Abstract
Background/Aims
An individual’s genotypes at a group of Single Nucleotide Polymorphisms (SNPs) can be used to predict that individual’s ethnicity, or ancestry. In medical studies, knowledge of a subject’s ancestry can minimize possible confounding, and in forensic applications, such knowledge can help direct investigations. Our goal is to select a small subset of SNPs, from the millions already identified in the human genome, that can predict ancestry with a minimal error rate.
Methods
The general form for this variable selection procedure is to estimate the expected error rates for sets of SNPs using a training dataset and consider those sets with the lowest error rates given their size. The quality of the estimate for the error rate determines the quality of the resulting SNPs. As the apparent error rate performs poorly when either the number of SNPs or the number of populations is large, we propose a new estimate, the Improved Bayesian Estimate.
Conclusions
We demonstrate that selection procedures based on this estimate produce small sets of SNPs that can accurately predict ancestry. We also provide a list of the 100 optimal SNPs for identifying ancestry.
R functions are available at http://bioinformatics.med.yale.edu/group/josh/index.html.
Keywords: Ancestry, Ethnicity, SNPs, Error Rate, Allele Frequency, Genotype, AIM, Bootstrap, FOSSIL
Introduction
An individual’s genotypes at a group of Single Nucleotide Polymorphisms (SNPs) can be used to predict that individual’s ethnicity, or ancestry [15, 22, 25, 28, 29, 31]. Identifying ancestry through this approach is often useful [2, 17, 27]. For example, subjects in a medical study may be genotyped because adjusting for precise ancestry can minimize one source of confounding [3, 11, 19]. Similarly, a sample from a crime scene may be genotyped so ancestry can be included in the description of a suspect [4, 6, 18]. Although millions of SNPs have been identified, only a small subset needs to be genotyped in order to accurately predict ancestry. Reducing the needed number to the 10’s or 100’s is still useful even in the era of SNP microarrays. First, genotyping only a few hundred SNPs, compared to the 100,000’s of SNPs on a microarray, should be less expensive [21,27]. Second, by basing predictions on only those informative SNPs, we can remove variability caused by considering SNPs with little information. In this article, we aim to describe a method for selecting an ‘optimal’ group of SNPs, or a group that has maximal predictive accuracy given its size.
The first set of methods for marker selection ranked SNPs individually by some measure of their ability to distinguish ancestries, such as the estimated values for FST, the allele frequencies, p, or the Informativeness for Assignment, In, calculated from a training dataset, and then the top ranked SNPs were selected [24]. See section 5.1 for formal definitions of In and FST. Obviously, this led to redundant markers, and the majority of markers separated African from European ancestries. Therefore, the next set of methods were more advanced [1,13,30], and included a) Selecting those SNPs that are the strongest contributors to the principal components [22], b) Selecting the 1000 SNPs with the highest FST and then, among those, using a genetic algorithm to jointly select the set with the largest In [16], and c) Selecting a set of SNPs with a greedy algorithm aimed to minimize the apparent error rate [23]. Method a) can still select redundant SNPs and may rank those SNPs that distinguish closely related populations relatively low. Method b) uses unnecessary surrogates, FST and In, for predictive accuracy. Method c) is the most promising, as it directly tries to minimize the error rate.
Our goal is to improve the latter method by selecting SNPs that minimize a better estimate of the error rate. A selection procedure based on our new estimate, which introduces an improved form for the error rate and an improved estimate for the allele frequencies, will result in a better group of SNPs. Although our focus is on SNP selection, our discussion should have broader appeal. We will introduce a parametric estimator for the error rate that can be applied to any prediction rule based on genotypes, or, even more generally, any prediction rule using logistic regression to discriminate categories. Unlike the apparent error, this method acknowledges that the true allele frequencies are unknown [5, 8]. Moreover, our new estimate of allele frequencies offer a means to reduce the variance of the maximum likelihood estimates whenever knowledge of the evolutionary tree is available [10, 26]. Instead of estimating the allele frequencies for an ancestry using only subjects from that ancestry, we will now average over all available subjects in the training dataset. The need for these improvements has only arrived as we now attempt to predict ancestry more accurately than continental origin. For a training dataset, we now have access to the Human Genome Diversity Project (HGDP), where 500,000+ SNPs have been genotyped on hundreds of subjects from 54 populations [14].
The order of the paper is as follows. In section 2, we introduce our new selection procedure. In Section 3, we apply our selection procedure to both simulated and HGDP data. In Section 4, we conclude with a short discussion.
Methods
2.1 Introduction to a New Selection Procedure
2.1.1 Overview
As discussed in the introduction, our goal is to define a procedure for choosing a small set of SNPs that, when genotyped, can be used to predict an individual’s ancestry. We focus our search to one specific group of candidate procedures. Each selection procedure considered here will first estimate the expected error rate for every set of SNPs and will then choose a set with the lowest estimated error rate given its size. Obviously, computational limits prevent truly searching over every set of SNPs, but we will deal with that technical issue later. The key point is that we need only define an estimate of the error rate to define a selection procedure.
2.1.2 Notation
Assume we select n individuals from a heterogeneous population containing ne distinct ancestries or ethnicities and that we denote the ancestry of individual i by Yi ∈ {1,… ne}. We let nk be the total number of subjects from ancestry k, nk = ∑i 1(Yi = k), where 1(Yi = k) = 1 if Yi = k and 0 otherwise. In the heterogeneous population, from which our sample was obtained, we denote the proportion of subjects from ethnicity k by . We will presume that π⃗* can be accurately estimated and treated as a known, fixed, quantity. Note that the asterisk, *, denotes the true value of a parameter.
