Abstract
Epigenetic inheritance, the transmission of gene expression states from parent to daughter cells, often involves methylation of DNA. In eukaryotes, cytosine methylation is a frequent component of epigenetic mechanisms. Failure to transmit faithfully a methylated or an unmethylated state of cytosine can lead to altered phenotypes in plants and animals. A central unresolved question in epigenetics concerns the mechanisms by which a locus maintains, or changes, its state of cytosine methylation. We developed “hairpin-bisulfite PCR” to analyze these mechanisms. This method reveals the extent of methylation symmetry between the complementary strands of individual DNA molecules. Using hairpin-bisulfite PCR, we determined the fidelity of methylation transmission in the CpG island of the FMR1 gene in human lymphocytes. For the hypermethylated CpG island of this gene, characteristic of inactive-X alleles, we estimate a maintenance methylation efficiency of ≈0.96 per site per cell division. For de novo methylation efficiency (Ed), remarkably different estimates were obtained for the hypermethylated CpG island (Ed = 0.17), compared with the hypomethylated island on the active-X chromosome (Ed < 0.01). These results clarify the mechanisms by which the alternative hypomethylated and hypermethylated states of CpG islands are stably maintained through many cell divisions. We also analyzed a region of human L1 transposable elements. These L1 data provide accurate methylation patterns for the complementary strand of each repeat sequence analyzed. Hairpin-bisulfite PCR will be a powerful tool in studying other processes for which genetic or epigenetic information differs on the two complementary strands of DNA.
The potential importance of DNA methylation for specifying epigenetic inheritance in eukaryotic cells was recognized soon after the discovery of the role that methylation plays in the modification and restriction of bacterial and bacteriophage DNA (1–5). In eukaryotic cells, inheritance of the methylated state usually involves 5-methylcytosine and predominantly depends on enzymatic recognition of CpG and CNG motifs. Base-pairing rules (6) ensure that these motifs are symmetrically located on complementary strands of DNA (for example, CpG/CpG dyads), thus providing the opportunity for the inheritance of cytosine methylation after DNA replication (7). In mammals, maintaining a methylated state of CpG cytosines is an important component of X-chromosome inactivation and genomic imprinting (8–10). The failure to maintain a methylated or an unmethylated state of key cytosines can lead to “epimutations”; such changes may alter cell and developmental pathways, resulting in new phenotypes (11–14) including disease (15–17). The mechanisms and fidelity of epigenetic inheritance are thus of crucial biological and medical importance.
A central issue in epigenetics concerns the mechanism by which a locus maintains a stable epigenetic state through many cell divisions. It appears that epigenetic mechanisms that use 5-methylcytosine within CpG dinucleotides have moderate to high fidelities of maintaining a methylated state of cytosine, after a transitory hemimethylation state during DNA replication (9, 18–23). Hemimethylated sites are also transitional states in developmental processes; active demethylation or de novo methylation may sometimes be involved in gene reactivation or inactivation (24–26). In a study to assess the dynamics of DNA methylation, Riggs and colleagues (9, 27), estimated the fidelity of maintenance methylation (Em) within partially methylated CpG islands to be >0.99 per methylated cytosine per cell division; de novo methylation efficiency (Ed) for unmethylated cytosines was estimated to be 0.05 per site per generation. This study, carried out with clones of tissue-culture cells in which methylation was perturbed with 5-azacytidine, also provides a useful mathematical model of the kinetics of DNA methylation (9).
Current inferences on epigenetic fidelities and transitional methylation states are based on data for single methylation sites or on patterns of methylation derived from populations of complementary strands. A major experimental limitation has been the difficulty in obtaining methylation patterns from the two complementary strands of an individual DNA molecule. If such a method were available, patterns of methylation fidelity, and detection of both gain and loss of methylation, could be studied relatively directly.
We have developed “hairpin-bisulfite PCR” for this purpose of analyzing patterns of cytosine methylation on complementary strands of individual DNA molecules. This method uses a hairpin linker, targeted and ligated to restriction-enzyme-cleaved genomic DNA, to maintain attachment of complementary strands during the subsequent denaturation steps required by bisulfite conversion (28, 29) and PCR amplification. We used this method to estimate the fidelity with which the epigenetic state of cytosine is transmitted in human lymphocytes, focusing on CpG cytosines in the promoter region of the fragile X gene, FMR1. We also present data on cytosine methylation in human L1 transposable elements to address a controversy on levels of hemimethylation of these sequences in fibroblasts.
