Abstract
Virus infection, double-stranded RNA, and lipopolysaccharide each induce the expression of genes encoding IFN-α and -β and chemokines, such as RANTES (regulated on activation, normal T cell expressed and secreted) and IP-10 (IFN-γ inducible protein 10). This induction requires the coordinate activation of several transcription factors, including IFN-regulatory factor 3 (IRF3). The signaling pathways leading to IRF3 activation are triggered by the binding of pathogen-specific products to Toll-like receptors and culminate in the phosphorylation of specific serine residues in the C terminus of IRF3. Recent studies of human cell lines in culture have implicated two noncanonical IκB kinase (IKK)-related kinases, IKK-ε and Traf family member-associated NF-κB activator (TANK)-binding kinase 1 (TBK1), in the phosphorylation of IRF3. Here, we show that purified recombinant IKK-ε and TBK1 directly phosphorylate the critical serine residues in IRF3. We have also examined the expression of IRF3-dependent genes in mouse embryonic fibroblasts (MEFs) derived from Tbk1-/- mice, and we show that TBK1 is required for the activation and nuclear translocation of IRF3 in these cells. Moreover, Tbk1-/- MEFs show marked defects in IFN-α and -β, IP-10, and RANTES gene expression after infection with either Sendai or Newcastle disease viruses or after engagement of the Toll-like receptors 3 and 4 by double-stranded RNA and lipopolysaccharide, respectively. Finally, TRIF (TIR domain-containing adapter-inducing IFN-β), fails to activate IRF3-dependent genes in Tbk1-/- MEFs. We conclude that TBK1 is essential for IRF3-dependent antiviral gene expression.
Host defense against infectious microbial pathogens requires the detection of invading microorganisms and the activation of the innate immune response (1). One way in which detection is achieved is by the binding of pathogen-specific molecules to extracellular receptors belonging to the Toll-like receptor (TLR) family (2-4). TLRs recognize microbial products derived from all of the major classes of microbes, including bacteria, viruses, yeast, and fungi. These recognition events trigger the activation of complex networks of signal transduction pathways, leading to the coordinate activation of several transcription factors that regulate the expression of antimicrobial genes, cytokines, chemokines, and costimulatory molecules (5).
The induction of type I IFN in response to virus, bacterial infection, double-stranded RNA (dsRNA), or lipopolysaccharide (LPS) provides an example of this phenomenon (5). dsRNA and LPS are detected by TLR-3 (6) and TLR-4 (7), respectively, although distinct viral ligands have been described also for additional members of the TLR family (8-11). Expression of the IFN-β gene in response to all of these inducers requires the coordinate activation of several transcription factors, including IRF3 (5, 12). Until recently, relatively little was known about the signaling pathways or upstream effector molecules required for IRF3 activation.
IRF3 is ubiquitously expressed and resides in the cytoplasm in an inactive state (13), and its activation requires phosphorylation of a cluster of serine/threonine residues in its C terminus (between residues 385 and 405) (14). This phosphorylation leads to its dimerization and association with the coactivators CBP-p300 (15). The IRF3 complex then translocates to the nucleus where it activates promoters containing IRF3 binding sites, such as the IFN-β, RANTES (regulated on activation, normal T cell expressed and secreted), and IP-10 (IFN-γ inducible protein 10) promoters. Recent studies have shown that the noncanonical IκB kinase (IKK)-like kinases IKK-ε (16, 17) and Traf family member-associated NF-κB activator (TANK) (tankyrase)-binding kinase 1 (TBK1) (18-20) are required for the activation of IRF3 and the induction of IFN-β gene expression in human cell lines in culture (21, 22). IKK-ε and TBK1, although highly homologous, appear to function in a nonredundant manner (22). Small interfering RNA silencing studies suggest that both of these kinases are essential components of an IRF3 signaling pathway activated by Sendai virus (SV) or dsRNA, which signals via TLR-3 (21, 22). Previous studies in cell extracts provided evidence that IKK-ε phosphorylates IRF3 directly (21). However, similar evidence for TBK1 has not been presented.
Although all TLRs activate NF-κB, not all TLRs activate IRF3 or induce type I IFN expression. TLR-3 and TLR-4 are the best characterized TLRs known to activate IRF3. TLR-3- (23) and TLR-4-mediated activation of IRF3 do not require MyD88 (24, 25) or the related adapter molecule Mal/TIRAP (26-28). Instead, these TLRs use alternative TIR-domain-containing adapter proteins to induce IRF3. For example, TLR-3 requires TRIF (28), known also as TICAM-1 (23), for IRF3 activation, whereas TLR-4 uses TRIF and TRAM [TRIF-related adapter molecule (29), known also as TIRP (30) or TICAM-2 (31)]. Recent studies with TRIF-deficient mice (32) and small interfering RNA silencing studies with TRAM (29) provide clear evidence that the recruitment of TRIF to TLR-3 or TRIF and TRAM to TLR-4 facilitates the activation of IRF3 and the induction of type I IFNs. TRIF-mediated induction of IRF3-dependent genes appears to require IKK-ε and TBK1 (22).
Here, we show that recombinant IKK-ε and TBK1 directly phosphorylate IRF3 on C-terminal residues known to be required for its activation. In addition, we show that IRF3 activation is defective in Tbk1-/- mouse embryonic fibroblasts (MEFs) compared with wild-type cells in response to virus infection or stimulation with dsRNA or LPS. Moreover, unlike MEFs derived from wild-type cells, overexpression of TRIF fails to induce the expression of IFN-β, RANTES, or IP-10 reporter genes in Tbk1-deficient cells. These data provide strong genetic evidence that TBK1 is an essential mediator of innate antiviral gene expression programs that require IRF3.
