Abstract
Epstein-Barr virus (EBV) is a herpesvirus that establishes a lifelong, persistent infection. It was first discovered in the tumor Burkitt's lymphoma (BL). Despite intensive study, the role of EBV in BL remains enigmatic. One striking feature of the tumor is the unique pattern of viral latent protein expression, which is restricted to EBV-encoded nuclear antigen (EBNA) 1. EBNA1 is required to maintain the viral genome but is not recognized by cytotoxic T cells. Consequently, it was proposed that this expression pattern was used by latently infected B cells in vivo. This would be the site of long-term, persistent infection by the virus and, by implication, the progenitor of BL. We now know that EBV persists in memory B cells in the peripheral blood and that BL is a tumor of memory cells. However, a normal B cell expressing EBNA1 alone has been elusive. Here we show that most infected cells in the blood express no detectable latent mRNA or proteins. The exception is that when infected cells divide they express EBNA1 only. This is the first detection of the BL viral phenotype in a normal, infected B cell in vivo. It suggests that BL may be a tumor of a latently infected memory B cell that is stuck proliferating because it is a tumor and, therefore, constitutively expressing only EBNA1.
It has long been thought that Burkitt's lymphoma (BL) may be a tumor derived from the cell in which Epstein-Barr virus (EBV) maintains persistent infection (1-3). Consistent with this idea are recent findings that BL has the Ig gene somatic mutations of a memory B cell (4) and that EBV persists in resting memory B cells in the blood (5, 6). BL also has a unique pattern of viral latent protein expression: Only the EBV-encoded nuclear antigen (EBNA) 1 protein, which is necessary for maintenance and replication of the viral genome (7), is detected (8). EBNA1 is produced from a transcript [EBNA1 (Q-K)] originating from a unique promoter, Qp (9). Because EBNA1 is not recognized by cytotoxic T lymphocytes, it was proposed that only EBNA1 would be expressed in the cell in which EBV persists in vivo. This cell type could be the progenitor of BL.
RT-PCR analysis has detected EBNA1 (Q-K) expression in tonsil B cell subsets (10, 11) but never alone. EBNA1 (Q-K) was always in association with latent membrane proteins (LMPs) 1 and 2. We call this the default transcription program (12), because LMP1 and LMP2 can deliver surrogate survival signals to B cells. By expressing the default program, the virus ensures that the B cell survives and the viral genome replicates. This program differs from the growth transcription program, which involves expression of all nine latent proteins. The growth program is responsible for the well known ability of EBV to transform normal resting B cells in vitro into proliferating lymphoblasts.
It is now established that EBV persists in resting memory B cells in the blood. We have provided evidence in support of a model that explains how these cells are produced and maintained. The essence of this model is that EBV-infected B cells recapitulate the normal processes that produce memory B cells, except that viral latent proteins provide the necessary signals (see refs. 12 and 13 for a detailed discussion). We have proposed, through analogy with the neurotropic herpesviruses (14) and contrary to the EBNA1-only model, that once the latently infected memory cells are produced all viral protein expression ceases (the latency transcription program). This would be possible because the latently infected memory cells are resting (12), and the consequence would be that the infected cells would remain undetected by the immune system while also becoming nonpathogenic, because the proliferation-associated genes are not expressed.
Attempts to identify EBNA1-only expression in infected peripheral blood memory B cells have also been unsuccessful (15-18). Based on RT-PCR results, several authors report detection of LMP2 in healthy carriers. In acute infectious mononucleosis (AIM) patients, the numbers of infected cells are much higher, allowing greater sensitivity for the detection of latent gene expression. In these studies, transcripts for several latent proteins, including EBNA1 (Q-K), were detected. All of these studies were nonquantitative; therefore, it was impossible to determine what fraction of cells was expressing any particular gene and whether different genes were expressed in the same or separate cells. Furthermore, it is unclear whether mRNAs were detected because of cells expressing discrete latency programs or whether these mRNAs were residual transcripts unrelated to functional latency programs.
Because of the inconclusive nature of RT-PCR results to date, we have carried out quantitative RT-PCR analyses and histochemical staining of infected cells in the peripheral blood to discover exactly which genes are expressed, what fraction of infected cells express them, and whether known latent transcription programs are used by individual cells. By using this approach, we hoped to resolve whether the EBNA1-only model or the latency program model was correct. We found that both models are correct. The cells are predominantly in a resting state and express no detectable latent proteins or mRNAs. However, on the rare occasion when the cells divide, they express EBNA1, presumably to allow the viral genome to divide with the cell.