Materials and Methods
Overview and Development of the Method. To apply hairpin-bisulfite PCR to genomic DNA, sequences within and adjacent to a CpG island of interest were scanned for restriction-endonuclease recognition sites. Preferred restriction enzymes are those that (i) leave staggered ends, (ii) have a cut site that is separate from the recognition site, and (iii) are not affected by a potentially methylated target sequence. Requirements i and ii increase the specificity of subsequent PCRs for the desired genomic region; requirement iii increases the likelihood that both hypermethylated and hypomethylated alleles are represented in the final PCR products and sequences. A hairpin-linker, 25 or 26 nt in length, was synthesized with staggered ends complementary to the targeted cut-site of genomic DNA. After restriction-enzyme cleavage of genomic DNA, the hairpin linker was attached to the DNA cleavage site by using DNA ligase, thus covalently connecting the complementary strands of individual DNA molecules. Hairpin-ligated genomic DNA was then subjected to bisulfite conversion and PCR amplification with appropriate primers, followed by conventional DNA sequencing of the PCR products. Primers were designed to anneal to genomic sequences within 150–200 nt of the linker; PCR products extending around the linker to the next primer site were thus in the range of 300–400 nt. Examples of restriction enzyme selection, and design of hairpin linkers and PCR primers, are given here for human FMR1 and L1 sequences.
To optimize conditions for bisulfite conversion, cloning, and sequencing of hairpin DNA, two 178-base hairpin oligonucleotides were synthesized, corresponding to a portion of the FMR1 CpG island. One oligonucleotide was hemimethylated at six CpG/CpG dyads (Fig. 1 A and B), whereas the other was unmethylated, thus providing a key test of our ability to detect hemimethylated dyads in DNA. Bisulfite conversion was used because it provides information on the methylation state of individual cytosines by converting cytosine (but not 5-methylcytosine) to uracil, and subsequently to thymine upon PCR amplification (28, 29). Bisulfite conversion also reduces base pair complementarity within the hairpin, a reduction that we found useful for PCR amplification, as well as for high-fidelity cloning and sequencing of hairpins containing unmethylated cytosines [as also recognized by Tanaka et al. (30), for natural hairpins]. Another approach to amplifying hairpin structures, recently reported by Kaur and Makrigiorgos (31), uses ligated primer binding sites. Our bisulfite conversion step makes primer sites available within the previously complementary strands (Fig. 4, which is published as supporting information on the PNAS web site).
Fig. 1.
Design of hairpin oligonucleotide, hemimethylated by synthesis and used for specificity tests of bisulfite conversion. A 178-bp oligonucleotide, modeling a 5′ portion of the CpG island region of FMR1, was synthesized in two steps: a 100-mer oligonucleotide was synthesized by Midland Scientific (Midland, TX), using 5-methyl–cytosine in CpGs on the lower strand (highlighted in red), and including a hairpin-forming sequence at the 3′ end (A); this oligonucleotide was extended by using Klenow DNA polymerase (46), incorporating unmethylated cytosines and the other three bases to complete the top strand; unmethylated CpGs are highlighted in blue (B). Primers were designed to amplify the converted sequences of the hairpin oligonucleotide (C) and its PCR copy (D). Hairpin sequences are folded into their original complementary patterns to illustrate the methylation states of CpG dyads.
Bisulfite conversion of cytosine to uracil is inhibited by complementary base-pairing with guanine, thus requiring well-denatured DNA molecules. The hairpin structure of our target DNAs (Fig. 1B) makes normal denaturation conditions inadequate because of the rapid fold-back renaturation of hairpin DNA. The primary modification of the method that proved successful for bisulfite conversion of the hairpin structure was the incorporation of up to five additional thermal-denaturation steps during the 6 h incubation of DNA in the presence of sodium bisulfite, rather than the single denaturation step that we used previously for conventional bisulfite conversion (32).
Considerable variation in conversion rates was obtained in different experiments and for individual molecules. It was useful to use the percentage of conversion of non-CpG cytosines as an index of overall bisulfite reaction efficiency. In control experiments with synthetic hairpin oligonucleotides, conversion rates of Cs to Us averaged lower than we observed previously with genomic DNA (>99%) (32), presumably because hairpin structures are difficult to denature completely. Incomplete conversion reduces discrimination between methylated and unmethylated cytosines.
Bisulfite conversion of hairpin DNA structures followed the protocol developed by Clark et al. (33) and further refined by Stöger et al. (32) with a few additional modifications. Hairpin DNA was denatured in 0.3 M NaOH at 42°C for 20 min, then heated to 100°C for 1 min before addition of sodium bisulfite and hydroquinone to 3.4 M and 1 mM, respectively. The reaction mixture was incubated for 6 h at 55°C, with additional thermal denaturation steps (95–99.9°C, three or six times over the 6 h); this was followed by a purification step using Microspin S-200 HR columns (Amersham Pharmacia Biosciences) and subsequent treatment with NaOH (final concentration 0.3 M) at 37°C for 20 min, and another purification using S-200 HR columns. Bisulfite conversion reduced the self-complementarity of the two strands, allowing the first-round primer to anneal to the top strand; for this first-round primer, R represents random incorporation of G and A, to accommodate either C (unconverted) or U (converted) in the complementary template position after bisulfite conversion of the oligonucleotide (Fig. 1C). After PCR synthesis around the hairpin to the 5′ end, a second-round primer (with Y representing random mixtures of C and T), was annealed to the PCR product (Fig. 1D), and the reaction continued through additional amplification rounds (data not shown). PCR conditions were as follows: Failsafe buffer C (Epicentre Technologies, Madison, WI) was used, with denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 30 sec, annealing at 60°C for 30 sec, and extension at 72°C for 1 min; a final extension was at 72°C for 7 min. All PCR products were analyzed by agarose gel electrophoresis; further cloning and sequencing of appropriately sized products was with TOPO vectors (Invitrogen Life Technologies); sequencing reactions were carried out with fluorescent dideoxy nucleotides (BIGDYE Terminator 3.0, Applied Biosystems), at the DNA Sequencing Facility, Department of Biochemistry, University of Washington. Each sequence was proofread against the electropherogram; errant base calling was corrected manually before being presented here. For purposes of analysis and presentation, the output sequence was folded, using word-processing software, into a hairpin conformation so that both strands align.
