Abstract
The GAL4-based Gene-Switch system has been engineered to regulate transgene expression in Drosophila in both time and space. We constructed a Gene-Switch transgene in which Gene-Switch expression is restricted spatially by a defined mushroom body enhancer. This system allows Gene-Switch to be active only in the mushroom bodies and only on administration of the pharmacological Gene-Switch ligand RU486. This line was used to drive the expression of a rutabaga cDNA in otherwise rutabaga mutant flies. Induction of the rutabaga cDNA in the mushroom bodies only during adulthood, or during adulthood along with the larval and pupal developmental stages, corrects the olfactory memory impairment found in rutabaga mutants. Induction of the cDNA only during the larval and pupal stages was inconsequential to performance in olfactory memory tasks. These data indicate that normal rutabaga function must be expressed in adulthood for normal memory and conclusively delimit the time and space expression requirements for correcting the rutabaga memory impairment. Such combined pharmacogenetic regulation of transgene expression now allows this time and space dissection to be made for other behavioral mutants.
Molecular genetic analyses of learning processes in Drosophila have identified several genes that support memory formation (1). Several of the identified genes encode components of the cAMP signaling pathway, and this has led to the fundamental conclusion that memories are formed, in part, by the activation of this signaling pathway (2). Included in this set of genes is the prototypic “learning” gene dunce, which codes for cAMP phosphodiesterase (3, 4), the rutabaga (rut) gene, which codes for a calcium:calmodulin-dependent adenylyl cyclase (5, 6), and DCO, which codes for protein kinase A (7, 8). Also included are amnesiac, which encodes a peptide regulator of adenylyl cyclase (9), NF1, which encodes an activator of adenylyl cyclase (10), and CREB, a transcription factor activated by protein kinase A (11). A second class of genes clearly involved in memory formation are members of the family of cell adhesion receptors, including Volado, which codes for an α-integrin (12), and fasII, which codes for a member of the Ig family of cell adhesion receptors (13).
Two critical questions that arise for each newly identified mutant that impairs memory formation are: (i) where in the nervous system is the gene product required for normal memories to form, and (ii) is the gene product required for the normal development of the nervous system or for the physiological processes in adult neurons that underlie memory formation? The answer to the first question for Drosophila olfactory conditioning has been answered to near certainty through combined studies showing the preferential expression of these genes in the mushroom bodies, the importance of normal mushroom body integrity and function for normal learning to occur, and tissue-specific gene rescue experiments (1, 2). The aforementioned genes dnc, rut, DCO, Volado, and fasII are all preferentially expressed in adult mushroom body neurons (1, 2), which highlights the importance of these gene products to the physiology of these neurons. In addition, olfactory learning is impaired in mutants that disrupt mushroom body development (14), by the disruption of intracellular signaling in mushroom body neurons through the targeted expression of a constitutively active Gαs (15), and after ablation of the mushroom bodies (16). Moreover, GAL4-directed expression of rut in the mushroom bodies rescues the learning deficit (17).
The second critical question has been addressed for some of the aforementioned genes but not all of them. Transgenes containing the heat shock promoter driving the expression of dunce (18), Volado (12), fasII (13), and NF1 (10) have been used to conditionally rescue the mutant phenotypes, with expression during adulthood being sufficient to rescue the impaired memory. These results indicate a physiological role in memory formation for these genes and their products. However, it is critically important to delimit the time window of expression necessary for the rescue for each mutant. This is because some mutants may produce a learning phenotype because of their developmental roles, as with the linotte/derailed gene (19). Others may have only physiological roles for plasticity of the adult brain. Yet, other genes may have both developmental roles and adult physiological roles for plasticity. For instance, the leonardo gene is required for development, because null alleles are lethal, yet hypomorphic alleles survive to adulthood and exhibit poor memory formation (20). The memory deficit, nevertheless, can be rescued by supplying gene activity only during adulthood, indicating a dual role for this gene in both developmental processes and in the physiology of the adult brain for memory formation (21).