Methods
Cells and Cell Lines. The EBV-negative BJAB cell line was used as a negative control. The lymphoblastoid cell line ER was used as positive controls for EBNA2, LMP1, LMP2, and EBV-encoded RNA (EBER) expression at the RNA and protein levels and for DNA PCR. The EBV+ BL line Rael was used as a positive control for EBNA1(Q-K) expression.
Adolescents (ages 17-24 years) presenting to the clinic at the University of Massachusetts Student Health Service (Amherst) with clinical symptoms consistent with AIM were recruited for this study. After obtaining informed consent, we collected blood from those who presented with symptoms. Diagnosis at the time of presentation to the clinic required a positive mononucleosis spot test and the presence of atypical lymphocytes (19). Confirmation of primary EBV infection required the detection of IgM antibodies to the EBV viral capsid antigen in patients' sera (20). These studies were approved by the Human Studies Committee at the University of Massachusetts Medical School (Worcester). All blood samples were diluted 1:1 in 1× PBS. Tonsils were provided by Massachusetts General Hospital. Blood and tonsil lymphocytes were isolated as described (5, 10).
Cell Separations. Negative selection was performed according to the manufacturer's instructions to isolate B cells by using the Stem Cell Technologies Stem Sep system. Isolated populations were analyzed for purity on a FACScan analyzer. In all cases purity levels were >90% and usually >95%. Memory B cells (IgD-, CD20+) were isolated by fluorescence-activated cell sorting as described (6).
Limiting Dilution DNA PCR. Limiting dilution analysis was used to determine the frequency of EBV-infected cells. The details of this assay have been published (5).
Limiting Dilution RT-PCR. Serial dilutions of isolated cell populations were prepared, and multiple aliquots of each dilution were placed into Eppendorf tubes. When necessary, EBV-negative tonsillar cells were added to each tube to bring the total number of cells to 5 × 106. As a positive control, 106 EBV+ LCL cells were used. RNA was isolated by the TRIzol method (Invitrogen), and cDNA was prepared as described (10).
PCR for the EBV genes EBNA2, EBNA1(Q-K), LMP1, LMP2, and EBERs were performed by taking 1/10th of the cDNA (20 μl) for each gene. This method allowed for simultaneous PCR for all of the genes on the same sample. Primers used were as follows: EBNA2, 5′-CATAGAAGAAGAAGAGGATAGAGA-3′ and 5′-GTAGGGAT TCGAGGGA ATTACTGA-3′ (15); EBNA1(Q-K), 5′-TGCCCCCTCGTCAGACATGATT-3′ and 5′-AGCGTGCGCTACCGGAT-3′ (21); LMP1, 5′-TTGGTGTACTCCTACTGATGATCACC-3′ and 5′-AGTAGATCCAGATACCTAAGACAAGT-3′ (15); LMP2, 5′-ATGACTCATCTCAACACATA-3′ and 5′-CATGTTAGGCAAATTGCAAA-3′ (17); and EBERs, 5′-AAAACATGCGGACCACCAGC-3′ and 5′-AGGACCTACGCTGCCCTAGA-3′ (17). Reactions were performed as described (10).
Immunofluorescence. The expression and localization patterns of the EBV latent proteins LMP1, LMP2, EBNA1, and EBNA2 were examined in purified memory B cells. Primary cells were resuspended to a concentration of 1 × 106 cells per ml, and cell lines were resuspended to a concentration of 5 × 105 cells per ml. Cytospins were prepared with 155 μl of cell suspension spun at 450 rpm for 10 min (Shandon Cytospin 2). Cells were fixed in 3% paraformaldehyde for 10 min at room temperature, followed by permeabilization in 0.1% Triton X-100 for 5 min at 4°C. Blocking was achieved through washing in 10% FBS in 1× PBS for 30 min at room temperature. Slides were transferred to a humidified chamber for staining. Primary staining was performed with 100 μl of antibody for 45 min at room temperature, followed by three 5-min washes in 1× PBS. Secondary antibodies were applied for 30 min at room temperature, followed by two 10-min washes in 1× PBS. DAPI (0.0001 μM; Molecular Probes) was used to localize the nucleus. The slides were then washed, mounted in 50% glycerol, and visualized with a fluorescence microscope. Dual staining for two proteins was carried out simultaneously. Primary and secondary antibody pairs are as follows: EBNA2, 1:8,000 mouse anti-EBNA2 (DAKO) and 1:8,000 goat anti-mouse Alexa Fluor 594 (Molecular Probes); EBNA1, 1:1,000 rabbit anti-EBNA1 (gift from J.M.) and 1:8,000 goat anti-rabbit Alexa Fluor 594 or 488; LMP1, 1:10,000 mouse anti-LMP1 (D.A.T.-L. lab) and 1:10,000 goat anti-mouse Alexa Fluor 594; LMP2, 1:10 rabbit anti-LMP2 (gift from J.M.) and 1:10,000 goat anti-rabbit Alexa Fluor 488.