Application to FMR1 and L1 Sequences in Genomic DNA. Conditions for hairpin-bisulfite PCR of human genomic FMR1 sequences were as follows: 2 μg of genomic DNA was cleaved by 40 units of restriction endonuclease DraIII for 1 h at 37°C, followed by enzyme inactivation at 65°C for 20 min. Ligation of the hairpin linker (5′P-AGCGATGCGTTCGAGCATCGCTTGA) to DraIII-cleaved genomic DNA was for 5 min at 20°C, using 1 unit of T4 ligase in 20 μl with 1× ligase buffer (GIBCO/BRL). The bisulfite conversion protocol was as described above. PCR conditions were Failsafe buffer C (Epicentre), with denaturation at 95°C for 5 min, followed by 35–40 cycles of denaturing at 95°C for 60 sec, annealing at 60°C for 2 min, and extension at 72°C for 3 min; this was followed by a final extension at 72°C for 7 min. Primers used were: first primer, CCTCTCTCTTCAAATAACCTAAAAAC, and second primer, GTTGYGGGTGTAAATATTGAAATTA.
Conditions for hairpin-bisulfite PCR of human L1 promoter sequences were as follows: 2 μg of genomic DNA (extracted from fibroblast cells: untransformed female line 8158A and transformed male line 82–6-h TERT), was cleaved by 10 units of restriction endonuclease BsmAI for 1 h at 55°C. Ligation of the hairpin linker (5′P-ACCAAGCGATGCGTTCGAGCA) to BsmAI-cleaved genomic DNA was for 5 min at 20°C, using 2 units of T4 DNA ligase in 20 μl with 1× ligase buffer (GIBCO/BRL). Bisulfite conversion was carried out according to the protocol outlined above, but with the following incubations after addition of sodium bisulfite: 55°C for 1 h, 99.9°C for 2 min, 55°C for 1 h, 99.9°C for 2 min, 55°C for 1 h, 99°C for 2 min, 55°C for 45 min, 99°C for 2 min, 55°C for 45 min, 99°C for 2 min, 55°C for 45 min, 95°C for 2min, and 55°C for 45 min. Postbisulfite PCR conditions were Failsafe buffer C (Epicentre), denaturation at 95°C for 5 min, followed by 35 cycles of denaturing at 95°C for 60 sec, annealing at 60°C for 2 min, and extension at 72°C for 3 min; this was followed by a final extension at 72°C for 7 min. Primers used for postbisulfite PCR were forward primer L1-HP2F = CAAACRAACATTACCTCACCT, and reverse primer L1-HP2Ra/b = (TT)GGGAAGYGTAAGGGGTTAG; to maintain 60°C annealing temperature, half of the reverse primer molecules contain a C at the position marked with a Y and lack the 5′-TT, and half contain a T at the Y position and include the 5′-TT. Instances of nucleotide variation at CpG dyads were subtracted from the total number of informative dyads, as these sites were no longer target sites for methylation.
Results and Discussion
Synthetic Oligonucleotides. Hairpin-bisulfite PCR correctly identified the hemimethylated CpG/CpG dyads in our hemimethylated oligonucleotide. In sequences with efficient conversion of non-CpG cytosines (>97% conversion), the four informative hemimethylated CpG/CpG dyads (i.e., those between the primer sites) showed no conversion of the methylated CpGs (0/24), and 100% conversion (24/24) of the four complementary, unmethylated CpGs (Fig. 5A, which is published as supporting information on the PNAS web site). The unmethylated FMR1 oligonucleotide (Fig. 5B) showed high conversion percentages for both CpG and non-CpG cytosines (100% and 98.7%, respectively), indicating that the methylated cytosines in the hemimethylated oligonucleotide were resistant to conversion because of their methylation rather than their positions in the oligonucleotide.