Assume a genome for an individual contains N SNPs and denote the three genotypes at each SNP by AA, AB, and BB. Denote the genotype for subject i at SNP j by Gij ∈ {AA, AB, BB} and the genotype at all SNPs by G⃗i. When referring to a subset, Ω ⊂ {1,…, N}, of those N SNPs, we denote the genotypes for those specific SNPs by G⃗i (Ω) ≡ {Gij : j ∈ Ω}. Let Xi = {G⃗i,Yi} be the genotype and ancestry information for subject i, and let X = {X1,…, Xn} be the training dataset.
The genotype frequencies at these N SNPs vary by population. For ancestry k, we denote the proportion of individuals with genotype g at SNP j by .
2.1.3 Maximum Likelihood Estimates
The maximum likelihood estimates, p̂ML, for p* will assume Hardy-Weinberg Equilibrium. Let
| (1) |
Then
| (2) |
Again, note that the MLE, p̂ML, are estimators of the true parameter p*.
2.1.4 Prediction Rule
For an individual i not in the training dataset, we would like to predict his ancestry using his genotype, G⃗i, and the training data, X. Our prediction rule, Ŷ, will assign the most likely ancestry to individual i assuming that p̂ML (X) were the true allele frequencies. This estimate is asymptotically optimal, in the sense that as n → ∞, this prediction rule will have the lowest possible error rate [24]. Because of its optimality, we chose this estimate over other options [12, 20].
To define the prediction rule formally, we need to provide equations for calculating the probability individual i has a specific genotype given his ethnicity and p*. The likelihood of the event can be written as
| (3) |
Next, we use Bayes theorem to define the probability that individual i is from a specific ancestry given his genotype, p*, and π⃗*,
| (4) |
If we knew p*, we would just classify individual i to the ancestry that maximized equation (4). However, without knowing the true value of p*, we replace p* with its estimate p̂ML. Then we can define our prediction rule, Ŷ, by
| (5) |
When clear, we may use one of the two abbreviation, Ŷi(X) or Ŷi(Ω). Similarly, when notation gets cumbersome, we omit π⃗* from P(Yi = k|G⃗i(Ω), p*, π⃗*).
2.1.5 Error Rate
There are two types of error rates for prediction. First, there is the expected error rate when p* is known. Second, there is the expected error rate when p* is unknown and an estimate, p̂ML, must be used in its place. These two error rates are distinct, and the error rate of interest is the second one. In the genetics literature, we are the first to propose a parametric estimate for this second, realistic, error rate.
Before describing our estimate, let us consider the example where we use known values of p* and π* to predict the ancestry of individual i, and another group aims to estimate our error rate. If this other group also knew the true parameters, they could accurately estimate our error rate by
| (6) |
Note that equation (6) is calculated as one minus the probability of correctly predicting the ancestry. Now, if this different group only knew p̂ML, their best prediction would be to plug p̂ML, π*, and Gi(Ω) into that same function. The result is the Apparent Error Rate
| (7) |
However, the true scenario, where our predictions are based only on estimates of p* presents a far more difficult challenge. There is no closed form equivalent to equation (6). Even if the other group knew the true parameters, they could not precisely calculate our error rate. In fact, we can only define a function, equation (13), that inputs the true parameters and outputs a consistent approximation of the error rate. The remainder of this section discusses the derivation of this equation.
We start by writing down a formula describing the error rate. Given our prediction rule and p*, the expected error rate for a given genotype, G⃗i(Ω), averaged over all possible training datasets can be described by equation (8). The probability of a training dataset is the probability of the observed genotypes given the ancestries (i.e. the product of equation 3 across all subjects). Note that in the prediction rule, we consider Ŷi to be a function of X and that Ŷi(X) is a random variable, as opposed to a fixed value. Here, the terms random and fixed refer to the training data.
| (8) |
Also, unless stated otherwise, we assume that ancestry is being predicted for an individual not in the training dataset so Ŷi(X) and Yi are independent. The probability of correctly predicting the ancestry is the sum, over all possible k, of the probability that the true ancestry is k multiplied by the probability that the predicted ancestry is k.
Unfortunately, equation (8) does not quite offer a way to calculate err(Gi(Ω), p*). For a subject with a specific genotype, we know how to calculate P(Yi = k|G⃗i(Ω), p*, π⃗*) using equation (4). However, we have yet to state a means to calculate P(Ŷi(X) = k|p*, π⃗*). Below, we suggest that P(Ŷi(X) = k|p*, π⃗*) can be approximated by the probability that a value drawn from one normal distribution is greater than k − 1 values drawn from other normal distributions. Unfortunately, there’s no closed form solution. The details of the derivation are left to the appendix, (section 5.3), and here we offer only a sketch of how to go from equation (8) to equation (12).
Consider the variable p̂ML(X). We can plug that variable into the function described by equation (4), to create a new variable P(Y = k|G⃗i(Ω), p̂ML(X), π⃗*). We can estimate the distribution of this continuous variable by
| (9) |
where
| (10) |
where κG⃗j = 1, 2, and 3 when Gij = AA, AB, and BB respectively and C is a constant independent of ancestry.