Results
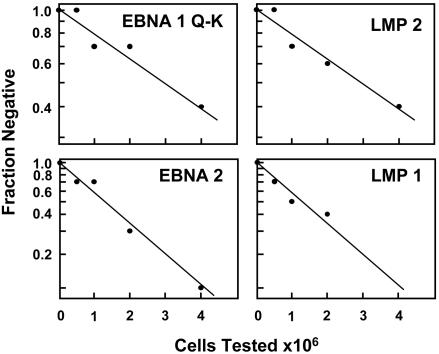
Limiting Dilution RT-PCR Analysis for Viral Gene Expression in Latently Infected Memory Cells in the Blood. To search for a memory B cell in the blood expressing only EBNA1, it was necessary to develop a quantitative test that distinguishes the different viral latent transcription programs (Table 1) in single cells. EBV potentially uses four discrete latency transcription programs, each characterized by the presence or absence of particular gene expression. The growth program is characterized by expression of all of the known latent proteins under the control of EBNA2. In the default program, only EBNA1 (expressed from Qp), LMP1, and LMP2 are used. EBNA2 is absent. In the putative EBNA1-only program, EBNA1 would be the only latent protein present, again expressed from Qp. In the putative latency program, no latent genes would be expressed. By testing for EBNA1(Q-K), EBNA2, LMP1, and LMP2, it is possible to distinguish the four different latency transcription programs (Table 1). To maximize the number of cells we could analyze, we took advantage of the fact that individuals presenting with AIM have elevated levels of infected B cells in their blood. Fig. 1 shows the result of a limiting dilution RT-PCR analysis for the expression of all four genes on a sample of peripheral blood mononuclear cells from an AIM patient. The results for all four genes demonstrate a linear relationship in a semilog plot of fraction of negative samples versus cell number tested. This indicates that the assays are able to detect single cells expressing any one of the genes in patient samples. Next, we measured the frequency of virus-infected memory B cells (Fig. 2) by using a limiting dilution DNA PCR assay that we have developed and described in detail (5). These measurements confirmed (Table 2) the presence of elevated numbers of infected cells and allowed us to calculate how many infected cells were tested when a similar serial dilution was set up for RT-PCR analysis. An example of one such analysis for all four of the marker genes is shown in Fig. 3.
Table 1.
| Program | Infected cells in vivo | EBNA1 (Q-K) | EBNA2 | LMP1 | LMP2 |
|---|---|---|---|---|---|
| Growth | Tonsil, naive B cells | — | + | + | + |
| Default | Tonsil, germinal center B cells | + | — | + | + |
| EBNA1* | Peripheral blood, dividing memory B cell | + | — | — | — |
| Latency* | Peripheral blood, resting memory B cell | — | — | — | — |
Only the four indicator genes used in this study are shown. +, detected. —, not detected.
As defined in this paper
Fig. 1.
Limiting dilution analysis of whole peripheral blood for the presence of latent viral transcripts. Limiting dilution analysis was performed on whole peripheral blood mononuclear cells from a patient with AIM. Ten replicates of each dilution were then tested for expression of the four indicated genes by RT-PCR. The results, plotted on a semilog scale, demonstrate that the dilution analysis follows a Poisson distribution.
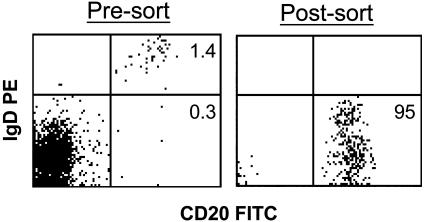
Fig. 2.
Fluorescence-activated cell sorting analysis of peripheral blood mononuclear cells from an AIM patient before and after purification of memory (CD20+, IgD-) cells. Note that the percentage of B cells is low, presumably because of the large T cell lymphocytosis characteristic of AIM. The purified memory B cells were always >90% pure, and often >95% pure, by fluorescence-activated cell sorting reanalysis. However, analysis of the expressed Ig genes of ≈120 single cells from such a sort revealed that all were memory cells, suggesting that the sorted memory cell population is >99% pure (T. Vorobyova and D.A.T.-L., unpublished observations).