Materials and Methods
Cells and Stimuli. MEFs from wild-type and Tbk1-deficient mice were provided by Wen-Chen Yeh (University of Toronto, Toronto) as described (19). SV was obtained from Charles River Breeding Laboratories. Newcastle Disease virus (NDV) was obtained from the American Type Culture Collection. LPS derived from Escherichia coli strain O11:B4 was obtained from Sigma and was prepared as described (33). Endotoxin-free poly(I-C) was obtained from Amersham Biosciences.
Reagents. The luciferase reporter genes IFN-β, IFN-β-PRDIII-I, and IFN-β-PRDII were generated as described (22). The ISG54 ISRE was obtained from Stratagene. The RANTES luciferase and IRF3-GFP were provided by John Hiscott (McGill University, Montreal), as described (34). The HIV LTR-κB-luc was obtained as described (26), and endothelial-leukocyte adhesion molecule (ELAM)-luc was provided by D. Golenbock (University of Massachusetts Medical School). The IP-10 reporter construct was provided by A. Luster (Massachusetts General Hospital, Boston). pEF-Bos-Flag murine TRIF was generated by PCR cloning from a RAW264.7 cDNA library. Hemagglutinin (HA)-IRF3ΔN was provided by T. Fujita (Tokyo Metropolitan Institute of Medical Science, Tokyo). ELISAs for measuring RANTES and IP-10 were obtained from R & D Systems. IFN-α ELISA was obtained from PBL Biomedical (Piscataway, NJ).
Recombinant Proteins. GST fusion proteins and baculovirus proteins were prepared as described in ref. 35.
Kinase Assays. In vitro kinase assays were performed as described (35) by using 0.5 μg of GST-IκBα substrates or 2.0 μg of GST-IRF3 substrates.
Transfection Assays. MEFs from Tbk1+/+and Tbk1-/- mice (7,500 cells per well) were plated in 24-well plates (100 μl per well) and transfected 24 h later with 0.25 μg of reporter genes, as indicated, by using FuGENE6 (Roche Diagnostics). Murine TRIF (0.5 μg) was cotransfected with reporter genes where indicated. In all cases, cells were transfected also with 0.25 μg of a thymidine kinase Renilla luciferase reporter gene (Promega). After transfection (24 h), some cells were infected with NDV [2 μl per well of American Type Culture Collection stock assayed at ≥108 chicken embryo infectious doses (50%)/0.2 ml, as confirmed by HA] or SV [(200 hemagglutinating units (HAU)], stimulated with LPS (100 ng/ml) or poly(dI-dC) (50 μg/ml) as indicated. Then, ≈16 h later cell lysates were prepared, luciferase reporter gene activity was measured, and data were normalized for transfection efficiency by using the Dual-Glo luciferase assay system (Promega). Data are expressed as mean relative stimulation ± SD for a representative experiment from a minimum of three separate experiments, each performed in triplicate. Whole-cell lysates were also subjected to immunoblotting to confirm expression of Flag-tagged TRIF (data not shown).
Confocal Microscopy. MEFs from Tbk1+/+ and Tbk1-/- mice were transiently transfected with an expression vector for IRF3-GFP, as indicated. After transfection (24 h), cells were infected with SV for 6 h and imaged by confocal microscopy with a TCS SP2 AOBS microscope (Leica, Deerfield, IL).
Immunoblotting. MEFs from Tbk1+/+and Tbk1-/- mice were infected with SV for the indicated times. Cells were lysed (50 mM Hepes, pH 7.5/100 mM NaCl/1 mM EDTA/10% glycerol/0.5% Nonidet P-40/protease inhibitors), separated by SDS/8.5% PAGE, and immunoblotted with antibodies to HA (Y-11; Santa Cruz Biotechnology) or calnexin (C-20; Santa Cruz Biotechnology), as indicated.
Northern Blot Analysis. MEFs from Tbk1+/+ and Tbk1-/- mice were infected with SV (200 HAU) for the indicated times. Total RNA was isolated by using TRIzol (Invitrogen) and run on glyoxal/dimethyl sulfoxide gels. Blots were hybridized with Ultrahyb (Ambion, Austin, TX) and visualized by autoradiography.
ELISA. MEFs from Tbk1+/+ and Tbk1-/- mice were stimulated with poly(I-C) (20 or 100 μg/ml) or LPS (20 or 100 ng/ml) or infected with SV (2 or 20 HAU) or NDV (0.5 or 5.0 μl of the American Type Culture Collection stock) for ≈16 h. For the analysis shown in Fig. 6D, 20 HAU of SV or 5.0 μl of NDV was used. Cell supernatants were harvested, and RANTES, IP-10, and IFN-α levels were measured according to the ELISA manufacturer's recommendations.
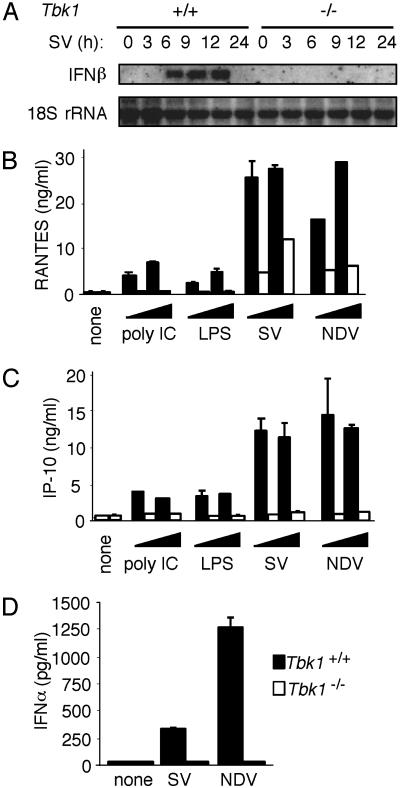
Fig. 6.