The prediction rule will output ancestry k, Ŷi = k, when P(Y = k|G⃗i(Ω), p̂ML(X)) is the largest among the probabilities for all ancestries. The probability of this event is the same as the probability that ZkG⃗i(p*) is greatest among {ZυG⃗i(p*)} where
| (11) |
Again, we exclude π⃗* to simplify notation. The Zυ are independent as the MLE for population k are derived only from the subjects within population k. Therefore, we take the following to be a satisfactory approximation of the error rate. Let
| (12) |
and as an approximation for the overall error rate, let
| (13) |
The foundation of this estimate is the ‘Prediction Focused Information Criteria’ described by Claeskens et. al. and Efron, et. al. for other scenarios [5, 8].
2.1.6 Estimated Error Rate and the Selection Procedure
We could estimate the error rate for a set of SNPs, Ω, by replacing p* with p̂ML in equation (13)
| (14) |
However, the maximum likelihood estimates, p̂ML, use only a small subset of the training data to estimate any given allele frequency. Estimates of are based only on subjects from ancestry k. We believe this to be wasteful because ancestries located near each other on an evolutionary tree should have similar allele frequencies. Therefore, we suggest estimating by an appropriate average of all p̂ML. Obviously, population k and its evolutionary neighbors will have the greatest weights in this average. In other words, we permit our estimates to be slightly biased when n is finite, and in return, our estimates will have much smaller variances. The details will be postponed to later, but we will create a Bayesian averaged estimate, p̂B, of p* and will ultimately suggest estimating the error rate by
| (15) |
Therefore our ideal selection procedure would be to estimate the error for every given set of SNPs, and then choose a set with the lowest error rate for its size. In practice, because it is computationally infeasible to search through all sets, we suggest using the greedy algorithm described in the appendix (section 5.4). Because our method does not naturally incorporate linkage disequilibrium (LD), we amend the standard greedy algorithm so that new SNPs cannot be added to the selected set if they are within approximately 75kb of any previously selected SNP.
2.2 Alternatives to
Instead of using the Improved Error Rates, we could have used the apparent error rate, , defined in equation (7). As the apparent error rate has been used for SNP selection previously [23], this will be one of the selection procedures discussed in our simulations and examples. As we will see, this estimate always underestimates the true error rate.
Because the apparent error performs poorly when dealing with 100,000’s of SNPs, we offer another selection procedure. This selection procedure uses the non-parametric 0.632+ bootstrap estimate for the error rate. We tried other non-parametric methods, including various methods of cross-validation, but in our simulations, we always found the 0.632+ bootstrap estimate to be the most accurate and precise. In general, the 0.632+ estimate outperforms other non-parametric estimates [7, 9]. Here, we briefly explain how to calculate this estimate.
A bootstrap sample, , is a randomly selected sample of n pairs of observations from {(G⃗1, Y1), (G⃗2, Y2),…, (G⃗n, Yn)}, with replacement. By chance, each bootstrap sample excludes some sets of observations. If we create our prediction rule based on , we can calculate the error rate for those excluded observations, leading to the leave-one-out estimate,
| (16) |
The bootstrap 0.632 estimator [7] combines and the observed error, ,
| (17) |
Note that uses the training data as test data as well. The bootstrap 0.632 estimate, , is defined as
| (18) |
The is a slight variation of this estimate, but for brevity we omit the details here and refer the reader elsewhere [9].
2.3 Calculating p̂B
As promised in section 2.1.6, we now describe p̂B. We found it best to propose a mathematical model that describes the development of the allele frequencies over time. We started with a single set of allele frequencies in one historical population, and then allowed these allele frequencies to change, in steps, as individuals spread around the globe and formed distinct populations. Given this model, we then calculate Bayesian estimates, p̂B, of the allele frequencies. We essentially obtain prior distributions for p* based on the evolutionary tree and then update these priors given the maximum likelihood estimates obtained in the training dataset. Therefore, our proposed estimate, given in equation (21), is E[p|p̂ML]. The remainder of this section shows the derivation of this estimate.