It has been assumed that one of the more central players in memory formation is the rut gene, because the adenylyl cyclase product of this gene has been posited to be a coincidence detector of sensory information (22). But, surprisingly, definitive evidence that this gene product is required for adult physiology rather than development has not been obtained. This finding is critical because the rut-encoded adenylyl cyclase and its mammalian homolog have clear developmental roles, opening the possibility that the cyclase may have nothing to do with physiological plasticity but may only affect the development of neural structures important for memory formation, such as mushroom bodies. The rut gene product is required, for example, for the normal development of the neuromuscular junction; rut mutants have a two-fold reduction in the number of synapses per bouton at this junction (23). The rut mutants appear to have a decrease in the number of mushroom body fibers compared to controls (24). Moreover, Corfas and Dudai (25) have shown that identified mechanosensory neurons in rut mutants have increased branching and varicosity number, demonstrating that rut is required for shaping normal neuronal connectivity. Developmental studies of the homologous adenylyl cyclase in the mouse also provide compelling evidence for developmental roles. The mutant barrelless lacks the normal barrel field patterning in the somatosensory cortex and is due to a mutation in the type 1 calcium:calmodulin-dependent adenylyl cyclase gene (26). Furthermore, barrelless mutants exhibit defects in the axonal projections of retinal ganglion neurons (27). Thus, mutations in the rut gene in Drosophila or its mammalian homolog clearly produce developmental abnormalities in neuronal wiring and pattern formation, but whether the structural changes of the nervous system due to the mutations cause the memory defect, or are separable from that defect, has remained unknown.
Recent technical advances now offer the possibility of addressing questions regarding gene expression requirements in both time and space simultaneously with transgenic gene expression systems. One of these advances uses a transcription factor named Gene-Switch, which has been engineered to contain the GAL4 DNA-binding domain, a human progesterone ligand-binding domain, and an activation domain from p65 (28). This transcription factor when inserted randomly into the Drosophila genome is activated in space by anonymous enhancers proximal to the transgene and in time by the administration of the antiprogestin RU486 (29). Administration of the ligand activates the chimeric transcription factor so that it binds to and promotes transcription from upstream activation sequence (UAS) sequences upstream of a target transgene.
We have generated a Drosophila Gene-Switch line [P{MB-Switch}] in which Gene-Switch is driven spatially from enhancer sequences that promote expression rather specifically in the mushroom bodies. We have used this line to drive the expression of the normal rut function in a rut mutant background, and show that RU486 administration only during adulthood is sufficient for complete rescue of the memory impairment of rut mutants. These data offer conclusive evidence that rut function is required in the adult mushroom bodies for normal olfactory memories to be formed.
Methods
Molecular Biology and Genetics. The 247-bp mushroom body enhancer from the Dmef2 gene was isolated by PCR using genomic DNA from P247 flies (17) as a template and was sequenced. This enhancer was cloned upstream of the minimal promoter of the hsp70Bb gene, which drives Gene-Switch in the enhancerless vector pP{wlo+hsGS} (30). P{MB-Switch} germ-line transformants were obtained by using standard methods. The UAS-rut on two or UAS-rut on three transgenes (17) were crossed into the rut2080 or rut2769 genomes by using a w;Sco/CyO; TM2/TM6 double balancer line and the appropriate genetic elements were selected phenotypically or by single-fly PCR assays designed to follow the UAS-rut transgenes and the insertion elements in the rut2080 and rut2769 alleles.
RU486 Administration. We used 500 μM RU486 in sucrose solution for all experiments except the developmental shift experiment, for which 200 μM RU486 in food was used. For administration with sucrose, one Kimwipe (11.4 × 21.3 cm) was wetted with 2 ml of either 500 μM RU486 in 2% sucrose or 2% sucrose without RU486. The Kimwipe was placed in a bottle or a vial and flies were added. Flies were transferred to regular food vials 2 h before training. Unless otherwise specified, flies were 1–5 days old at the time of collection, and were fed RU486 in sucrose or sucrose without RU486 for 2 days before training. For administration with fly food, the appropriate amount of a concentrated RU486 stock solution was mixed into molten food at 65°C to a final concentration of 200 μM, and the food was poured and allowed to solidify.
Olfactory Classical Conditioning. Olfactory conditioning was performed under dim red light at 60% relative humidity and at 25°C. Benzaldehyde (0.3% in mineral oil) and 3-octanol (0.6% in mineral oil) were used as odorants. Twelve pulses of electrical shock at 90 V over 1 min were presented as the unconditioned stimulus. Flies were transferred to an electrifiable shock tube and sequentially presented with (i) fresh air for 30 sec, (ii) one odor (CS+) paired with electrical shock for 1 min, (iii) fresh air for 30 sec, (iv) the other odor (CS-) with no shock for 1 min, and (v) fresh air for 45 sec. Flies were then loaded into a T-maze. After a 1-min rest period, flies were allowed to choose between two arms of the T-maze for 2 min, with one arm carrying the CS+ and the other carrying the CS-. Data were analyzed by using STATVIEW software. ANOVA and Bonferroni–Dunn post hoc tests were used to examine statistical significance.