Single-Copy FMR1 Alleles from Genomic DNA. After successful identification of hemimethylated sites in our synthetic hairpin oligonucleotide, we applied hairpin-bisulfite PCR to genomic DNA. For a single-copy locus, we chose a portion of the CpG island of human FMR1, a gene that in mutant form can become abnormally hypermethylated, often leading to mental retardation (34, 35). DNA from lymphocytes of a normal female was used because each female cell contains a hypomethylated allele and a hypermethylated allele of FMR1, representing those on the active and inactive X chromosomes, respectively (32). After cleavage with DraIII restriction endonuclease (Fig. 2A and Fig. 6, which is published as supporting information on the PNAS web site), a linker was ligated to the 3′ TCA overhang at the DraIII site in the FMR1 region (Fig. 2 B and C). After bisulfite conversion, primers were used to amplify a region containing 22 CpG dyads in the 5′ half of the CpG island.
Fig. 2.
Inferred methylation states of CpG-island sequences of FMR1 from a normal female, using hairpin-bisulfite PCR. (A) A portion of the FMR1 sequence (from normal female DNA A80) before bisulfite conversion. (B) Cleavage site for restriction endonuclease DraIII (lime green), and a hairpin linker designed with a TGA tail at the 3′ end. (C) Hairpin-linked FMR1 sequence before bisulfite conversion. (D and E) Sequences recovered after hairpin-bisulfite PCR with ≥99% conversion of non-CpG cytosines. Among sequences that met this conversion criterion, hypermethylated (D) and hypomethylated (E) sequences averaged 99.5% and 99.6% conversion of non-CpG cytosines, respectively. Other examples of hypomethylated sequences with ≥99% conversion are presented in Fig. 7. Conversion efficiencies for non-CpG cytosines are shown at the right. Other highlighting shows: hairpin linker, gray; unconverted CpG dyads, red; converted CpG dyads, blue; unconverted non-CpG cytosines, magenta; CpG dyads that are uninformative because of PCR or sequencing errors, as well as ambiguous sequence polymorphisms, yellow; PCR errors, green. The ability to detect PCR errors is inherent in hairpin-bisulfite PCR because information in both strands is maintained throughout the PCR.
Alleles of FMR1 exhibiting high levels of conversion were obtained from this lymphocyte DNA; we further analyzed sequences of PCR products where conversion efficiency of non-CpG cytosines was ≥99% (Fig. 2 D and E). Considering first the methylation status of CpG cytosines on the top, or nontranscribed, strand [to which our previous data correspond (32)], all of these alleles could be placed into the same two categories previously described. Alleles with ≥16 of 22 possible CpG methylation sites were classified as hypermethylated; those with ≤6 sites methylated were classified as hypomethylated.
Hypermethylated FMR1 Alleles. Methylation patterns of hypermethylated alleles obtained by hairpin-bisulfite PCR are very similar to those we obtained previously with conventional bisulfite PCR. The frequency of methylation of top-strand CpG cytosines was 0.82 (Fig. 2D), which is not significantly different from the 0.85 previously observed for normal female lymphocyte DNA (0.4 ≤ P ≤ 0.6; χ2 = 0.48, 1 df) (32). Among bisulfite-converted FMR1 alleles from female lymphocytes, the hypermethylated and hypomethylated classes were recovered at frequencies of 0.3:0.7, instead of the 0.5:0.5 frequencies expected from the relationship between X-chromosome inactivation and hypermethylation. A similar bias (0.27:0.73), the basis of which has not been systematically investigated, was noted in our previous study using conventional, upper-strand-only PCR (32). These results indicate that our genomic hairpin sequences are drawn from the same pool of sequences that were previously recovered by conventional single-strand PCR.
With hairpin-bisulfite PCR, we can assess the methylation status on the bottom strand of each hypermethylated allele for which we have top-strand data. In particular, we can distinguish between symmetrical and asymmetrical patterns of methylation for each of the CpG/CpG dyads. Alleles from a normal female demonstrated symmetrical methylation status for 103 of 110 dyads, with 86 of those being fully methylated (78.2%), and 17 being fully unmethylated (15.4%). Asymmetrical methylation status, i.e., hemimethylation, was observed for seven dyads, representing 6.4% of the total (Fig. 2D).
Fully methylated or fully unmethylated CpG/CpG dyads reflect accurate transmission of the epigenetic signal of methylated cytosine or unmethylated cytosine after the DNA replication event that gave rise to the double-stranded DNA molecule. In contrast, hemimethylated dyads represent the failure of epigenetic fidelity after the last replication event. Our data for hypermethylated alleles may be used to estimate methylation fidelity by assigning each of the seven hemimethylated dyads alternatively to one of two classes: (i) the failure to methylate a CpG in the daughter strand after replication of a methylated CpG (M → U), or (ii) de novo methylation of a daughter-strand CpG after replication of an unmethylated CpG (U → M). The designations of these two classes can be made by assigning parent-strand status alternatively to the top and the bottom strands, and then calculating the maximum and minimum estimates of epigenetic fidelity by appropriate groupings.