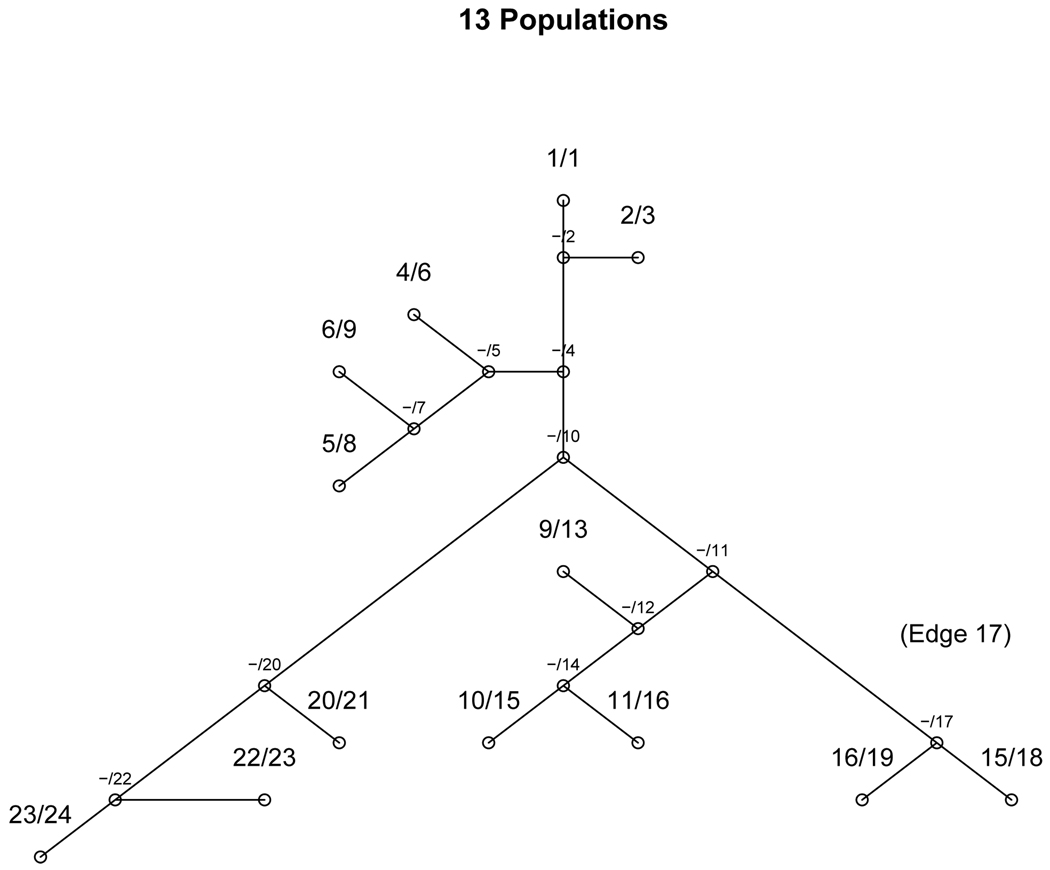
The new estimate, p̂B, takes advantage of a known evolutionary tree. Assume the tree has nn nodes (see figures 1, 2, and 3 for examples). There are ne terminal nodes, each representing an observed population, and nn − ne interior nodes, each representing a historical, combined, population. Label the nodes 1, 2, ‥, nn. Label the edges by the attached node, 2,…, nn, where the edge acquires the label of the larger of the two attached nodes.
Figure 1.
evolutionary Tree - 25 nodes. An example of an evolutionary tree with 25 nodes (nn = 25) and 20 populations (ne = 20). Next to each population node is the population identifier (k).
Figure 2.
evolutionary Tree - 24 nodes. An example of an evolutionary tree with 46 nodes (nn = 46) and 24 populations (ne = 24). Next to each population node is the population identifier (k). See appendix for population names.
Figure 3.
evolutionary Tree - 13 nodes. An example of an evolutionary tree with 24 nodes (nn = 24) and 13 populations (ne = 13). Next to each node is both the node identifier (node) and the population identifier (k), listed as k/node. Internal nodes are listed –/node. Edge 17 is labeled for discussion in the text.
In the actual model, we will assume that groups of populations share a common allele frequency at each SNP. We will introduce a vector S⃗ which identifies those ancestries sharing the same allele frequency. For SNP j, 1 ≤ j ≤ N we create an nn − 1 length vector of binary variables, S⃗j ≡ {S2j,…, Snnj}. If S⃗j = 0⃗ then pkj is the same for all populations. If Sυ1j = 1, but Sυj = 0 for all other υ ≠ υ1, then the populations prior to edge υ1 will share a common allele frequency, and the populations following edge υ1 will share a different common allele frequency. In figure 3 we label edge 17 for an example. If S17j = 1, but Sυj = 0 for all other υ, then populations 15 and 16 would share a common allele frequency, and all other populations would share a different frequency. Since Sυj = 1 allows allele frequencies to vary, we refer to it as a ‘bottleneck event’. In general, if two nodes, or populations, k1 and k2, can be connected by a set of edges, V, and Sυj = 0 ∀ υ ∈ V, then pk1j = pk2j. We place a prior distribution on Sυj, P(Sυj = 1) = αυ, where αυ can be an increasing function of the distance between the nodes adjacent to edge υ.
The number of unique allele frequencies, NSj, will be much smaller than the total number of ancestries, ne.
| (19) |
We denote the NSj unique allele frequencies by p(1)j,…, p(NSj)j, and we denote the number of subjects in populations with each of those NSj unique allele frequencies by n(1)j,…, n(NSj)j.
| (20) |
The Bayesian hierarchical model can be described as follows. We place a uniform prior on S. Given S, the distribution of p is f(p|S) = 1 for any set of allele frequencies consistent with S. Given p, we know the distributions of the training dataset. After, we perform the appropriate integrations, we can calculate the posterior means for p.
| (21) |
where nkj1 is the number of A alleles in population k, n(κ)j1 is the number of A alleles in populations with the κth unique allele frequency, nkj0 and n(κ)j0 are the respective quantities for the a allele. To minimize confusion, we note that although nkj and n(κ)j are numbers of subjects, nkj0, n(κ)j0, nkj1 and n(κ)j1 are numbers of alleles. Furthermore,
| (22) |
Note that p̂B requires specification of the hyperparameter {α2,…, αnn}. Derivation of this equation is in the appendix.