Table 2. Expression of EBV latent genes in infected memory B cells from the blood of AIM patients.
| % infected cells expressing gene
|
|||||
|---|---|---|---|---|---|
| Patients | % B cells infected | EBNA1 (Q-K) | EBNA2 | LMP1 | LMP2 |
| 1 | 17.0 | 0.06 | 0.1 | 0.1 | 0.07 |
| 2 | 5.0 | 0.01 | 0.04 | 0.7 | 0.1 |
| 3 | 4.7 | 0.01 | 0.04 | 0.06 | 0.04 |
| 4 | 0.6 | 0.1 | 0.3 | 0.6 | 0.6 |
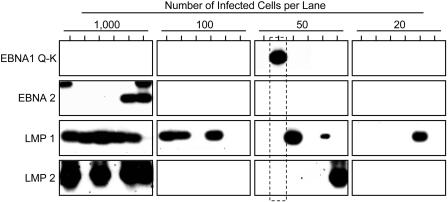
Fig. 3.
Limiting dilution RT-PCR analysis of EBV latent gene expression. Purified B cells were serially diluted, and multiple replicates of each dilution were used to prepare cDNA. Each cDNA was tested for the four indicator genes EBNA1 (Q-K), EBNA2, LMP1, and LMP2. The number of infected cells was measured in parallel by limiting dilution DNA PCR (refs. 5 and 33 and data not shown). Each sample is labeled with the number of infected cells tested, not the absolute number of cells. In control experiments, each RT-PCR assay was able to detect expression of the gene in a single infected cell from an EBV-positive cell line (data not shown). The RT-PCR products were separated on agarose gels and identified by Southern blotting with specific probes. A single example of a cell expressing only EBNA1 from the Qp promoter is indicated by the dotted box. One concern is that the signal is seen at one of the higher dilutions, whereas none are seen at the lower dilutions; however, the following calculation can explain this apparent discrepancy: In this particular experiment, six replicates were tested containing 1,000, 100, 50, and 20 infected cells, for a total of 7,020 cells, of which 1 was positive. There were 420 cells tested in total in the dilutions containing 50 and 20 cells; therefore, the chance of finding the single positive cell in one of these dilutions is ≈420/7,020 or ≈1 in 15 (i.e., possible simply by chance).
For the samples in which no viral gene expression was detected, it was important to establish that this was not due to a technical artifact. To eliminate this possibility, the presence of the ubiquitous, small, noncoding viral EBER transcript (22) was confirmed. Detecting EBER served as a control in several important aspects. Most importantly, detection of EBER demonstrates that the quality of the RNA is good enough for PCR in samples in which no latent viral protein transcripts were detected. EBER RNA is a better control than cellular RNA, because every experimental sample was brought up to a concentration of 5 × 106 cells with EBV-negative tonsil filler cells before RNA isolation (see Methods). Therefore, any cellular RNA used as a positive control would probably be present in hundreds of millions of copies. By comparison, although EBERs potentially have a high copy number per cell, the number of infected cells per sample is small; therefore, the EBER RNA copy number would be lower per sample than any cellular RNA control. Thus, EBER RNA is a more sensitive marker of mRNA degradation. EBER has two further advantages: First, it is an RNA from the infected cells; therefore, it attests to the quality of RNA from the infected cells themselves, whereas a cellular RNA would not distinguish infected cells and filler cell. Lastly, EBER allows us to say that there were indeed infected cells present in samples that were negative for the viral latent protein RNAs.
By using the premeasured frequency of infected cells based on DNA PCR and the frequency of cells expressing each gene based on RT-PCR, it was then possible to estimate the percentage of infected cells expressing each gene. The results for this patient (patient 1) are summarized in Table 2 along with data from three other patients. It is apparent from this study that <1% of the infected B cells are expressing any of the four latent genes.
