholar]
- 45.Levy, D. E., Kessler, D. S., Pine, R. & Darnell, J. E., Jr. (1989) Genes Dev. 3, 1362-1371. [DOI] [PubMed] [Google Scholar]
- 46.Kessler, D. S., Veals, S. A., Fu, X. Y. & Levy, D. E. (1990) Genes Dev. 4, 1753-1765. [DOI] [PubMed] [Google Scholar]
- 47.Kaelin, W. G., Jr., Pallas, D. C., DeCaprio, J. A., Kaye, F. J. & Livingston, D. M. (1991) Cell 64, 521-532. [DOI] [PubMed] [Google Scholar]
- 48.Pear, W. S., Nolan, G. P., Scott, M. L. & Baltimore, D. (1993) Proc. Natl. Acad. Sci. USA 90, 8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schouten, G. J., Vertegaal, A. C., Whiteside, S. T., Israel, A., Toebes, M., Dorsman, J. C., Van der Eb, A. J. & Zantema, A. (1997) EMBO J. 16, 3133-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feo, S., Arcuri, D., Piddini, E., Passantino, R. & Giallongo, A. (2000) FEBS Lett. 473, 47-52. [DOI] [PubMed] [Google Scholar]
- 51.Subramanian, A. & Miller, D. M. (2000) J. Biol. Chem. 275, 5958-5965. [DOI] [PubMed] [Google Scholar]
- 52.Beraud, C., Henzel, W. J. & Baeuerle, P. A. (1999) Proc. Natl. Acad. Sci. USA 96, 429-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arlt, A., Vorndamm, J., Muerkoster, S., Yu, H., Schmidt, W. E., Folsch, U. R. & Schafer, H. (2002) Cancer Res. 62, 910-916. [PubMed] [Google Scholar]
- 54.Lu, T., Sathe, S. S., Swiatkowski, S. M., Hampole, C. V. & Stark, G. R. (2003) Oncogene, in press. [DOI] [PubMed]
- 55.Bargou, R. C., Emmerich, F., Krappmann, D., Bommert, K., Mapara, M. Y., Arnold, W., Royer, H. D., Grinstein, E., Greiner, A., Scheidereit, C., et al. (1997) J. Clin. Invest. 100, 2961-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bargou, R. C., Leng, C., Krappmann, D., Emmerich, F., Mapara, M. Y., Bommert, K., Royer, H. D., Scheidereit, C. & Dorken, B. (1996) Blood 87, 4340-4347. [PubMed] [Google Scholar]