The model is an obvious simplification for the development of allele frequencies. In history, bottlenecks are actually rare events and allele frequencies change gradually over evolutionary development. Therefore, allele frequencies for neighboring populations should be highly correlated. For any given S·j, the abrupt bottlenecks may distort the estimated allele frequencies. However, by averaging over multiple S·j, we observe allele frequencies that vary smoothly across the evolutionary tree. Therefore, although we could try to incorporate a correlation structure into f(p|S) and allow for more events, these additions did not improve our estimates. We found our proposed model to perform as well as any of the more complex models examined.
2.4 Data and Simulation
2.4.1 Simulations: Aims
Our main objective is to understand and compare the selection procedures. We start with three sets of simulations, designed to answer three questions:
Which is a better estimate of p: p̂B or p̂ML?
Which is a better estimate of the error rate: ?
Which error rate is best for our selection procedure: ?
2.4.2 Simulations: Common Framework
The three sets of simulations examining these questions share a common framework. There are ne ancestries and these ancestries are related by an evolutionary tree with all edges of equal length. In these simulations, ne ∈ {13, 20, 24} and the possible evolutionary trees are illustrated in figures 1, 2, and 3. The evolutionary trees with 13 and 24 populations were trimmed versions of the tree that described the relationships among the HGDP populations [14]. Details of the tree follow in section 2.4.4. We assume that all populations are equally common and let the training dataset contain an equal number of subjects, 5, 10, or 20, from each population. Simulation results were always based 10,000 datasets.
For simulation sets 2 and 3, we introduce a new type of error. Recall that the expected error rate, err(Gi(Ω), p*) (equation (8)), is averaged over all possible training datasets. Now, we let errX(Gi(Ω), p*) be the error that would be observed given a specific value of p̂ML or a specific training dataset, X.
2.4.3 Simulations: Description
Simulation 1
General: For each dataset containing a group of subjects with one genotyped SNP, calculate p̂B and p̂ML and compare them to p*.
Specifics: Let ne = 20 and N = 1. To fairly compare p̂B and p̂ML, we examine three possible sets of allele frequencies, (1) No variation: for all k, (2) Inter-continental variation: for k ∈ {1,…, 5}, for k ∈ {6,…, 10}, for k ∈ {11,…, 15}, and for k ∈ {16,…, 20}, and (3) Intra-continental variation: , where d1 = 0.2 and d2 = 0.067.
Simulation 2
General: For each dataset, calculate . With the additional use of a test set containing 100,000 individuals, calculate errX. Compare the three estimated error rates with errX.
Specifics: Let ne ∈ {13, 24} and N ∈ {10, 40, 80}. We generate allele frequencies from the more complex evolutionary trees according to the Bayesian model described in section 2.3. For each SNP, we first generate S⃗j = {S2j,…, SV j}. For ne = 13, P(∑υ Sυj = t) = 1/3 for t ∈ {1,…, 3}. For ne = 24, P(∑υ Sυj = t) = 1/4 for t ∈ {1,…, 4}. Sυj are iid ∀υ. Allele frequencies for each connected set of populations are generated from a uniform[0.05,0.95] distribution.
Simulation 3
General: For each dataset, select the top 40 SNPs according to . Then using those SNPs and a training dataset, calculate . We compare these three error rates to see which is the lowest.
Specifics: Let ne ∈ {13, 20, 24} and N ∈ {1000, 10000}. Here, P(∑υ Sυj = 0) = 0.9 and the remaining probability is split evenly over ∑υ Sυj ∈ {1,…, 3} events when ne = 13 and ∑υ Sυj ∈ {1,…, 4} events when ne ∈ {20, 24}, where Sυj are iid ∀υ.
2.4.4 HGDP Data
Data 1
The Human Genome Diversity Project (HGDP) data set is more than an example. As it is the data set that will likely be used for selecting SNPs, the performance of the three possible selection procedures on this specific data set is of primary importance. As the HGDP grows and changes, the rankings of the three methods will need to be reevaluated. For this comparison, we use only a subset of the data, containing 400 subjects in 24 populations, from the HGDP (population names given in appendix). We limit our focus to those subjects with easily available data [14]. The evolutionary tree for these groups was based on pairwise allele-sharing distance among populations and had been previously estimated by Jakobsson [14]. We split the data into 50 sets of 10,000 SNPs. For each set of SNPs, we select the top 40 using the greedy algorithm with either on 80% of the data. Then we estimate the true error rate using using the remaining 20%. These error rates are then averaged over all 50 sets of data. Splitting the data into smaller sets was a necessity to decide whether the improvement in the set of SNPs selected by is statistically significant. In the supplementary material, we show the results from selecting SNPs according to a different set of methods. In these methods, the top ranked SNPs, where rankings are by FST, In, or the Optimal Rate of Correct Assignment (ORCA), are selected.
Data 2
We use the entire HGDP data set to select an optimal group of 100 SNPs for distinguishing ancestry. We start by selecting a candidate group of 5,000 SNPs. This group includes the 2000 SNPs (40 SNPs × 50 test sets) chosen from our initial 10,000-SNP searches. We then repeat the analysis described for data set 1 focusing on populations within each continent separately. Here, we select the top 20, as opposed to the top 40 SNPs. These chosen SNPs comprise the remaining 3,000 SNPs (3 continental regions × 20 SNPs × 50 data sets). The top 100 SNPs are selected from this set of 5,000 SNPs and listed in the supplementary material.