Results
Construction and Characterization of an MB-Switch Line. We considered two strategies for bringing Gene-Switch under the control of DNA elements that would restrict expression to the mushroom bodies. The first strategy was to isolate new P{Switch} enhancer detector elements (29) showing preferential mushroom body expression by histochemical screening of adult head cryosections (31). This strategy requires new enhancer detector elements. The second strategy was to bring Gene-Switch under the control of enhancer sequences that have been shown to provide restricted expression in the mushroom bodies. A 247-bp fragment from the 5′ flanking region of the Dmef2 gene (17) has previously been shown to provide such enhancer activity by cloning the fragment upstream of a minimal promoter driving GAL4. This GAL4 line, named P247, provides rather specific spatial expression in the adult mushroom bodies and has been used to drive UAS transgene expression in several prior studies (17, 32). However, the temporal expression of the element is fixed by the sequences inherent to the enhancer.
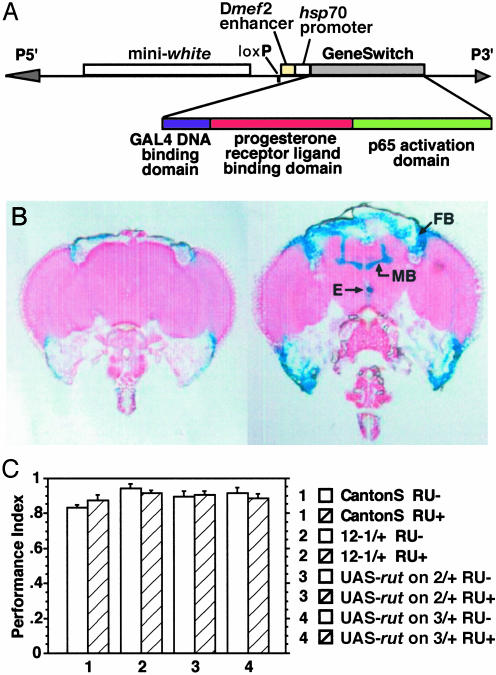
The 247-bp fragment was isolated and cloned upstream of the hsp70Bb minimal promoter and transcription start site in the Gene-Switch vector, pP{wlo+hsGS} (ref. 30 and Fig. 1A). Several Drosophila transformants were obtained and first tested for preferential mushroom body expression by examining the expression pattern of the reporter, UAS-lacZ (Fig. 1B). Only faint expression of lacZ was observed in fat body tissue lining the head cuticle in flies withheld from the Gene-Switch inducer RU486. In contrast, flies fed RU486 for either 24 or 48 h showed robust expression of lacZ in the fat bodies, the esophagus, and the calyx, peduncle, and lobes of the mushroom bodies. Detectable expression in mushroom bodies was visible as early as 90 min after feeding on RU486.
Fig. 1.
Construction and characterization of the P{MB-Switch}12-1. (A) The 247-bp Dmef2 enhancer sequence was cloned into pP{wlo+hsGS} vector. This vector utilizes mini-white+ as a selectable marker. The Gene-Switch protein, which contains the GAL4 DNA-binding domain, a mutant progesterone receptor ligand-binding domain, and an activation domain from p65, is expressed from the minimal promoter region of hsp70Bb. (B) RU486-induced expression of the reporter UAS-lacZ in frontal head sections of P{MB-Switch}12-1/P{UAS-lacZ.B}. (Left) Cryosection of a fly fed 2% sucrose for 48 h before sectioning. (Right) Cryosection of a fly fed 500 μM RU486 in 2% sucrose for 24 h before sectioning. Robust lacZ expression was detectable in all areas of the mushroom bodies (MB) as well as the fat bodies (FB) and esophagus (E). (C) Normal memory in flies of four different genotypes fed (+) or unfed (-) RU486. Two days of feeding on 500 μM RU486 in 2% sucrose did not affect performance of Canton-S, P{MB-Switch}12-1/+, UAS-rut on 2/+, or UAS-rut on 3/+ flies. One-way ANOVA showed no significant difference among the eight groups (n = 6 per group). The P{MB-Switch}12-1 line contains two transgenes, one on the second and one on the third chromosome. For simplicity, the genotypic designation in this and subsequent figures lists only one of the two elements.
We previously failed to find any effect of RU486 administration on viability and several different types of behavior, including phototaxis, geotaxis, locomotion, and escape responses (29). To test whether RU486 administration affects memory, we tested flies fed or unfed RU486 after discriminative, negatively reinforced, olfactory classical conditioning. In this assay, flies are trained with exposure to one odor and simultaneously given electric shocks, and are then exposed to a counterodor in the absence of shock. Memory is assayed by measuring avoidance of the shocked odor. There was no effect of the RU486 administration on memory, as evidenced by nearly identical performance scores among four genotypes fed or unfed RU486. Furthermore, there was no effect on memory of heterozygosity of two different UAS-rut transgenes, or one MB-Switch line named P{MB-Switch}12-1 (Fig. 1C), which we selected for subsequent use. The P{MB-Switch}12-1 line contains two Gene-Switch transgenes, one on the second chromosome and one on the third.