Induction of IFN-α and -β, RANTES, and IP-10 is TBK1-dependent. (A) MEFs from Tbk1+/+ (Left) and Tbk1-/- (Right) mice were infected with SV for the indicated times. Total RNA (5 μg) was loaded in each lane, and Northern blot analysis was performed by using a mouse IFN-β-specific probe. (B-D) MEFs from Tbk1+/+ (filled bars) and Tbk1-/- (empty bars) mice were stimulated with poly(I-C) or LPS, infected with SV or NDV as indicated, or left untreated (none) for ≈16 h. Cell supernatants were harvested, and RANTES, IP-10, or IFN-α levels were measured by ELISA.
Results
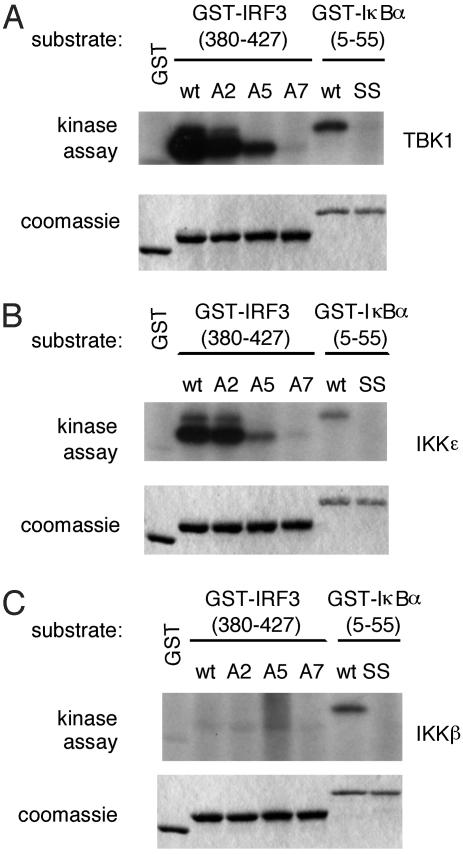
IKK-ε and TBK1 Directly Phosphorylate IRF3 in Vitro. To determine whether IKK-ε and TBK1 directly phosphorylate IRF3, we purified recombinant IKK-ε, TBK1, and IKK-β and performed in vitro kinase assays by using the C terminus of IRF3, fused in-frame to GST, as a substrate. Two distinct groups of serine residues have been implicated in the activation of IRF3. Group 1 includes serines 385 and 386, whereas group 2 includes serines 396, 398, 402, and 405 and threonine 404. As shown in Fig. 1A, purified baculovirus-expressed TBK1 or IKK-ε phosphorylated wild-type IRF3, and replacement of group 2 residues with alanine (A5) led to a significant decrease in phosphorylation compared with wild-type IRF3. By contrast, only a slight decrease in phosphorylation was observed with IRF3 in which group 1 serine residues were replaced by alanine (A2; Fig. 1 A and B). No phosphorylation of IRF3 was observed when both groups of residues were replaced by alanines (A7; Fig. 1 A and B). Thus, it appears that the group 2 serines are the primary targets of phosphorylation by IKK-ε and TBK1. This finding is consistent with mass spectrometry analysis of full-length IRF3 phosphorylated by TBK1 (unpublished data) and with previous studies implicating group 2 residues as the primary targets of phosphorylation (21).
Fig. 1.
Purified IKK-ε and TBK1 phosphorylate the C terminus of IRF3. Wild-type (wt) TBK1 (A), IKK-ε (B), or IKK-β (C) was expressed in insect cells by using baculovirus vectors. Kinase activity of the purified proteins was assayed by using various GST-IRF3-(380-427) or GST-IκBα-(5-55) substrates as indicated. A2, S385A, S386A IRF3 mutant; A5, S396A, S398A, S402A, T404A, S405A IRF3 mutant; A7, S385A, S386A, S396A, S398A, S402A, T404A, S405A IRF3 mutant; SS, S32A, S36A IκBα mutant.
As expected from previous studies (21), recombinant IKK-β failed to phosphorylate IRF3 (Fig. 1C). As a positive control for IKK-β kinase activity, we showed that recombinant IKK-β could phosphorylate IκBα specifically (Fig. 1C). We showed also that recombinant IKK-ε and TBK1 could phosphorylate IκBα (Fig. 1 A and B). However, as reported, this phosphorylation occurred on serine 36 only and did not result in the degradation of IκBα (36). The functional significance, if any, of this phosphorylation event is not understood. Taken together, these observations suggest that IKK-ε and TBK1 phosphorylate the C terminus of IRF3 directly.
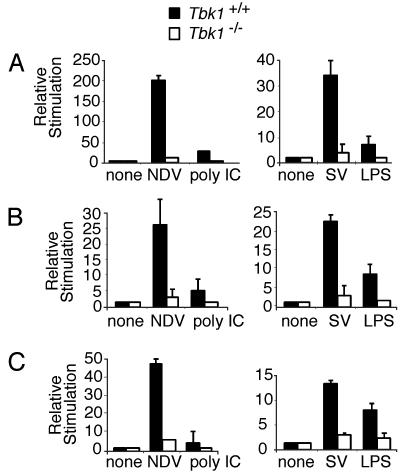
TBK1 Is Required for IRF3-Dependent Gene Expression in MEFs. The role of TBK1 in IRF3 activation was investigated by using a previously reported Tbk1-/- knockout mouse (19). Previous studies showed that Tbk1-deficient mice die at approximately embryonic day 14.5 from massive tumor necrosis factor (TNF)α-induced liver apoptosis (19). Based on the similarity of this phenotype with that observed with RelA-/- mice, TBK1 was proposed to play an essential role in the NF-κB pathway (19). The role of TBK1 in the activation of other transcription factors was not investigated. We, therefore, carried out studies to determine the effect of Tbk1 deficiency on IRF3 activation in MEFs. As shown in Fig. 2A, induction of an IFN-β reporter gene in response to infection with SV was completely defective in the Tbk1-deficient MEFs. When NDV, another well characterized inducer of type I IFN expression, was examined, IFN-β reporter gene expression was completely abrogated in Tbk1-deficient cells also (Fig. 2 A). We also investigated the role of TBK1 in signaling by TLR-3 and TLR-4, after stimulation with poly(I-C) or LPS, respectively. Induction of the IFN-β reporter gene was reduced significantly in Tbk1-deficient cells after stimulation with either poly(I-C) or LPS (Fig. 2 A). Furthermore, induction of both the RANTES and IP-10 reporter genes also showed similar defects in Tbk1-deficient cells with all tested stimuli (Fig. 2 B and C). Together, these results show that type I IFN and related genes require TBK1 for their expression.