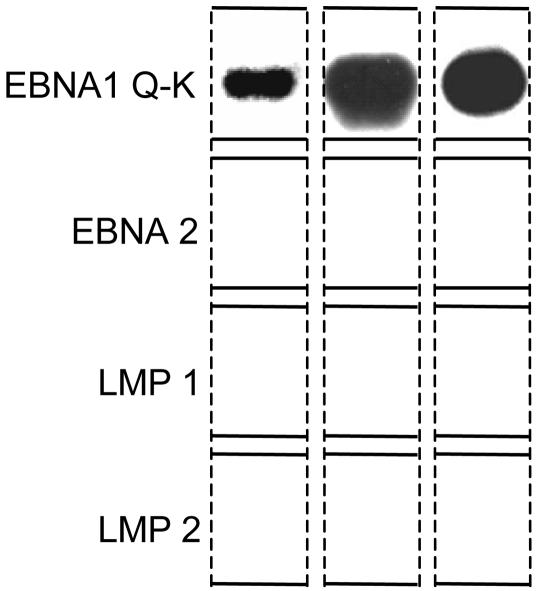
Detection of Cells Expressing Only EBNA1 (Q-K). In BL, EBNA1 is transcribed from the Qp promoter. The number of cells expressing the Qp transcript [EBNA1(Q-K)] in the blood of the four patients (Table 2) was low and not obviously different from those expressing the other genes we tested. Furthermore, the results as calculated in Table 2 indicate only what fraction of infected cells express each gene; they do not reveal whether one or multiple latent genes are expressed in any given cell. This information can be derived by looking at the gene expression profile of each sample, because each sample was simultaneously analyzed for all four genes. By performing this analysis on samples at the limit dilution for gene expression, we can minimize the chance of fortuitously detecting gene expression from two or more cells in the same sample; thus, we can also define the gene expression profile of individual infected cells. Therefore, to more closely examine the question of whether EBNA1-only latency may be used in the blood, we analyzed viral latent gene expression by RT-PCR at the limit dilution for detection of the four viral transcripts studied. We took as our cut-off point the dilution in which one-third or less of the samples expressed any one gene. Cells were identified that expressed only one of the latent genes, whereas others were found that expressed two or more. The analysis showed no clear patterns for the expression of EBNA2, LMP1, and LMP2 in single cells consistent with any of the defined transcription programs. They were all detected singly and in various combinations with each other. This would suggest that we were picking up signals from residual transcripts of these genes that are not part of defined transcription programs. The exception was EBNA1(Q-K), which was always detected alone. One example is shown in Fig. 3 (dotted box). Three examples, cropped from blots such as the one in Fig. 3, are collated in Fig. 4. We conclude that rare cells in the peripheral blood do express EBNA1 only at the level of mRNA. This result does not address whether these mRNAs represent a true EBNA1-only transcription pattern or are residual transcripts of no biological significance, as we have suggested for cells expressing mRNA for the other latent genes.
Fig. 4.
Detection of single cells expressing the EBNA1 (Q-K) transcript alone. Cells at the limit dilution were simultaneously tested by RT-PCR for the four genes indicated (see the legend to Fig. 3). The results shown are cropped from films such as that shown in Fig. 3. The sequences for the primers used to amplify these products are taken from ref. 21. The 5′ primer extends from +25 to +41 relative to the major Qp transcription initiation site, and the 3′ primer is at the 5′ end of the K (coding) exon (coordinates in the EBV sequence are 62,437-62,453 and 107,967-107,947, respectively).
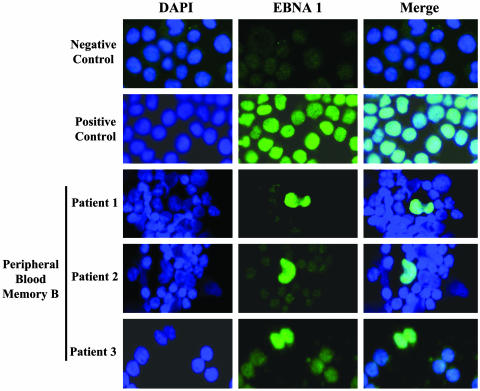
Immunofluorescence Analysis for Viral Gene Expression in Latently Infected Memory Cells in the Blood. Immunofluorescence staining for expression of the four indicator genes was carried out for two reasons. First, we wanted a general test to see whether the number of cells expressing the latent proteins agreed with the mRNA results; a discrepancy could arise if the half-life of the proteins was markedly longer than the half-life of the mRNAs. Second, we wanted to specifically test whether we could find evidence of cells expressing EBNA1 alone. Due to the high frequency of infected cells present in AIM, we were able to screen multiple stained slides containing at least 1,000 infected cells from three patients. For these experiments, staining was performed for the four latent proteins in various pairwise combinations. Although precise quantitation is difficult to achieve with these techniques, clear staining was seen for EBNA1 in a small fraction of the infected B cells from all three patients tested. In most cases the staining was observed in dividing cells (Fig. 5). EBNA2-positive cells were never detected. Extremely rare positive cells (one or two per smear) were found for LMP1 and LMP2. In one cell, coexpression of LMP1 and LMP2 was seen, and, in one other, LMP1 was detected along with EBNA1. In every case, the staining was limited to nondividing cells and was atypical compared with cell line controls, so it was difficult to be sure that the staining was specific. Therefore, given the limits of sensitivity of the techniques, it appears that dividing, latently infected cells express only EBNA1, the classic viral phenotype of BL.
Fig. 5.