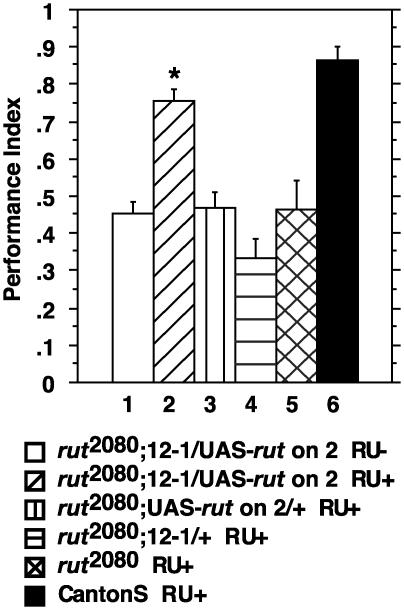
Rescue of rut2080 with MB-Switch. We constructed rut2080 mutants (6) that carried both a UAS-rut transgene on the second chromosome and the P{MB-Switch}12-1 transgene to test whether the rut memory impairment could be rescued with RU486 administration to adult animals. Such rescue, if observed, would offer three critical insights. First, it would demonstrate conclusively that the rut-encoded adenylyl cyclase is required only in the adult nervous system for complete memory rescue. The rut2080 allele is either a very strong hypomorph or an amorph (6) so it provides a very stringent test of the system. Second, it would indicate that any developmental abnormality in the mutant animals, if present, due to the absence of the cyclase at earlier stages is inconsequential for olfactory classical conditioning. Third, it would demonstrate the utility of the Gene-Switch system for combined pharmacogenetic therapy.
Adult rut2080 animals, along with several different control genotypes, were classically conditioned to odors and tested for three min memory (Fig. 2). The rut2080, rut2080 with UAS-rut, and rut2080 with P{MB-Switch}12-1 groups performed indistinguishably at mutant levels (Fig. 2). This memory impairment was observed in flies fed for 2 days on 500 μM RU486 in 2% sucrose before conditioning and in unfed flies of the same genotypes (data not shown). Thus, RU486 administration by itself alters neither wild-type (Fig. 1C) nor mutant performance. Moreover, rut2080 flies carrying both the UAS-rut and P{MB-Switch} transgenes and fed only sucrose performed at the same mutant level. Importantly, administration of RU486 to flies of the latter genotype corrected the memory impairment to a level indistinguishable from the Canton-S control (Fig. 2). Therefore, the expression of the adenylyl cyclase only in the adult mushroom bodies, which is contingent on RU486 administration, completely corrects the otherwise mutant memory of these animals.
Fig. 2.
Memory impairment of rut2080 is rescued by using MB-Switch transgenes. The performance of rut2080;UAS-rut on 2/P{MB-Switch}12-1 flies was rescued from mutant levels to wild-type levels after feeding for 2 days on RU486. All other groups performed at mutant levels. ANOVA followed by Bonferroni–Dunn post hoc tests indicated that groups 1, 3, 4, and 5 are not significantly different from each other, and that groups 2 and 6 are not significantly different from each other. Group 2 was significantly different from group 1 (P = 0.0002), group 3 (P = 0.0004), group 4 (P < 0.0001), and group 5 (P = 0.0003). Group 6 was significantly different from group 1 (P < 0.0001), group 3 (P < 0.0001), group 4 (P < 0.0001), and group 5 (P < 0.0001). n = 6 for all groups.