Fig. 2.
Reduced IFN-β reporter gene activity in the absence of TBK1. MEFs from Tbk1+/+(filled bars) and Tbk1-/- (empty bars) mice were transfected with IFN-β (A), RANTES (B), or IP-10 (C) luciferase reporter genes. Cells were left untreated (none), infected with NDV or SV, or stimulated with poly(I-C) or LPS. Lysates were assayed for luciferase activity.
A common feature of the IFN-β, RANTES, and IP-10 genes is that multiple transcriptional regulatory elements are required for their activation. For example, the coordinate activation of the transcriptional activators NF-κB, IRF3, and ATF2/cJun are required for IFN-β gene expression after virus infection (5). Various studies have established an essential role for IRF3 in LPS- and poly(I-C)-induced IFN-β, RANTES, and IP-10 gene expression (37, 38). The transcription enhancer of the IFN-β gene contains four positive regulatory domains (PRDs), PRDI-IV. NF-κB binds to the PRDII element, IRF3 binds to adjacent PRDIII and PRDI elements (referred to collectively as PRDIII-I), and the heterodimeric transcription factor ATF2/cJun binds to PRDIV. Each of these promoter elements, when present in multiple copies, can be activated by virus infection or by transfected IKK-ε or TBK1 (5, 22, 39).
We examined the effect of SV, NDV, poly(I-C), and LPS on transcription from these individual IFN-β regulatory elements. NDV, SV, poly(I-C), and LPS induced transcription of a multimerized PRDIII-I reporter gene in wild-type cells. However, the reporter gene was not expressed in Tbk1-deficient cells with all tested stimuli (Fig. 3A). Consistent with these observations, SV and LPS induced the activation of another IRF3-dependent promoter element, the IFN-stimulated regulatory element (ISRE) from the ISG-54 gene. The expression of this reporter gene was also defective in Tbk1-deficient cells (Fig. 3A Right). We also examined the effects of SV, NDV, poly(I-C), and LPS on the NF-κB-regulated IFN-β-PRDII reporter gene. All of these inducers activated expression of the NF-κB reporter gene, but in contrast to the dramatic defects on PRDIII-I and ISG54-ISRE expression in Tbk1-deficient cells, induction of PRDII was unaffected in these cells (Fig. 3B). We also examined the induction of two additional NF-κB-regulated genes, a reporter gene consisting of the HIV LTR, which contains five copies of the NF-κB consensus sequence in tandem (HIV LTR-κB) and the ELAM (E-selectin) reporter gene. Levels of SV-, poly(I-C)-, and LPS-dependent induction of these reporters were similar in wild-type and Tbk1-deficient cells (Fig. 3B Lower). We conclude that TBK1 is essential for IFN-β, RANTES, and IP-10 reporter gene expression, and we show that TBK1 activates IRF3 but is not required for virus-, poly(I-C)- or LPS-induced NF-κB activation.
Fig. 3.
Deficiency of TBK1 leads to impaired induction of IRF3-dependent reporters and normal induction of NF-κB-dependent reporters. MEFs from Tbk1+/+ (filled bars) and Tbk1-/- (empty bars) mice were transfected with IFN-β PRDIII-I or ISG-54 ISRE reporter genes (A), IFN-β PRDII (B), or HIV LTR-κB or ELAM luciferase reporter genes (B Lower). Cells were left untreated (none), infected with SV or NDV, or stimulated with poly(I-C) or LPS. Lysates were assayed for luciferase activity.
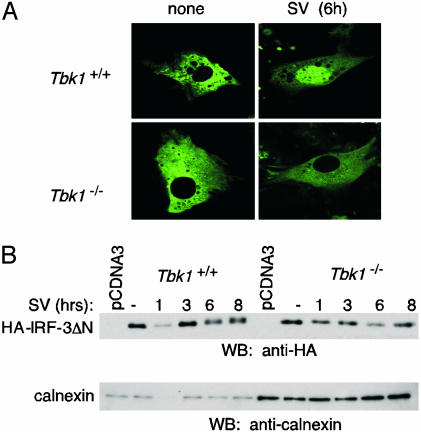
TBK1 Is Essential for IRF3 Activation. The inhibition of IFN-β-PRDIII-I and ISRE-, but not NF-κB-containing, promoters in Tbk1-deficient mice suggested that in agreement with our observations (22), TBK1 mediates its effects by means of IRF3. We, therefore, examined the activation of IRF3 in Tbk1-deficient cells. Viral infection leads to the phosphorylation of IRF3, followed by its dimerization, nuclear translocation, and interaction with the coactivators CBP-p300 (15). Embryonic fibroblasts from wild-type and Tbk1-deficient mice were cotransfected with a vector encoding an IRF3-GFP fusion protein, and nuclear translocation of IRF3 was examined after virus infection. In uninfected cells, IRF3-GFP localized to the cytosol of either wild-type or Tbk1-deficient cells. In both cases, the nucleus was nearly devoid of GFP fusion protein (Fig. 4A). However, when cells were infected with SV for 6 h, IRF3-GFP clearly translocated to the nucleus in wild-type cells. In contrast, IRF3-GFP remained in the cytosol in Tbk1-deficient cells after virus infection (Fig. 4A).