The most informative sequences are those that contain two hemimethylated dyads in which the methylated CpG is on the top strand for one dyad, and on the bottom strand for the other dyad (Fig. 2D, sequences AC 262 and AC 308). For these sequences, the assignment of parent strand status is irrelevant, because in both cases one dyad represents the loss of methylation after replication, and the other dyad represents a gain of methylation after replication. For example, sequence AC 262 has, in addition to two hemimethylated dyads, 18 dyads that are fully methylated, and two that are fully unmethylated. Thus, 18 of 19 methylated CpGs in the parent strand became methylated in the complementary CpG in the daughter strand (fidelity = 0.95); 2 of 3 unmethylated CpGs in the parent strand retained the unmethylated state in the complementary CpG in the daughter strand (fidelity = 0.67). Because of strand-placement symmetry of the two hemimethylated dyads, these fidelity estimates are precise for this molecule even though the number of dyads is small. The low fidelity of transmitting the unmethylated state in this hypermethylated allele contrasts with the much higher fidelity of transmitting the methylated state. As will be seen below, this result mirrors the average fidelity estimates obtained for the set of hypermethylated alleles.
The means of maximum and minimum values are reasonable estimates of fidelities because all hairpin-bisulfite PCR sequences will have information from both a parent strand and a daughter strand, with reference to the preceding event of DNA replication. For any collection of sequences, half of the top strands will correspond to parent strands, and half to daughter strands. The range and mean of estimates so obtained for the hypermethylated alleles are 0.95–0.98 (mean = 0.96) for the fidelity of inheritance of methylcytosine from methylcytosine, and 0.77–0.89 (mean = 0.83) for inheritance of unmethylated cytosine from unmethylated cytosine (Table 1). Similar values were obtained for a set of hypermethylated alleles from another normal female (data not shown).
Table 1. Estimates of methylation fidelities in CpG islands of FMR1 in normal female lymphocytes.
| CpG Island | No. informative dyads (sequences) | M→M Em mean (min, max; no. dyads) | U→U Eu mean (min, max; no. dyads) | U→M Ed mean (min, max; no. dyads) |
|---|---|---|---|---|
| Hypermethylated FMR1 (inactive X) | 110 (5) | 0.96 (0.95—0.98; 93) | 0.83 (0.77—0.89; 24) | 0.17 (0.11—0.23; 24) |
| Hypomethylated FMR1 (active X) | 625 (29) | 0 (0—0; 3) | >0.99 (0.995—1.0; 625) | <0.01 (0.00—0.005; 625) |
Estimated methylation fidelities for parent-strand methyl-cytosine giving rise to methyl-cytosine in the daughter strand [Em = P (M→M)], parent-strand unmethylated cytosine giving rise to unmethylated cytosine in the daughter strand [Eu = P(U→U)], and parent-strand unmethylated cytosine giving rise to methyl-cytosine in the daughter strand, i.e., de novo methylation [Ed = P(U → M) = 1—P(U→U)]. The number of informative CpG dyads, the number of allelic sequences from which they are derived, the means and ranges of estimates, and the number of informative dyads critical for each estimate are given. For the hypomethylated alleles, the expected number of nonconverted CpG cytosines (five) resulting from the frequency of incomplete bisulfite conversion (0.004), was substracted from the observed number (eight) to provide our estimates for this class of alleles.
These estimates of epigenetic fidelity in lymphocytes are relevant to the analysis of methylation density at equilibrium, i.e., when the density of methylation within a CpG island remains constant through many cell divisions (9, 19). In particular, Pfeiffer et al. (9) relate the equilibrium level of methylation density for a CpG island to the probability of Em (which corresponds to our estimated fidelity of inheriting the methylated state of cytosine, 0.96), and the probability of Ed (which corresponds to 1 - the fidelity of transmitting the unmethylated state of cytosine, or 0.17). Using equation 4 of Pfeiffer et al. (9), and our estimates of Em and Ed, we calculate an expected methylation density of 0.81 at equilibrium for our hypermethylated FMR1 alleles from lymphocytes. The observed value of 0.805 obtained from our data (Fig. 2D), indicates that methylation of the hypermethylated FMR1 CpG island is indeed at equilibrium in the female lymphocytes from which DNA was obtained. This result also lends credence to our estimates of Em and Ed, and in particular indicates that unmethylated CpG cytosines that are within hypermethylated CpG islands in lymphocytes can have a relatively high probability of de novo methylation, 0.17 per site per replication event.
De novo methylation after DNA replication may be important in maintaining high methylation density in hypermethylated islands, thereby preventing their gradual conversion to a less densely methylated or unmethylated island (9, 19). Our results provide a relatively direct measure of this de novo methylation rate. Reduction or loss of CpG island methylation by changes in de novo methylation could increase the probability of transcriptional reactivation either directly or by altering replication timing (36). Recent discovery and manipulation of DNA methyltransferases will also provide information on methylation transitions (37–39). Understanding these transitions may be useful in designing treatment strategies for epigenetic diseases (40).