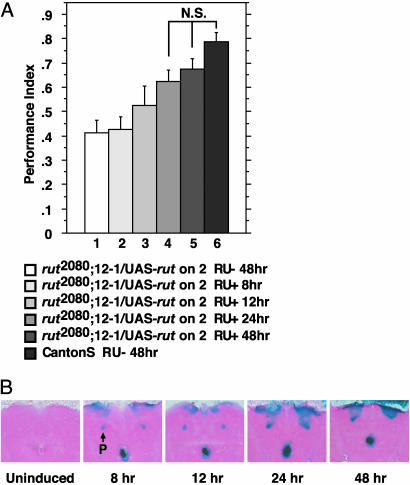
Two additional experiments were performed to better understand the parameters that might affect the use of the Gene-Switch system. First, we examined the effect of temperature on rescue. Induction by RU486 was usually carried out at 25°C, and so we compared the effect of induction at 30°C with that at 25°C. The results (data not shown) showed that induction at 25°C or 30°C both rescued performance of rut2080;UAS-rut/P{MB-Switch}12-1 to wild-type levels, and that the performance scores of flies induced at 25°C was not significantly different from flies induced at 30°C. Second, we determined the time of induction necessary to produce wild-type levels of performance. We fed rut2080;UAS-rut/P{MB-Switch}12-1 flies on 500 μM RU486 for 8, 12, 24, or 48 h before olfactory conditioning. The performance improved with longer durations of feeding, reaching wild-type levels only with a 24- to 48-h feeding period (Fig. 3A). Only the 24- and 48-h time groups showed performance scores statistically indistinguishable from the wild-type control. A parallel experiment was performed to monitor the level of transgene expression during RU486 administration. Flies carrying P{MB-Switch}12-1 and UAS-lacZ were fed RU486 for various periods of time and then lacZ activity was assayed in head cryosections, with the assay time for lacZ remaining constant for all groups. LacZ activity was more robust with longer feeding periods and reached maximal levels at 24–48 h (Fig. 3B). These experiments indicate that robust levels of expression are observed 1–2 days after initiating induction with RU486.
Fig. 3.
Kinetics of Gene-Switch-induced behavioral rescue. (A) Flies were fed 500 μM RU486 in 2% sucrose (RU+) for 8, 12, 24, or 48 h, or were fed 2% sucrose (RU-) for 48 h before olfactory classical conditioning. Twenty-four to 48 h of RU486 administration was required for rut2080;UAS-rut/P{MB-Switch}12-1 flies to perform indistinguishably from the Canton-S control. n = 6 for all groups except group 3, which was n = 3. ANOVA followed by Bonferroni–Dunn post hoc tests revealed that group 6 is significantly different from group 1 (P < 0.0001), group 2 (P < 0.0032), and group 3 (P < 0.0032), but was not significantly different from groups 4 or 5. NS, not significant. (B) P{MB-Switch}12-1/UAS-lacZ flies were fed as in A and assayed for lacZ activity by histochemical staining. Robust lacZ activity is observed 24–48 h after induction. P, peduncle of mushroom bodies.
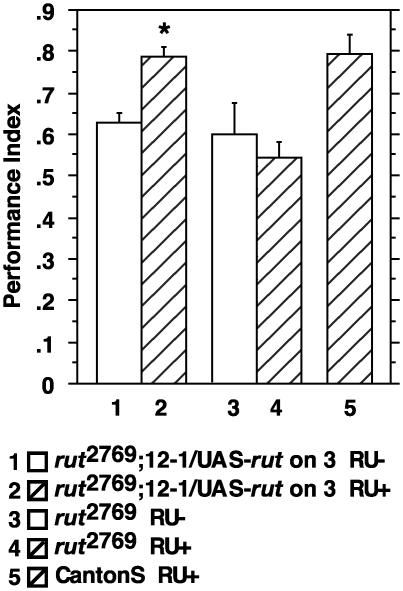
MB-Switch Rescue Is Not Specific for rut2080. We also constructed flies by combining a UAS-rut transgene on the third chromosome, the transgene P{MB-Switch}12-1, and the rut allele, rut2769. The rut2769 allele is weaker than rut2080, and we wanted to ensure that the pharmacogenetic rescuing effect of P{MBSwitch}12-1 was not an unusual, allele-specific effect. Memory after training of rut2769;P{MB-Switch}12-1/+;UAS-rut on 3/+ flies without RU486 was not significantly different from that of rut2769 mutant flies without any transgenes (Fig. 4). However, as expected, we confirmed our prior observations (6) that rut2769 is behaviorally a weaker allele than rut2080, with 3-min memory scores averaging ≈0.57 compared with 0.46 for rut2080. The two alleles both have insertions of the enhancer detector element, PlArB in the promoter region of rut, but the insertion in rut2769 is 13 base pairs more distal than that in rut2080 (6). The administration of RU486 again did not affect the performance of mutant flies, but it rescued the performance of rut2769; P{MB-Switch}12-1/+; UAS-rut on 3/+ flies to a level statistically indistinguishable from that of CantonS (Fig. 4). These results demonstrated that the pharmacogenetic rescuing effect of P{MB-Switch}12-1 is not allele-specific, but is generalizable to loss-of-function alleles.
Fig. 4.
The impairment of 3-min memory of rut2769 is rescued by using MB-Switch. Some groups of flies were fed RU486 for 2 days before training. This treatment, along with the presence of the two transgenes, rescued the performance of rut2769; P{MB-Switch}12-1/+;UAS-rut on three flies to wild-type levels. ANOVA followed by Bonferroni–Dunn post hoc tests revealed that groups 1, 3, and 4 were not significantly different from each other, groups 2 and 5 were not significantly different from each other, and groups 2 and 5 were significantly different from groups 1, 3, and 4 (P < 0.004 for all six comparisons). n = 5–10 for all groups.