Fig. 4.
Deficiency of TBK1 leads to defective virus-induced nuclear translocation and phosphorylation of IRF3. (A) MEFs from Tbk1+/+and Tbk1-/- mice were transfected with 1.0 μg of an IRF3-GFP fusion protein. Samples were left untreated (none) or infected with 100 HAU of SV and visualized by confocal microscopy. (B) MEFs from Tbk1+/+and Tbk1-/- mice were transfected with 500 ng of HA-IRF-3ΔN. Cells were infected 24 h later with SV for the indicated times. Whole-cell lysates were analyzed by Western blotting (WB) with anti-HA antibodies. As a loading control, membranes were stripped and reprobed with anti-calnexin antibodies.
To examine the requirement of TBK1 for SV-induced IRF3 phosphorylation, extracts from SV-induced wild-type and Tbk1-deficient MEFs overexpressing HA-tagged IRF3 were assayed by Western blotting using an anti-HA antibody. A more slowly migrating IRF3 band was reproducibly detected after infection with SV for 6 or 8 h (Fig. 4B). The SV-induced phosphorylation of IRF3 was not observed in Tbk1-deficient cells.
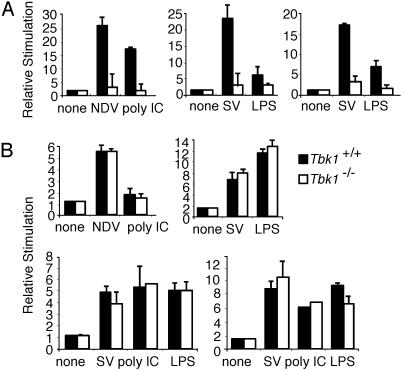
TBK1 Is Essential for IRF3 Activation Mediated by TRIF. A TIR domain-containing adapter molecule, TRIF, has been shown recently to activate IRF3 and induce expression of IRF3 target genes, such as IFN-β, RANTES, and IP-10 (22, 23, 32). Recent studies with Trif-deficient mice confirm its essential role in type I IFN induction after engagement of TLR-3 and TLR-4 by poly(I-C) and LPS, respectively (10, 32). Furthermore, Trif mutant mice, derived by chemical mutagenesis, are hypersusceptible to murine cytomegalovirus infection because of a failure to produce type I IFNs (10). Taken together, these observations suggest that TRIF is an essential shared component of the pathways by which TLRs and viruses induce type I IFN.
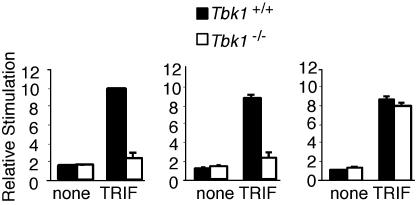
We have demonstrated previously by using small interfering RNA knockdown of IKK-ε or TBK1 that the induction of type I IFN and related genes by TRIF depends on these kinases (22). Furthermore, immunoprecipitation studies suggested that TRIF interacts with IKK-ε or TBK1 and that this interaction is abolished by deletion of the N- or C-terminal non-TIR domain portion of the molecule (22). IKK-ε and TBK1 have also been shown to phosphorylate TRIF (40). Overexpression of TRIF induces IFN-β, IP-10, and RANTES reporter gene expression in HEK293 cells (22). We examined the induction of IFN-β, IP-10, and RANTES reporter genes by TRIF in wild-type and Tbk1-deficient MEFs. Induction of IFN-β, IP-10, and RANTES reporter genes by TRIF was completely abrogated in Tbk1-deficient cells, confirming the importance of TBK1 in mediating TRIF signaling to these target genes (Fig. 5). In contrast, TRIF-induced expression of the NF-κB-dependent ELAM reporter gene was normal in Tbk1-deficient cells (Fig. 5 Right).
Fig. 5.
TRIF induces IFN-β and RANTES reporter genes by means of TBK1. MEFs from Tbk1+/+ (filled bars) and Tbk1-/- (empty bars) mice were transfected with IFN-β (Left), RANTES (Center), or ELAM (Right) luciferase reporter genes and cotransfected with a murine TRIF expression vector or left untreated (none). Lysates were assayed for luciferase activity.
TBK1 Is Essential for the Induction of Type I IFN Expression. We next examined the kinetics of endogenous IFN-β gene induction in wild-type and Tbk1-deficient cells. Infection of cells with SV for at least 6 h induced IFN-β mRNA levels. In contrast, IFN-β mRNA was not induced by SV in cells from Tbk1-deficient MEFs (Fig. 6A). Consistent with the dramatic defect in IP-10 and RANTES reporter gene expression in Tbk1-deficient cells, induction of endogenous RANTES and IP-10 expression after exposure to poly(I-C), LPS, SV, and NDV was also impaired severely in Tbk1-deficient mice. The induction of RANTES was attenuated severely, whereas IP-10 was completely defective in the absence of TBK1 (Fig. 6 B and C). Stimulation of both wild-type and Tbk1-deficient cells with TNFα also induced similar levels of RANTES expression (although the levels of induction were considerably lower than with the other stimuli), suggesting that TBK1 is not required for TNF-α-induced RANTES induction (data not shown). Induction of IFN-α by SV and NDV was also completely abrogated in Tbk1-deficient cells (Fig. 6D).