Our estimated fidelity of inheritance of the methylated state of each cytosine in a hypermethylated CpG island in uncultured lymphocytes (0.96) falls between previous estimates primarily made on cultured cells. Estimates from cultured cells ranged from 0.94 to >0.99 (23, 41, 42); an estimate for ribosomal DNA from uncultured cells was ≥0.98 (18). It is known that establishing cells in culture often alters methylation patterns (43), so it is likely that our estimates from uncultured lymphocytes more accurately reflect methylation fidelities in the organism.
Our estimate of 0.83 (Ed = 0.17) for the fidelity of inheritance of the unmethylated state of cytosine in a hypermethylated island is an estimate of this important process in noncultured cells. This estimate differs substantially from the 0.95 estimate of Pfeifer et al. (Ed = 0.05) inferred from human-hamster hybrid cells (42). This difference could also arise from our use of DNA from fresh lymphocytes rather than from the cultured cells used by those authors. Additional hairpin-bisulfite PCR data will be useful in refining these estimates for different loci, obtained from different cell types and individuals.
Hypomethylated FMR1 Alleles. The hypomethylated alleles from our lymphocyte FMR1 sequences (Fig. 2E and Fig. 7, which is published as supporting information on the PNAS web site) provide additional information. From these alleles we can estimate the probability of CpG cytosines of a hypomethylated CpG island remaining unmethylated after replication. Maintaining CpG islands on the active-X chromosome essentially free of methylation is important for transcriptionally active and early replicating alleles (44).
Among our hypomethylated FMR1 alleles (Figs. 2D and 7), very few CpG cytosines (8/1,250; 0.006) were unconverted. The eight unconverted CpG cytosines occurred at about the same low frequency as observed for the non-CpG cytosines (10/2,348; 0.004) that remained unconverted in these sequences. The low level of unconverted, non-CpG cytosines likely reflects the background level of conversion failure in our experiments, although occasional methylation of non-CpG cytosines at FMR1 has not been excluded (32). This small difference in frequency between conversion of CpG and non-CpG cytosines is not statistically significant (0.5 ≤ P ≤ 0.7; χ2 = 0.29, 1 df), indicating that the fidelity of inheritance of the unmethylated state of cytosine in this sample of alleles was >0.99 (Table 1).
This fidelity estimate of >0.99 is considerably more than our fidelity estimate of 0.83 for transmitting the unmethylated state of cytosine when embedded in hypermethylated CpG islands. This result confirms the prediction of Pfeifer et al. (9) based on their modeling of methylation kinetics and agrees with estimates of Ushijima et al. (23) using conventional PCR of DNA from cultured cells. Our finding of no significant level of hemimethylation in hypomethylated FMR1 alleles of lymphocytes, and therefore of no significant level of de novo methylation at these sites (Ed of <0.01), suggests that these alleles remain essentially free of methylation by inhibiting de novo methylation. Such inhibition would clearly have to be selective for hypomethylated alleles, because, as described above, de novo methylation is occurring in these same cells at a significant rate on hypermethylated FMR1 alleles.
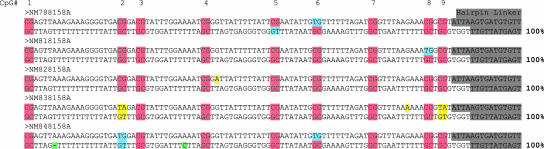
Hairpin-Bisulfite PCR of Repeated Sequences: L1 Elements. Hairpin-bisulfite PCR also offers an important advance in the study of methylation of repeated sequences: information is gained on the exact complementary strand of each analyzed sequence, in contrast to the population averages that are obtained from conventional, single-strand PCR. With repeated sequences, variation among repeat family members is superimposed on variations between cells; this makes methylation patterns derived from population averages difficult to interpret precisely. We analyzed a portion of the CpG island of human L1 sequences to address a controversy over the level of hemimethylation in fibroblasts (Fig. 3). Using hairpin-bisulfite PCR, the observed level of hemimethylation for this region averaged 12.1% for DNAs from two different adult fibroblast lines (14.1% and 10.0%). Although this average value is higher than the 6.4% hemimethylation reported above for the hypermethylated alleles of FMR1 in lymphocytes, it is strikingly lower than the 58% estimate derived from figure 3 of ref. 21 for this region of the L1 promoter in cultured human fibroblasts. Our value is, however, in accord with results of Hansen (45). Final resolution of this discrepancy will likely require hairpin-bisulfite PCR on DNAs from embryonic fibroblasts used by Woodcock et al. (21).
Fig. 3.
Inferred methylation states of CpGs within the human L1 promoter. DNA from an untransformed female fibroblast cell line, 8158A, was analyzed by hairpin-bisulfite PCR of L1 elements, representing nucleotides 219–323 (21). The first five analyzed L1 sequences showing ≥99% conversion of non-CpG cytosines are presented here. Highlighting is as described for Fig. 2, except for yellow, which represents sequence variation of individual L1 repeats. Because the presence/absence of complementarity within hairpin sequences can be used to distinguish between evolutionary mutations and PCR errors, the non-CpG cytosine in sequence NM848158A was counted as a PCR error rather than as a nonconversion.