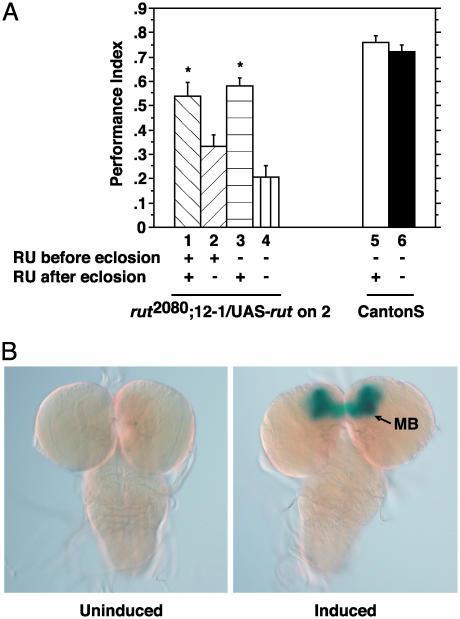
Temporal Requirement of rut Expression for Learning. The above data indicate that the induced expression of a rut cDNA in the mushroom bodies just before training in adulthood was able to rescue the memory impairment of rut mutant flies to wild-type levels. One interpretation for these results is that the memory impairment of rut mutants is due to a disruption in physiological processes engaged at the time of training, rather than from a developmental defect. An alternative possibility is that the adenylyl cyclase is required for the brain to acquire the competence to form memories, and this competence could be gained from adenylyl cyclase expression either during development or during adulthood. To further reveal the role of the cyclase, we compared the memory after olfactory conditioning of rut2080;UAS-rut/P{MB-Switch}12-1 flies under four different induction paradigms. In the ON-ON paradigm, flies were reared on 200 μM RU486-containing food during development and after eclosion. In the OFF-OFF paradigm, flies were reared on regular food during development and after eclosion. In the ON-OFF paradigm, flies were reared on RU486-food and transferred onto regular food on eclosion. And in the OFF-ON paradigm, flies were reared on regular food and transferred onto RU486-containing food on eclosion. All flies were conditioned 7–10 days after eclosion (Fig. 5A).
Fig. 5.
Effects of inducing rut expression during development or in adulthood. (A) Flies were fed during development and/or in adulthood on food with (+) or without (-) 200 μM RU486 and were conditioned 7–10 days after eclosion. Induction in adulthood was necessary and sufficient for rescue to occur. Induction during development was unimportant. One-way ANOVA followed by Bonferroni–Dunn post hoc tests revealed that groups 1 and 3 were not significantly different from each other. Groups 2 and 4 were not significantly different from each other. Group 1 was significantly different from group 2 (P = 0.0032), group 4 (P < 0.0001), and group 5 (P = 0.0013) but was not significantly different from group 6. Group 3 was significantly different from groups 2 (P = 0.0004) and 4 (P < 0.0001) but was not significantly different from groups 5 or 6. (n = 4–6 to six for all groups.) (B) Histochemical assay for lacZ in P{MB-Switch}12-1/UAS-lacZ larvae showing expression in the mushroom bodies (MB) of the third-instar larva.
Both the ON-ON group and the OFF-ON group performed significantly better than the ON-OFF group and the OFF-OFF group. The performance scores of the ON-ON and OFF-ON groups were not significantly different from each other, nor were the performance scores of the ON-OFF and OFF-OFF groups significantly different from each other. As a control, Canton-S flies also underwent the OFF-ON and the OFF-OFF induction paradigms and tested for olfactory memory. The results indicate that the 7–10 days feeding on RU486 in adulthood did not alter performance of wild-type flies. Most importantly, the performance scores of the ON-ON and OFF-ON mutant rescue groups were not significantly different from the Canton-S groups. Interestingly, there was a significant difference between the performance scores of the OFF-ON group and the ON-OFF group, despite the fact that histochemical assays using UAS-lacZ as a target for MBSwitch indicate that this element promotes expression in the mushroom bodies during larval stages of brain development as well as in the adult (Fig. 5B). These results indicate that the induction of rut expression only in the adult mushroom bodies is sufficient for full rescue of the impaired memory of rut mutants. Expression of the adenylyl cyclase during development is of no consequence to odor memory during adulthood.