Discussion
Induction of type I IFN gene expression, a central event in establishing the innate antiviral response, requires cooperative activation of the transcription factors NF-κB and IRF3. Here, we make use of Tbk1-deficient MEFs to demonstrate definitively that the induction of type I IFN (and related genes, such as RANTES and IP-10) depends on TBK1. We show also that TBK1 directly phosphorylates the C-terminal serine residues of IRF3 that are required for its activation. This observation confirms and extends the findings of previous studies showing that TBK1-containing whole-cell extracts phosphorylate the group 2 residues of IRF3 (21).
At present, the relationship between TBK1-IKK-ε and the classical IKK-α-IKK-β-IKK-γ complex is unclear. The primary target of the IKK-α-IKK-β-IKK-γ complex is the NF-κB inhibitor, IκBα (41). A variety of inducers, including virus infection, LPS, or cytokines, such as TNF-α or IL-1, activate the IKK-α-IKK-β -IKK-γ complex. The activated kinase phosphorylates IκBα on serines 32 and 36, leading to its ubiquitination and degradation by the proteosome (41). This degradation results in the release of the NF-κB heterodimer, its nuclear translocation, and the activation of NF-κB-regulated genes. Although TBK1 and IKK-ε share significant sequence similarity with IKK-α and IKK-β, they have been shown to phosphorylate serine 36, but not serine 32, of IKK-α (16-18, 20). TBK1 and IKK-ε are, therefore, unable to activate NF-κB by the degradation of IκBα because such degradation requires phosphorylation of both serine residues of IκBα (41). However, overexpression of either of these noncanonical IKKs resulted in the activation of NF-κB reporter genes (16-18, 20). Other overexpression studies led to the conclusion that TBK1 functions as an IKK kinase (20).
Consistent with this possibility, the phenotype of Tbk1-/- mice was indistinguishable from that of IKK-γ-/-, IKK-β-/-, and RelA-/- mice, namely, death at embryonic day 14.5 from profound liver degeneration and apoptosis (42-46). However, IκBα degradation was unaffected in TNF-α-stimulated MEFs from the Tbk1-deficient mice (19). Thus, TBK1 is not required for the activation of IKK-α-IKK-β-IKK-γ. Surprisingly, however, additional studies indicated that the expression of two NF-κB-regulated genes, namely TLR-2 and ICAM-1, was defective in Tbk1-deficient MEFs after stimulation with IL-1β or TNF-α (19). Based on these and other observations, it was suggested that TBK1 regulates a subset of NF-κB-dependent genes, possibly by the phosphorylation of the p65 subunit of NF-κB rather than by the phosphorylation of IκBα. In contrast to these earlier studies, we were unable to detect a role for TBK1 in activation of three different NF-κB-dependent promoters in response to virus infection or stimulation with TLR ligand. In addition, we were unable to observe an effect of TBK1 deficiency on TNF-α-induced activation of these NF-κB reporters (data not shown). At present, we cannot explain this discrepancy, but we note that the observed effect of TBK1 deficiency on TLR-2 and intercellular adhesion molecule 1 (ICAM-1) expression may be indirect. For example, the murine TLR-2 promoter contains a putative STAT (signal transducer and activator of transcription) binding site in addition to NF-κB binding sites (47). Thus, defects in TLR-2 expression in Tbk1-deficient MEFs could be an indirect consequence of the loss of IRF3 activation. Additional studies will be necessary to determine the mechanism by which TBK1 regulates the expression of certain NF-κB target genes in addition to its effects on IRF3 activation.
IKK-ε is highly homologous to TBK1, and both kinases have been implicated in the activation of IRF3 (21, 22). Here, we show that both kinases can directly phosphorylate serine residues that are critical for IRF3 activation. In previous studies, we found that both IKK-ε and TBK1 are required for IRF3 activation in human HEK293 cells in culture (22). This result was surprising, considering that TBK1 is ubiquitously expressed, whereas IKK-ε is expressed primarily in cells of lymphoid origin and inducible in other cell types. Whether this apparent nonredundancy of IKK-ε and TBK1 observed in HEK293 cells is a general phenomenon in primary cells is unclear. The relative roles of IKK-ε and TBK1 can be examined by using single- or double-deficient mice.
TBK1 was originally identified by virtue of its association with the TANK protein [TRAF (tumor necrosis factor receptor-associated factor) family member associated with NF-κB activator, also called I-TRAF] (18, 48). Subsequently, IKK-ε was shown to associate with TANK. Although the functional significance of this interaction is not understood, TANK has been shown to facilitate the association of TBK1 or IKK-ε with the IKK-α-IKK-β-IKK-γ complex (48).
Recently, the TLR-associated adapter molecule TRIF was shown to be a potent activator of IRF3 signaling, and TRIF-deficient mice fail to induce type I IFNs after LPS dsRNA stimulation, or infection with vaccinia virus (10, 32). Here, we show that TRIF-dependent activation of IRF3 requires TBK1. TRIF associates with TBK1 (22), and this association has been mapped to the N-terminal domain of TRIF (40). Moreover, the association of TRIF with TBK1 appears to require the kinase activity of TBK1 and the phosphorylation of TRIF (40). TRIF has also been shown to be essential for NF-κB activation by its association with TRAF6 (40). Thus, TRIF appears to function as an adapter protein for both the NF-κB and IRF3 signaling pathways.
Thus, both the classical and noncanonical IKKs play critical roles in establishing the innate antiviral immune response. Although the classical IKK-α-IKK-β-IKK-γ kinase functions primarily by the phosphorylation of IκBα, an important target of the noncanonical “IKKs” is IRF3. Further studies may reveal crosstalk between the two pathways and additional targets for both types of kinase complexes.