Identification of memory B cells expressing only EBNA1. Immunofluorescence staining identified the expression of EBNA1 in dividing cells. The cells were counterstained with 4′,6-diamidino-2-phenylindole to highlight the nucleus.
Discussion
In this study we have used quantitative approaches to solve the question of which viral proteins are expressed in latently infected cells in the blood. Most (>99%) of the cells express no mRNAs for latent proteins, which is reflected at the level of protein expression (the latency program; ref. 12). The exception is that when the cells divide they express only EBNA1 (EBNA1 program). This demonstrates that the elusive EBNA1-only form of latency, previously seen only in BL, is used by normal, infected B cells in vivo. These results raise the possibility that BL may be a tumor of latently infected memory B cells. Such cells would normally express the latency program but in BL would be forced into constitutive EBNA1 expression, because the tumor cells are continuously driven to proliferate by the deregulated c-Myc. This hypothesis is supported by Ig gene analysis, which shows that BL is derived from a memory cell, but this theory is not consistent with the surface phenotype of BL, which more closely resembles a germinal center cell (23). It is important to emphasize the difficulty in deciphering the origin of tumors like BL based on the phenotype of the end-stage tumor. It is known that tumor-igenesis is a multistep process involving the selection of a single malignant clone from large numbers of premalignant precursors over long periods of time. It is therefore virtually impossible to know how directly the final cellular or viral phenotype of BL relates to the original infected precursor.
One possible interpretation of our results could be that the very rare cells expressing EBNA1 came from a nonmemory population. However, from six independent estimates of the level of virus-infected cells in the nonmemory compartment, we find the level to be no more than 0.0025% (assuming a 1% cross-contamination between the two populations). Therefore, even if the population tested were pure nonmemory cells, there would not be enough infected cells to account for the EBNA1-positive cells we observed.
We detected EBNA1 in dividing cells in the blood. In retrospect this is not surprising; if EBNA1 were not expressed, the viral genomes would be gradually lost. These cells were not driven to divide by the virus, because they do not express the growth transcription program. This finding suggests that the virus is simply “along for the ride” when the latently infected memory B cells, like the rest of the B cells, divide occasionally to maintain homeostatic levels, a phenomenon we have described previously (24). It has been reported that the Qp promoter becomes activated when infected cells enter into S phase (25). That report is consistent with our finding that EBNA1 protein is predominantly expressed in dividing cells. However, we found very few EBNA1-positive resting B cells. This finding was surprising, because it is thought that EBNA1 may be a relatively stable protein. However, it is apparent from rodent studies that memory B cells divide only once every few months (26), which is consistent with our observations that only a small fraction of the infected cells are dividing. Such a long period between divisions would mean that, even if EBNA1 survived for 1 or 2 days, it would be absent for the vast majority of time that the cells were resting, and the strongest staining would be seen in dividing cells.
It was long believed that EBNA1 would be the only viral latent protein expressed at the site of latent persistence (1-3). It now appears that this idea is correct, but in a modified way (Fig. 6). EBV at the site of persistence expresses no proteins (the latency program). The virus does not need to, because the cells are resting. EBNA1 is used only when the cells divide. Ultimately, EBV is like other members of the herpesvirus family (14) in that it shuts off all viral protein-encoding genes when it reaches the site of latent persistence. The unique feature of EBV is that it uses a series of discrete latency transcription programs to convert the newly infected cell into a resting memory cell (12) before switching off transcription of the protein-encoding genes. Thus, the virus has found a perfect niche for long-term persistence: latency in long-lived memory cells. These cells are invisible to the immune response, because no cytotoxic T lymphocyte targets are present, and are not pathogenic to the host, because the growth-promoting genes are no longer expressed.
Fig. 6.
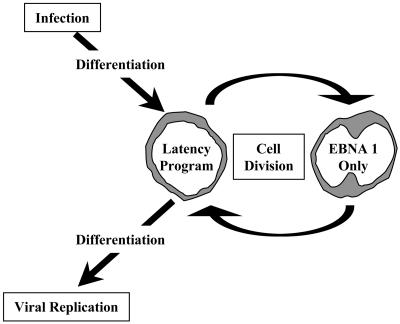
Schematic representation of EBV persistence in the peripheral blood. Cells latently infected with EBV are thought to be produced in Waldeyer's ring (10, 12) and to enter the peripheral circulation (34) as resting memory B cells. In this study, we have shown that these cells do not express viral latent proteins (the latency program). Occasionally these cells divide as part of the homeostatic regulation of the B cell pool. When they divide, they express only EBNA1. This is the putative precursor cell of BL.