Discussion
Rutabaga is the most thoroughly characterized of all Drosophila memory mutants. Previous studies (5) have shown that the gene encodes for a calcium:calmodulin-dependent adenylyl cyclase and that this cyclase is preferentially expressed in the axons of mushroom body neurons (6). These observations, combined with the knowledge that the products of other learning genes like dunce show preferential expression in the mushroom bodies (1, 2), prompted two major and important questions. First, are mushroom bodies the site of action of the adenylyl cyclase in terms of promoting memory formation? This possibility was strongly predicted to be the case, given the preferential expression in these neurons and the fact that other gene products important for learning exhibit a similar expression pattern. This prediction was confirmed through standard GAL4-mediated rescue experiments (17). The second question, whose answer, surprisingly, was unknown, was whether the adenylyl cyclase is important for the development of brain structures such as mushroom bodies that are required for memory, or whether the requirement of the cyclase is restricted to adult stages. The possibility that developmental defects underlie the impairment in memory was made stronger by studies that demonstrated that the mutants do have an altered nervous system (23–25, 33) and the discovery that mutation of the rut homolog in the mouse produces an alteration in barrel fields of the somatosensory cortex (26) and in the refinement of axonal projections from retinal ganglion cells (27).
We have provided through the Gene-Switch pharmacogenetic approach described here, an answer to this question. Expression of rut only in the adult mushroom bodies is sufficient for the rescue of the memory impairment of rut mutants, and expression of rut in the mushroom bodies during development is inconsequential to adult odor memory formation. This result was achieved in part through the construction of an RU486-inducible, mushroom body Gene-Switch line P{MB-Switch}12-1. This line should be very valuable for similar pharmacogenetic studies in which the experimenter requires control over the timing of gene expression in the mushroom bodies. Moreover, we have also obtained nearly identical results with a second system for controlling transgene expression in time and space (34). This system, named TARGET, utilizes a temperature-sensitive repressor of GAL4 (GAL80ts) to modulate GAL4 activity with temperature shifts.
Even though the requirement for adenylyl cyclase activity for odor learning has been delimited to the adult brain, its specific role in learning processes is still uncertain. The most attractive possibility is that the adenylyl cyclase is coupled to neurotransmitter receptors (31, 35) that register the unconditioned stimulus during learning, or provide a neuromodulatory role for the actual association of the conditioned stimulus and the unconditioned stimulus. An alternative idea is that the adenylyl cyclase is required for the maturation of some other physiological process or cellular components and, in its absence, the neurons remain incompetent to support memory formation.
The Gene-Switch system, like TARGET, requires several hours to days for complete induction (29, 34). This time course is to be expected for any transcriptionally based system, which requires an applied inducer to alter the activity of a transcription factor to initiate the subsequent processes of transcription, RNA processing, translation, posttranslational modification, and cellular targeting. The system should be extremely effective for answering questions regarding broad temporal expression requirements such as the one posed here. Questions that require more acute induction and deinduction, such as whether any given gene product is required for the formation of memories, their stability, or their retrieval, will likely require the development of gene expression systems that operate at the posttranslational level, to turn on or turn off the activity of any given protein.
Acknowledgments
This work was supported by National Institutes of Health Grants NS19904 and GM63929. R.L.D. is the R. P. Doherty-Welch Chair in Science at the Baylor College of Medicine.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: UAS, upstream activation sequence.
References
- 1.Roman, G. & Davis, R. L. (2001) BioEssays 23, 571-581. [DOI] [PubMed] [Google Scholar]