Acknowledgments
We thank Wen-Chen Yeh for providing Tbk1-/- MEFs; Sha-Mei Liao for providing TBK1 protein; E. Latz for assistance with confocal studies; and Neal Silverman, Charles Lin, and Russell Vance for critical reading of the manuscript. This work was supported by National Institutes of Health Grant AI20642 (to S.M.M. and T.M.), a Cancer Research Institute Fellowship (to S.M.M.), and the Wellcome Trust (K.A.F.).
Abbreviations: IRF3, IFN regulatory factor 3; IKK, IκB kinase; MEF, mouse embryonic fibroblast; NDV, Newcastle disease virus; ELAM, endothelial-leukocyte adhesion molecule; TLR, Toll-like receptor; dsRNA, double-stranded RNA; LPS, lipopolysaccharide; TANK, Traf family member-associated NF-κB activator; TBK1, TANK-binding kinase 1; SV, Sendai virus; HA, hemagglutinin; HAU, hemagglutinating units; TNF, tumor necrosis factor; PRD, positive regulatory domain; ISRE, IFN-stimulated regulatory element.
References
- 1.Janeway, C. A., Jr., & Medzhitov, R. (2002) Annu. Rev. Immunol. 20, 197-216. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov, R., Preston-Hurlburt, P. & Janeway, C. A., Jr. (1997) Nature 388, 394-397. [DOI] [PubMed] [Google Scholar]
- 3.Akira, S. (2001) Adv. Immunol. 78, 1-56. [DOI] [PubMed] [Google Scholar]
- 4.Dunne, A. & O'Neill, L. A. (2003) Sci. STKE 2003, re3. [DOI] [PubMed] [Google Scholar]
- 5.Maniatis, T. (1986) Harvey Lect. 82, 71-104. [PubMed] [Google Scholar]
- 6.Alexopoulou, L., Holt, A. C., Medzhitov, R. & Flavell, R. A. (2001) Nature 413, 732-738. [DOI] [PubMed] [Google Scholar]
- 7.Poltorak, A., He, X., Smirnova, I., Liu, M. Y., Van Huffel, C., Du, X., Birdwell, D., Alejos, E., Silva, M., Galanos, C., et al. (1998) Science 282, 2085-2088. [DOI] [PubMed] [Google Scholar]
- 8.Kurt-Jones, E. A., Popova, L., Kwinn, L., Haynes, L. M., Jones, L. P., Tripp, R. A., Walsh, E. E., Freeman, M. W., Golenbock, D. T., Anderson, L. J. & Finberg, R. W. (2000) Nat. Immun. 1, 398-401. [DOI] [PubMed] [Google Scholar]
- 9.Lund, J., Sato, A., Akira, S., Medzhitov, R. & Iwasaki, A. (2003) J. Exp. Med. 198, 513-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoebe, K., Du, X., Georgel, P., Janssen, E., Tabeta, K., Kim, S. O., Goode, J., Lin, P., Mann, N., Mudd, S., et al. (2003) Nature 424, 743-748. [DOI] [PubMed] [Google Scholar]
- 11.Vaidya, S. A. & Cheng, G. (2003) Curr. Opin. Immunol. 15, 402-407. [DOI] [PubMed] [Google Scholar]
- 12.Sato, M., Suemori, H., Hata, N., Asagiri, M., Ogasawara, K., Nakao, K., Nakaya, T., Katsuki, M., Noguchi, S., Tanaka, N. & Taniguchi, T. (2000) Immunity 13, 539-548. [DOI] [PubMed] [Google Scholar]
- 13.Au, W. C., Moore, P. A., Lowther, W., Juang, Y. T. & Pitha, P. M. (1995) Proc. Natl. Acad. Sci. USA 92, 11657-11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoneyama, M., Suhara, W. & Fujita, T. (2002) J. Interferon Cytokine Res. 22, 73-76. [DOI] [PubMed] [Google Scholar]
- 15.Hiscott, J., Pitha, P., Genin, P., Nguyen, H., Heylbroeck, C., Mamane, Y., Algarte, M. & Lin, R. (1999) J. Interferon Cytokine Res. 19, 1-13. [DOI] [PubMed] [Google Scholar]
- 16.Peters, R. T., Liao, S. M. & Maniatis, T. (2000) Mol. Cell 5, 513-522. [DOI] [PubMed] [Google Scholar]
- 17.Shimada, T., Kawai, T., Takeda, K., Matsumoto, M., Inoue, J., Tatsumi, Y., Kanamaru, A. & Akira, S. (1999) Int. Immunol. 11, 1357-1362. [DOI] [PubMed] [Google Scholar]
- 18.Pomerantz, J. L. & Baltimore, D. (1999) EMBO J. 18, 6694-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonnard, M., Mirtsos, C., Suzuki, S., Graham, K., Huang, J., Ng, M., Itie, A., Wakeham, A., Shahinian, A., Henzel, W. J., et al. (2000) EMBO J. 19, 4976-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tojima, Y., Fujimoto, A., Delhase, M., Chen, Y., Hatakeyama, S., Nakayama, K., Kaneko, Y., Nimura, Y., Motoyama, N., Ikeda, K., et al. (2000) Nature 404, 778-782. [DOI] [PubMed] [Google Scholar]
- 21.Sharma, S., tenOever, B. R., Grandvaux, N., Zhou, G. P., Lin, R. & Hiscott, J. (2003) Science 300, 1148-1151. [DOI] [PubMed] [Google Scholar]
- 22.Fitzgerald, K. A., McWhirter, S. M., Faia, K. L., Rowe, D. C., Latz, E., Golenbock, D. T., Coyle, A. J., Liao, S. M. & Maniatis, T. (2003) Nat. Immun. 4, 491-496. [DOI] [PubMed] [Google Scholar]