Our analysis of sequences from the L1 family also provides an important validation of our method. We have analyzed >200 well-converted hairpin-bisulfite PCR sequences from the promoter region of L1 elements. In all cases the sequences were fully consistent with complementarity between the upper and lower strands of the PCR products, as expected for accurate hairpin-bisulfite PCR (see Fig. 3). No sequence showed evidence of cross-over PCR, which could have brought two divergent members of the L1 family together in one hairpin sequence. Thus, the information that we recover in a genomic hairpin sequence accurately reflects sequence and methylation status on the complementary strands of an individual DNA molecule.
Concluding Remarks
We demonstrate here that hairpin-bisulfite PCR can be used to assess patterns of cytosine methylation on both strands of individual DNA molecules from single-copy and repeated sequences in human genomic DNA. Our results provide estimates of the epigenetic fidelity of cytosine methylation, and thus provide information on how stable epigenetic states are inherited through many cell divisions. Two other major problems of epigenetics, the establishment of epigenetic marks, and the programmed or abnormal transitions from one epigenetic state to another, may also be approached with this method.
Supplementary Material
Acknowledgments
We thank Alice Burden, Lisa Chakrabarti, Susan Clark, Arthur Riggs, Stanley Gartler, Brooks Miner, Larry Loeb, Tom Daniel, and three anonymous reviewers for suggestions. This project was initiated while C.D.L., R.S., J.C.V., and T.M. were at the Hutchinson Cancer Center, Seattle. C.D.L. also thanks the Department of Zoology, University of Bergen, Bergen, Norway, for its hospitality during a final stage of data analysis and writing. Support was provided by a fellowship grant from the FRAXA Foundation and National Institutes of Health Grants GM 53805, HD16659, and GM83805.
Abbreviations: Em, maintenance methylation efficiency; Ed, de novo methylation efficiency.
See Commentary on page 4.
References
- 1.Meselson, M. & Yuan, R. (1968) Nature 217, 1110-1114. [DOI] [PubMed] [Google Scholar]
- 2.Arber, W. (1974) Prog. Nucleic Acid Res. Mol. Biol. 14, 1-37. [DOI] [PubMed] [Google Scholar]
- 3.Sager, R. & Kitchin, R. (1975) Science 189, 426-433. [PubMed] [Google Scholar]
- 4.Riggs, A. D. (1975) Cytogenet. Cell Genet. 14, 9-25. [DOI] [PubMed] [Google Scholar]
- 5.Holliday, R. & Pugh, J. E. (1975) Science 187, 226-232. [PubMed] [Google Scholar]
- 6.Watson, J. D. & Crick, F. H. C. (1953) Nature 171, 737-738. [DOI] [PubMed] [Google Scholar]
- 7.Bird, A. (1992) Cell 70, 5-8. [DOI] [PubMed] [Google Scholar]
- 8.Swain, J. L., Stewart, T. A. & Leder, P. (1987) Cell 50, 719-727. [DOI] [PubMed] [Google Scholar]
- 9.Pfeifer, G. P., Steigerwald, S. D., Hansen, R. S., Gartler, S. M. & Riggs, A. D. (1990) Proc. Natl. Acad. Sci. USA 87, 8252-8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stöger, R., Kubicka, P., Liu, C.-G., Kafri, T., Razin, A., Cedar, H. & Barlow, D. P. (1993) Cell 73, 61-71. [DOI] [PubMed] [Google Scholar]
- 11.Jacobsen, S. E. & Meyerowitz, E. M. (1997) Science 277, 1100-1103. [DOI] [PubMed] [Google Scholar]
- 12.Cubas, P., Vincent, C. & Coen, E. (1999) Nature 401, 157-161. [DOI] [PubMed] [Google Scholar]
- 13.Freitag, M., Williams, R. L., Kothe, G. O. & Selker, E. U. (2002) Proc. Natl. Acad. Sci. USA 99, 8802-8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLoia, J. A. & Solter, D. (1990) Development (Cambridge, U.K.) Suppl. 73-79. [PubMed]
- 15.Hansen, R. S., Gartler, S. M., Scott, C. R., Chen, S.-H. & Laird, C. D. (1992) Hum. Mol. Genet. 1, 571-578. [DOI] [PubMed] [Google Scholar]
- 16.Baylin, S. B., Herman, J. G., Graff, J. R., Vertino, P. M. & Issa, J. P. (1998) Adv. Cancer Res. 72, 141-196. [PubMed] [Google Scholar]