- 2.Davis, R. L. (1993) Neuron 11, 1-4. [DOI] [PubMed] [Google Scholar]
- 3.Chen, C. N., Denome, S. & Davis, R. L. (1986) Proc. Natl. Acad. Sci. USA 83, 9313-9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiu, Y. & Davis, R. L. (1993) Genes Dev. 7, 1447-1458. [DOI] [PubMed] [Google Scholar]
- 5.Levin, L. R., Han, P. L., Hwang, P. M., Feinstein, P. G., Davis, R. L. & Reed, R. R. (1992) Cell 68, 479-489. [DOI] [PubMed] [Google Scholar]
- 6.Han, P. L., Levin, L. R., Reed, R. R. & Davis, R. L. (1992) Neuron 9, 619-627. [DOI] [PubMed] [Google Scholar]
- 7.Drain, P., Folkers, E. & Quinn, W. G. (1991) Neuron 6, 71-82. [DOI] [PubMed] [Google Scholar]
- 8.Skoulakis, E., Kalderon, D. & Davis, R. L. (1993) Neuron 11, 197-208. [DOI] [PubMed] [Google Scholar]
- 9.Waddell, S., Armstrong, J. D., Kitamoto, T., Kaiser, K. & Quinn, W. G. (2000) Cell 103, 805-813. [DOI] [PubMed] [Google Scholar]
- 10.Guo, H. F., Tong, J., Hannan, F., Luo, L. & Zhong, Y. (2000) Nature 403, 895-898. [DOI] [PubMed] [Google Scholar]
- 11.Yin, J. C., Wallach, J. S., Del Vecchio, M., Wilder, E. L., Zhou, H., Quinn, W. G. & Tully, T. (1994) Cell 79, 49-58. [DOI] [PubMed] [Google Scholar]
- 12.Grotewiel, M. S., Beck, C. D., Wu, K. H., Zhu, X. R. & Davis, R. L. (1998) Nature 391, 455-460. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, Y., Endo, K., Wu, K., Rodan, A. R., Heberlein, U. & Davis, R. L. (2001) Cell 105, 757-768. [DOI] [PubMed] [Google Scholar]
- 14.Heisenberg, M., Borst, A., Wagner, S. & Byers, D. (1985) J. Neurogenet. 2, 1-30. [DOI] [PubMed] [Google Scholar]
- 15.Connolly, J. B., Roberts, I. J., Armstrong, J. D., Kaiser, K., Forte, M., Tully, T. & O'Kane, C. J. (1996) Science 274, 2104-2107. [DOI] [PubMed] [Google Scholar]
- 16.DeBelle, J. S. & Heisenberg, M. (1994) Science 263, 692-695. [DOI] [PubMed] [Google Scholar]
- 17.Zars, T., Fischer, M., Schulz, R. & Heisenberg, M. (2000) Science 288, 672-675. [DOI] [PubMed] [Google Scholar]
- 18.Dauwalder, B. & Davis, R. L. (1995) J. Neurosci. 15, 3490-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreau-Fauvarque, C., Taillebourg, E., Preat, T. & Dura, J. M. (2002) NeuroReport 13, 2309-2312. [DOI] [PubMed] [Google Scholar]
- 20.Skoulakis, E. M. & Davis, R. L. (1996) Neuron 17, 931-944. [DOI] [PubMed] [Google Scholar]
- 21.Philip, N., Acevedo, S. F. & Skoulakis, E. M. (2001) J. Neurosci. 21, 8417-8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kandel, E. R., Abrams, T., Bernier, T., Carew, T. J., Hawkins, R. D. & Schwartz, J. H. (1983) Cold Spring Harbor Symp. Quant. Biol. 48, 820-830. [DOI] [PubMed] [Google Scholar]
- 23.Renger, J. J., Ueda, A., Atwood, H. L., Govind, C. K. & Wu, C. F. (2001) J. Neurosci. 20, 3980-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balling, A., Technau, G. M. & Heisenberg, M. (1987) J. Neurogenet. 4, 65-73. [PubMed] [Google Scholar]
- 25.Corfas, G. & Dudai, Y. (1991) Proc. Natl. Acad. Sci. USA 88, 7252-7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abdel-Majid, R. M., Leong, W. L., Schalkwyk, L. C., Smallman, D. S., Wong, S. T., Storm, D. R., Fine, A., Dobson, M. J., Guernsey, D. L. & Neumann, P. E. (1998) Nat. Genet. 19, 289-291. [DOI] [PubMed] [Google Scholar]
- 27.Ravary, A., Muzerelle, A., Herve, D., Pascoli, V., Nguyen Ba-Charvet, K., Girault, J. A., Welker, E. & Gaspar, P. (2003) J. Neurosci. 23, 2228-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burcin, M. M., Schiedner, G., Kochanek, S., Tsai, S. Y. & O'Malley, B. W. (1999) Proc. Natl. Acad. Sci. USA 96, 355-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roman, G., Endo, K., Zong, L. & Davis, R. L. (2001) Proc. Natl. Acad. Sci. USA 98, 12602-12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roman, G. & Davis, R. L. (2002) Genesis 34, 127-131. [DOI] [PubMed] [Google Scholar]
- 31.Han, P. L., Meller, V. H. & Davis, R. L. (1996) J. Neurobiol. 31, 88-102. [DOI] [PubMed] [Google Scholar]
- 32.McGuire, S. E., Le, P. T. & Davis, R. L. (2001) Science 293, 1330-1333. [DOI] [PubMed] [Google Scholar]
- 33.Hitier, R., Heisenberg, M. & Preat, T. (1998) NeuroReport 9, 2717-2729. [DOI] [PubMed] [Google Scholar]
- 34.McGuire, S. E., Le, P. T., Osborn, A. J., Matsumoto, K. & Davis R. L. (2003) Science 302, 1765-1768. [DOI] [PubMed] [Google Scholar]
- 35.Han, K. A., Millar, N. S. & Davis, R. L. (1998) J. Neurosci. 18, 3650-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]