- 23.Oshiumi, H., Matsumoto, M., Funami, K., Akazawa, T. & Seya, T. (2003) Nat. Immun. 4, 161-167. [DOI] [PubMed] [Google Scholar]
- 24.Kawai, T., Takeuchi, O., Fujita, T., Inoue, J., Muhlradt, P. F., Sato, S., Hoshino, K. & Akira, S. (2001) J. Immunol. 167, 5887-5894. [DOI] [PubMed] [Google Scholar]
- 25.Toshchakov, V., Jones, B. W., Perera, P. Y., Thomas, K., Cody, M. J., Zhang, S., Williams, B. R., Major, J., Hamilton, T. A., Fenton, M. J. & Vogel, S. N. (2002) Nat. Immun. 3, 392-398. [DOI] [PubMed] [Google Scholar]
- 26.Fitzgerald, K. A., Palsson-McDermott, E. M., Bowie, A. G., Jefferies, C. A., Mansell, A. S., Brady, G., Brint, E., Dunne, A., Gray, P., Harte, M. T., et al. (2001) Nature 413, 78-83. [DOI] [PubMed] [Google Scholar]
- 27.Horng, T., Barton, G. M. & Medzhitov, R. (2001) Nat. Immun. 2, 835-841. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto, M., Sato, S., Mori, K., Hoshino, K., Takeuchi, O., Takeda, K. & Akira, S. (2002) J. Immunol. 169, 6668-6672. [DOI] [PubMed] [Google Scholar]
- 29.Fitzgerald, K. A., Rowe, D. C., Barnes, B. J., Caffrey, D. R., Visintin, A., Latz, E., Monks, B., Pitha, P. M. & Golenbock, D. T. (2003) J. Exp. Med. 198, 1043-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bin, L. H., Xu, L. G. & Shu, H. B. (2003) J. Biol. Chem. 278, 24526-24532. [DOI] [PubMed] [Google Scholar]
- 31.Oshiumi, H., Sasai, M., Shida, K., Fujita, T., Matsumoto, M. & Seya, T. (2003) J. Biol. Chem. 278, 49751-49762. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto, M., Sato, S., Hemmi, H., Hoshino, K., Kaisho, T., Sanjo, H., Takeuchi, O., Sugiyama, M., Okabe, M., Takeda, K. & Akira, S. (2003) Science 301, 640-643. [DOI] [PubMed] [Google Scholar]
- 33.Hirschfeld, M., Ma, Y., Weis, J. H., Vogel, S. N. & Weis, J. J. (2000) J. Immunol. 165, 618-622. [DOI] [PubMed] [Google Scholar]
- 34.Genin, P., Algarte, M., Roof, P., Lin, R. & Hiscott, J. (2000) J. Immunol. 164, 5352-5361. [DOI] [PubMed] [Google Scholar]
- 35.Lee, F. S., Peters, R. T., Dang, L. C. & Maniatis, T. (1998) Proc. Natl. Acad. Sci. USA 95, 9319-9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peters, R. T. & Maniatis, T. (2001) Biochim. Biophys. Acta 1471, M57-M62. [DOI] [PubMed] [Google Scholar]
- 37.Doyle, S., Vaidya, S., O'Connell, R., Dadgostar, H., Dempsey, P., Wu, T., Rao, G., Sun, R., Haberland, M., Modlin, R. & Cheng, G. (2002) Immunity 17, 251-263. [DOI] [PubMed] [Google Scholar]
- 38.Sakaguchi, S., Negishi, H., Asagiri, M., Nakajima, C., Mizutani, T., Takaoka, A., Honda, K. & Taniguchi, T. (2003) Biochem. Biophys. Res. Commun. 306, 860-866. [DOI] [PubMed] [Google Scholar]
- 39.Wathelet, M. G., Lin, C. H., Parekh, B. S., Ronco, L. V., Howley, P. M. & Maniatis, T. (1998) Mol. Cell 1, 507-518. [DOI] [PubMed] [Google Scholar]
- 40.Sato, S., Sugiyama, M., Yamamoto, M., Watanabe, Y., Kawai, T., Takeda, K. & Akira, S. (2003) J. Immunol. 171, 4304-4310. [DOI] [PubMed] [Google Scholar]
- 41.Karin, M. & Ben-Neriah, Y. (2000) Annu. Rev. Immunol. 18, 621-663. [DOI] [PubMed] [Google Scholar]
- 42.Li, Q., Van Antwerp, D., Mercurio, F., Lee, K. F. & Verma, I. M. (1999) Science 284, 321-325. [DOI] [PubMed] [Google Scholar]
- 43.Li, Z. W., Chu, W., Hu, Y., Delhase, M., Deerinck, T., Ellisman, M., Johnson, R. & Karin, M. (1999) J. Exp. Med. 189, 1839-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beg, A. A., Sha, W. C., Bronson, R. T., Ghosh, S. & Baltimore, D. (1995) Nature 376, 167-170. [DOI] [PubMed] [Google Scholar]
- 45.Rudolph, D., Yeh, W. C., Wakeham, A., Rudolph, B., Nallainathan, D., Potter, J., Elia, A. J. & Mak, T. W. (2000) Genes Dev. 14, 854-862. [PMC free article] [PubMed] [Google Scholar]
- 46.Tanaka, M., Fuentes, M. E., Yamaguchi, K., Durnin, M. H., Dalrymple, S. A., Hardy, K. L. & Goeddel, D. V. (1999) Immunity 10, 421-429. [DOI] [PubMed] [Google Scholar]
- 47.Musikacharoen, T., Matsuguchi, T., Kikuchi, T. & Yoshikai, Y. (2001) J. Immunol. 166, 4516-4524. [DOI] [PubMed] [Google Scholar]
- 48.Chariot, A., Leonardi, A., Muller, J., Bonif, M., Brown, K. & Siebenlist, U. (2002) J. Biol. Chem. 277, 37029-37036. [DOI] [PubMed] [Google Scholar]