Earlier studies have reported EBNA expression in the blood B cells of AIM patients (27, 28). One report claimed extremely high levels (up to 20%) of all B cells being EBNA-positive. These results have been challenged by Crawford et al. (29), who claimed that all of the infected cells in AIM blood are EBNA-negative. Our findings suggest that the truth lies somewhere between these two extremes, namely that EBNA1-positive cells are detectable, but rare, with the vast majority of infected cells (>99%) being EBNA1-negative. In agreement with our results, many of the EBNA-positive cells described in the earlier studies were dividing (27). The earlier interpretation that those dividing cells were EBV-driven lymphoblasts is probably incorrect, however, because the only latency state known at that time was the growth program. We have now shown that EBNA2, LMP1, and LMP2 are not present; thus, the cells are not proliferating because of the viral growth program.
One potential limitation of our study is that we did not test all of the known viral proteins. Our RT-PCR and in situ fluorescence studies used targets that define the known latency transcription programs (Table 1) and for which sensitive RT-PCR and protein staining techniques were available. Nevertheless, there may be novel forms of infection in vivo that have not been detected in vitro. Therefore, it is formally possible that other viral proteins could be expressed at high levels by novel transcriptional mechanisms or that unique proteins, not found to be detected in latent infection, are expressed in the blood.
We have proposed a model of EBV persistence whereby the EBV transcription programs drive the activation and differentiation of B cells into resting memory cells (12). One prediction of this model is that virus-driven growth is self-regulated because the virus itself pushes the cells into a resting state. A crucial observation in the establishment of this model was that tonsil cells bearing germinal center or memory markers express the default program, and memory cells in the blood express no detectable latent proteins. These ideas in turn imply that the EBV-driven lymphomas associated with immunosuppression occur because of inappropriate infection of B cells, i.e., in a form or location where they cannot differentiate out of the proliferative state, an event we refer to as bystander infection. Such events are seen in acute AIM patients' tonsils, which contain infected memory and germinal center cells clonally expanding under the influence of EBNA2 (30, 31). These types of cells are not present in healthy carriers (10, 11) and, therefore, do not relate to the biology of persistence. Nevertheless, they are important, because they have the potential to give rise to EBV lymphoma. Destroying these cells is a critical role for the cellular immune response that allows the survival of both host and virus.
In conclusion, we have shown that memory cells latently infected with EBV in the blood are predominantly resting and express no viral latent proteins, with the exception that only EBNA1 is expressed when the cells divide. Our findings demonstrate that the BL viral phenotype is used by normal, infected cells in vivo.
Acknowledgments
We thank Rose Ciccarelli (nurse practitioner), clinical research coordinator for the University of Massachusetts Amherst Health Services; Allen Parmelee (Tufts University School of Medicine) for flow cytometry; and Robin Brody and Jim Coderre (University of Massachusetts Medical School) for technical support. This work was supported by Public Health Service Grants AI 18757 and CA 65883 (to D.A.T.-L.) and AI 49320 (to K.L. and J.L.S.) and University of Massachusetts Center for AIDS Research Clinical Investigation Core Public Health Services Grant AI 42845.
Abbreviations: EBV, Epstein-Barr virus; BL, Burkitt's lymphoma; EBNA, EBV-encoded nuclear antigen; LMP, latent membrane protein; AIM, acute infectious mononucleosis; EBER, EBV-encoded RNA.