Results
3.1 Simulations
Simulation 1
The MLE, , are the most commonly used approximations for . The Bayesian estimates, , shrink the MLE toward the average value from neighboring populations. Therefore, if the truth is that neighboring populations share a common ‘A’ allele frequency, , at SNP j, then the mean square error, MSEML, for the MLE, should be larger than the MSEB for the Bayesian estimates, where . The first two columns in table 1 show that the improvement can be quite high when all populations in the study share a single . When populations can have different allele frequencies, the extent of the advantage or disadvantage depends on the evolutionary tree. The tradeoff between maximum likelihood and Bayesian estimates is a tradeoff between variance and bias. p̂B can be biased, but will have lower variance. In general, as the the number of subjects per population decreases, the MSEB : MSEML decreases, favoring estimation by p̂B (table 1).
Table 1.
The mean-square error , between the MLE and the truth, and the , between the Bayesian estimate and the truth. These MSE, averaged over all simulations, are listed for different sets of p, where a) all populations share a common p (No Var) b) all populations within a continent share a common p (Inter-Continental Var) c) all populations have a unique p (Intra-Continental Var)
| No Var | Inter-Continental Var | Intra-Continental Var | ||||
|---|---|---|---|---|---|---|
| MLE | Bayes | MLE | Bayes | MLE | Bayes | |
| 5 | 0.025 | 0.002 | 0.02 | 0.016 | 0.019 | 0.013 |
| 10 | 0.013 | 0.001 | 0.01 | 0.011 | 0.01 | 0.008 |
| 15 | 0.008 | 0.001 | 0.007 | 0.009 | 0.007 | 0.007 |
Simulation 2
We compared three options, for the 13 and 24 population examples (table 2). Clearly, greatly underestimates the true error, and the , where nsim is the number of simulations, is an order of magnitude larger than the MSE for either of the other estimates. The ratios, increase as the number of informative SNPs or the number of populations increases. In these simulations, tends to be lower than , but the order reverses as N grows large. The , with its default settings for α⃗, slightly overestimates the true value, but when calculating the MSE, this bias is offset by lower variance and a higher correlation between and errX.
Table 2.
A comparison of the three methods for estimating error rates . The first results column, errX, is the true unconditional error. The second set of results’ columns is the MSE from comparing the estimated errors with errx̂. The third set of columns is the standard deviation of each estimate. The fourth set of columns is the correlation between each estimate and errx̂.
| 13 Populations | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| errX | MSE | ||||||||||||||||
| nk | N | AE | 0.632+ | IBE | AE | 0.632+ | IBE | AE | 0.632+ | IBE | AE | 0.632+ | IBE | ||||