- 17.Hansen, R. S., Wijmenga, C., Luo, P., Stanek, A. M., Canfield, T. K., Weemaes, C. M. & Gartler, S. M. (1999) Proc. Natl. Acad. Sci. USA 96, 14412-14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bird, A. P. (1978) J. Mol. Biol. 118, 49-60. [DOI] [PubMed] [Google Scholar]
- 19.Otto, S. P. & Walbot, V. (1990) Genetics 124, 429-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steigerwald, S. D., Pfeifer, G. P. & Riggs, A. D. (1990) Nucleic Acids Res. 18, 1435-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodcock, D. M., Lawler, C. B., Linsenmeyer, M. E., Doherty, J. P. & Warren, W. D. (1997) J. Biol. Chem. 272, 7810-7816. [DOI] [PubMed] [Google Scholar]
- 22.Liang, G., Chan, M. F., Tomigahara, Y., Tsai, Y. C., Gonzales, F. A., Li, E., Laird, P. W. & Jones, P. A. (2002) Mol. Cell. Biol. 22, 480-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ushijima, T., Watanabe, N., Okochi, E., Kaneda, A., Sugimura, T. & Miyamoto, K. (2003) Genome Res. 13, 868-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith, S. S. (1991) Mol. Carcinog. 4, 91-92. [DOI] [PubMed] [Google Scholar]
- 25.Weiss, A., Keshet, I., Razin, A. & Cedar, H. (1996) Cell 86, 709-718. [DOI] [PubMed] [Google Scholar]
- 26.Jost, J. P., Fremont, M., Siegmann, M. & Hofsteenge, J. (1997) Nucleic Acids Res. 25, 4545-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riggs, A. D., Xiong, Z., Wang, L. & LeBon, J. M. (1998) Novartis Found. Symp. 214, 214-225; discussion 225-232. [DOI] [PubMed] [Google Scholar]
- 28.Clark, S. J., Harrison, J. & Frommer, M. (1995) Nat. Genet. 10, 20-27. [DOI] [PubMed] [Google Scholar]
- 29.Frommer, M., McDonald, L. E., Millar, D. S., Collis, C. M., Watt, F., Grigg, G. W., Molloy, P. L. & Paul, C. L. (1992) Proc. Natl. Acad. Sci. USA 89, 1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka, H., Tapscott, S. J., Trask, B. J. & Yao, M. C. (2002) Proc. Natl. Acad. Sci. USA 99, 8772-8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaur, M. & Makrigiorgos, G. M. (2003) Nucleic Acids Res. 31, e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stöger, R., Kajimura, T. M., Brown, W. T. & Laird, C. D. (1997) Hum. Mol. Genet. 6, 1791-1801. [DOI] [PubMed] [Google Scholar]
- 33.Clark, S. J., Harrison, J., Paul, C. L. & Frommer, M. (1994) Nucleic Acids Res. 22, 2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oberlé, I., Rousseau, F., Heitz, D., Kretz, C., Devys, D., Hanauer, A., Boué, J., Bertheas, M. F. & Mandel, J. L. (1991) Science 252, 1097-1102. [DOI] [PubMed] [Google Scholar]
- 35.Verkerk, A. J. M. H., Pieretti, M., Sutcliffe, J. S., Fu, Y.-H., Kuhl, D. P. A., Pizzuti, A., Reiner, O., Richards, S., Victoria, M. F., Zhang, R., et al. (1991) Cell 65, 904-914. [DOI] [PubMed] [Google Scholar]
- 36.Hansen, R. S., Stöger, R., Wijmenga, C., Stanek, A. M., Canfield, T. K., Luo, P., Matarazzo, M. R., D'Esposito, M., Feil, R., Gimelli, G., et al. (2000) Hum. Mol. Genet. 9, 2575-2587. [DOI] [PubMed] [Google Scholar]
- 37.Bestor, T. H. (2000) Hum. Mol. Genet. 9, 2395-2402. [DOI] [PubMed] [Google Scholar]
- 38.Jaenisch, R. & Bird, A. (2003) Nat. Genet. 33, Suppl., 245-254. [DOI] [PubMed] [Google Scholar]
- 39.Chen, T., Ueda, Y., Dodge, J. E., Wang, Z. & Li, E. (2003) Mol. Cell. Biol. 23, 5594-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laird, C. D. (1987) Genetics 117, 587-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wigler, M., Levy, D. & Perucho, M. (1981) Cell 24, 33-40. [DOI] [PubMed] [Google Scholar]
- 42.Pfeifer, G. P., Tanguay, R. L., Steigerwald, S. D. & Riggs, A. D. (1990) Genes Dev. 4, 1277-1287. [DOI] [PubMed] [Google Scholar]
- 43.Antequera, F., Boyes, J. & Bird, A. (1990) Cell 62, 503-514. [DOI] [PubMed] [Google Scholar]
- 44.Holliday, R. & Ho, T. (1991) Somat. Cell Mol. Genet. 17, 537-542. [DOI] [PubMed] [Google Scholar]
- 45.Hansen, R. S. (2003) Hum. Mol. Genet. 12, 2559-2567. [DOI] [PubMed] [Google Scholar]
- 46.Maniatis, T., Fritsch, E. F. & Sambrook, J. (1982) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.