References
- 1.Klein, G. (1994) Cell 77, 791-793. [DOI] [PubMed] [Google Scholar]
- 2.Masucci, M. G. & Ernberg, I. (1994) Trends Microbiol. 2, 125-130. [DOI] [PubMed] [Google Scholar]
- 3.Rickinson, A. B. & Kieff, E. (1996) in Fields Virology, eds. Fields, B. N., Knipe, D. M. & Howley, P. M. (Lippincott-Raven, Philadelphia), Vol. 2, pp. 2397-2446. [Google Scholar]
- 4.Klein, U., Klein, G., Ehlin-Henriksson, B., Rajewsky, K. & Kuppers, R. (1995) Mol. Med. 1, 495-505. [PMC free article] [PubMed] [Google Scholar]
- 5.Babcock, G. J., Decker, L. L., Volk, M. & Thorley-Lawson, D. A. (1998) Immunity 9, 395-404. [DOI] [PubMed] [Google Scholar]
- 6.Joseph, A. M., Babcock, G. J. & Thorley-Lawson, D. A. (2000) J. Immunol. 165, 2975-2981. [DOI] [PubMed] [Google Scholar]
- 7.Yates, J. L., Warren, N. & Sugden, B. (1985) Nature 313, 812-815. [DOI] [PubMed] [Google Scholar]
- 8.Gregory, C. D., Rowe, M. & Rickinson, A. B. (1990) J. Gen. Virol. 71, 1481-1495. [DOI] [PubMed] [Google Scholar]
- 9.Schaefer, B. C., Strominger, J. L. & Speck, S. H. (1995) Proc. Natl. Acad. Sci. USA 92, 10565-10569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babcock, G. J., Hochberg, D. & Thorley-Lawson, D. A. (2000) Immunity 13, 497-506. [DOI] [PubMed] [Google Scholar]
- 11.Babcock, G. J. & Thorley-Lawson, D. A. (2000) Proc. Natl. Acad. Sci. USA 97, 12250-12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thorley-Lawson, D. A. (2001) Nat. Rev. Immunol. 1, 75-82. [DOI] [PubMed] [Google Scholar]
- 13.Thorley-Lawson, D. A. & Babcock, G. J. (1999) Life Sci. 65, 1433-1453. [DOI] [PubMed] [Google Scholar]
- 14.Roizman, B. & Knipe, D. M. (2001) in Fields Virology, eds. Fields, B. N., Howley, P. M., Griffin, D. E., Lamb, R. A., Martin, M. A., Roizman, B., Straus, S. E. & Knipe, D. M. (Lippincott, Philadelphia), Vol. 2, pp. 2399-2459. [Google Scholar]
- 15.Chen, F., Zou, J. Z., di, R. L., Winberg, G., Hu, L. F., Klein, E., Klein, G. & Ernberg, I. (1995) J. Virol. 69, 3752-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu, L. & Rowe, D. T. (1992) J. Virol. 66, 3715-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tierney, R. J., Steven, N., Young, L. S. & Rickinson, A. B. (1994) J. Virol. 68, 7374-7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babcock, G. J., Decker, L. L., Freeman, R. B. & Thorley-Lawson, D. A. (1999) J. Exp. Med. 190, 567-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoagland, R. J. (1967) Infectious Mononucleosis (Grune & Stratton, New York).
- 20.Henle, W. & Henle, G. (1979) in The Epstein-Barr Virus, ed. Achong, B. G. (Springer, Berlin), pp. 61-78.
- 21.Schaefer, B. C., Strominger, J. L. & Speck, S. H. (1996) J. Virol. 70, 8204-8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arrand, J. J. & Rymo, L. (1982) J. Virol. 41, 376-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregory, C. D., Tursz, T., Edwards, C. F., Tetaud, C., Talbot, M., Caillou, B., Rickinson, A. B. & Lipinski, M. (1987) J. Immunol. 139, 313-318. [PubMed] [Google Scholar]
- 24.Miyashita, E. M., Yang, B., Babcock, G. J. & Thorley-Lawson, D. A. (1997) J. Virol. 71, 4882-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davenport, M. G. & Pagano, J. S. (1999) J. Virol. 73, 3154-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schittek, B. & Rajewsky, K. (1990) Nature 346, 749-751. [DOI] [PubMed] [Google Scholar]
- 27.Robinson, J., Smith, D. & Niederman, J. (1980) Nature 287, 334-335. [DOI] [PubMed] [Google Scholar]
- 28.Robinson, J. E., Smith, D. & Niederman, J. (1981) J. Exp. Med. 153, 235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crawford, D. H., Rickinson, A. B., Finerty, S. & Epstein, M. A. (1978) J. Gen. Virol. 38, 449-460. [DOI] [PubMed] [Google Scholar]
- 30.Kurosaki, T. (1999) Annu. Rev. Immunol. 17, 555-592. [DOI] [PubMed] [Google Scholar]
- 31.Kurth, J., Perniok, A., Schmitz, R., Iking-Konert, C., Chiorazzi, N., Thompson, K. M., Winkler, T., Rajewsky, K. & Kuppers, R. (2002) Eur. J. Immunol. 32, 3785-3792. [DOI] [PubMed] [Google Scholar]
- 32.Thorley-Lawson, D. A. & Miyashita, E. M. (1996) Trends Microbiol. 4, 204-208. [DOI] [PubMed] [Google Scholar]
- 33.Miyashita, E. M., Yang, B., Lam, K. M., Crawford, D. H. & Thorley-Lawson, D. A. (1995) Cell 80, 593-601. [DOI] [PubMed] [Google Scholar]
- 34.Laichalk, L. L., Hochberg, D., Babcock, G. J., Freeman, R. B. & Thorley-Lawson, D. A. (2002) Immunity 16, 745-754. [DOI] [PubMed] [Google Scholar]