| 5 | 10 | 0.776 | 0.1311 | 0.0269 | 0.0204 | 0.6481 | 0.7662 | 0.786 | 0.0488 | 0.0477 | 0.0449 | 0.805 | 0.851 | 0.9178 | |||
| 5 | 40 | 0.4489 | 0.2669 | 0.0311 | 0.0278 | 0.1844 | 0.4461 | 0.4548 | 0.0403 | 0.0653 | 0.0684 | 0.8212 | 0.8815 | 0.9185 | |||
| 5 | 80 | 0.2357 | 0.2038 | 0.0287 | 0.023 | 0.0352 | 0.2486 | 0.2361 | 0.0144 | 0.0477 | 0.048 | 0.591 | 0.8456 | 0.8776 | |||
| 10 | 10 | 0.7797 | 0.133 | 0.0274 | 0.0197 | 0.6499 | 0.7684 | 0.7876 | 0.0505 | 0.0502 | 0.0467 | 0.8146 | 0.8674 | 0.9219 | |||
| 10 | 40 | 0.4426 | 0.263 | 0.03 | 0.0282 | 0.1819 | 0.4386 | 0.4476 | 0.0383 | 0.0625 | 0.0643 | 0.7828 | 0.8796 | 0.9027 | |||
| 10 | 80 | 0.2284 | 0.1987 | 0.0274 | 0.0221 | 0.0332 | 0.2415 | 0.2286 | 0.0134 | 0.0482 | 0.0494 | 0.5882 | 0.8676 | 0.8945 | |||
| 24 Populations | |||||||||||||||||
| errX | MSE |
|
|||||||||||||||
| nk | N | AE | 0.632+ | IBE | AE | 0.632+ | IBE | AE | 0.632+ | IBE | AE | 0.632+ | IBE | ||||
| 5 | 10 | 0.9037 | 0.1033 | 0.016 | 0.0099 | 0.8021 | 0.8962 | 0.9084 | 0.0306 | 0.0265 | 0.0219 | 0.802 | 0.8474 | 0.9178 | |||
| 5 | 40 | 0.7792 | 0.3432 | 0.0226 | 0.0207 | 0.4368 | 0.7704 | 0.7928 | 0.046 | 0.0455 | 0.0384 | 0.8553 | 0.8907 | 0.9174 | |||
| 5 | 80 | 0.6601 | 0.4851 | 0.0227 | 0.0257 | 0.1758 | 0.6573 | 0.6775 | 0.0307 | 0.0507 | 0.0484 | 0.8149 | 0.8952 | 0.9214 | |||
| 10 | 10 | 0.9044 | 0.1017 | 0.0154 | 0.0096 | 0.8044 | 0.8985 | 0.9081 | 0.0302 | 0.0267 | 0.0216 | 0.8032 | 0.852 | 0.9109 | |||
| 10 | 40 | 0.7814 | 0.3436 | 0.0215 | 0.0207 | 0.4386 | 0.7722 | 0.7949 | 0.0459 | 0.046 | 0.0413 | 0.863 | 0.907 | 0.9267 | |||
| 10 | 80 | 0.6542 | 0.4816 | 0.0227 | 0.0259 | 0.1734 | 0.6508 | 0.6699 | 0.0305 | 0.0488 | 0.0483 | 0.8016 | 0.8879 | 0.9043 | |||
Simulation 3
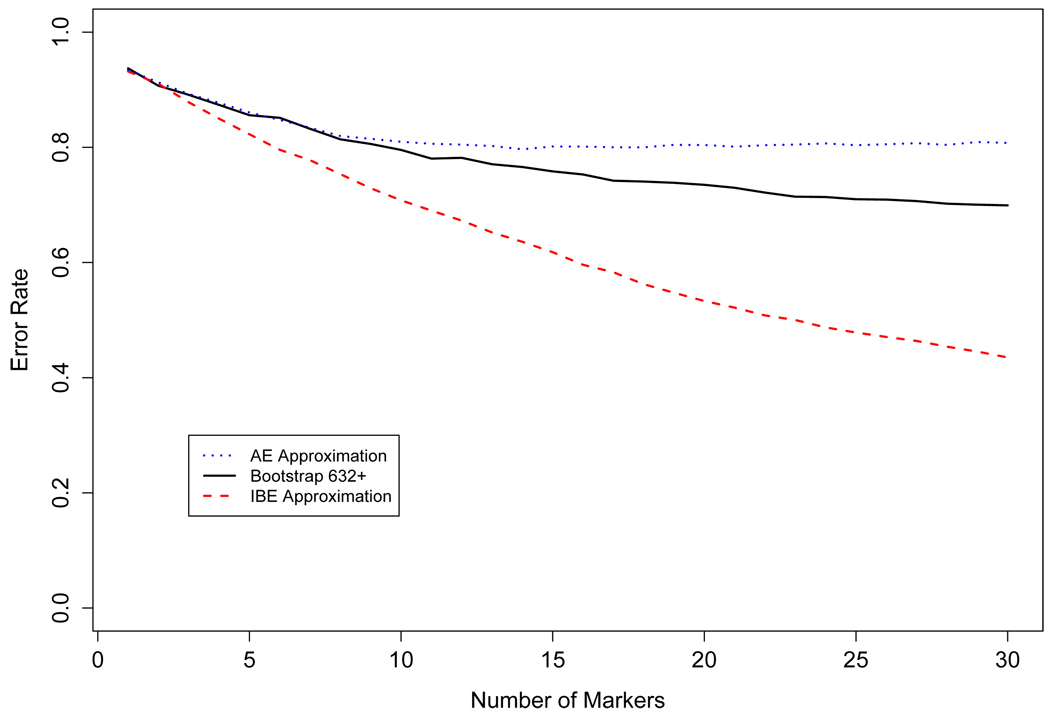
SNPs were selected by the greedy algorithm aimed to minimize either . For each group, the selected SNPs were ordered by the step in which they were added. Therefore, SNP 1 is essentially the most informative and SNP 40 is the least informative. For each group, the error rate was calculated (via simulation) when the top T SNPs were used, T ∈ {1,…, 40} and is illustrated in figure 4. The main point is that when more than 3 SNPs were used, the SNPs in ΩAE (the set chosen using the apparent error) proved to be poor predictors of the true ancestries. Selection based on resulted in lower error rates, and selection based on resulted in the lowest error rates. Therefore, these simulations clearly suggest that the use of is extremely inefficient and the use of can be the most efficient. However, as the simulation model unfairly favors , we hold off general statements about the -based selection procedure until we see the results for the HGDP data.
Figure 4.
Comparison of the error rates when the markers are chosen by er̂r632+, er̂rIBE, and er̂rAE using simulated data.
3.2 Data
Data 1
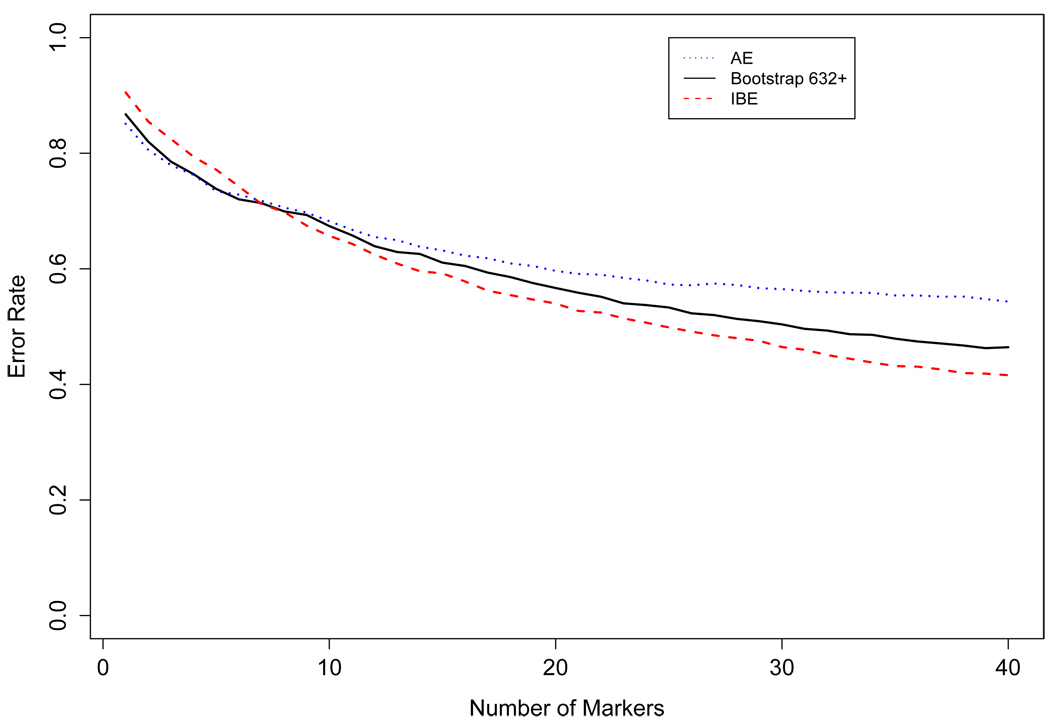
Selecting from groups of 10,000 SNPs, we denoted the resulting sets of 40 SNPs by . These SNPs and their corresponding p̂ML were then used to predict the ancestry for the 80 subjects in the separate test dataset, resulting in three sets of error rates errAE, err0.632+, and errIBE. These error rates were then averaged over all 50 sets of 10,000 SNPs to produce figure 5. The results are similar to those from the simulations, showing that SNP selection by outperformed both of the other selection procedures so long as there were more than 8 SNPs. Using only 8 SNPs, 77% of the subjects were assigned to a population in the correct continental region. Additional SNPs were selected to distinguish intra-continental populations. At this stage in the selection procedure, differences in allele frequencies due to random chance could rival informative differences, and because is designed to remove those that occur by chance, it starts to perform better.
Figure 5.
Comparison of the error rates when the markers are chosen by er̂r632+, er̂rIBE and er̂rAE using HGDP data.
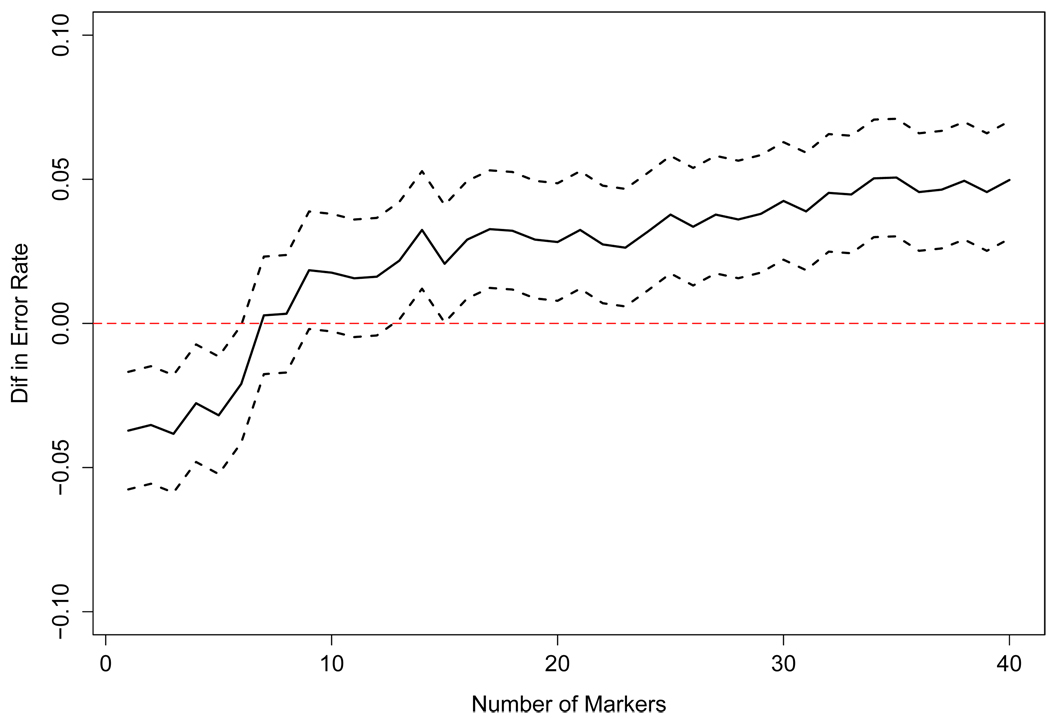
We then compared the 0.632+ and IBE methods to see whether selection by produced a statistically significantly better set of SNPs than selection by . Figure 6 shows the difference in error rate, , and a point-wise 95% CI, using the sample variance of the 50 values and assuming normality. The improvement was statistically significant. Although training sets contained only 80% of the data, we presume this benefit persists when selecting SNPs using all individuals. For future studies, we recommend splitting the data into training and test sets or using a cross-validation approach to choose the optimal method for selecting SNPs and, when desirable, to tune the hyper-parameter α⃗. Here, using simulations as our guide, we let (nn − 1)αυ = 7 ∀ υ.
Figure 6.
The difference between the error rates when the markers are chosen by er̂r632+ and er̂rIBE. The solid line is the difference, error rate using er̂r632+ - error rate using er̂rIBE, and the dotted lines are the point-wise 95% confidence intervals.
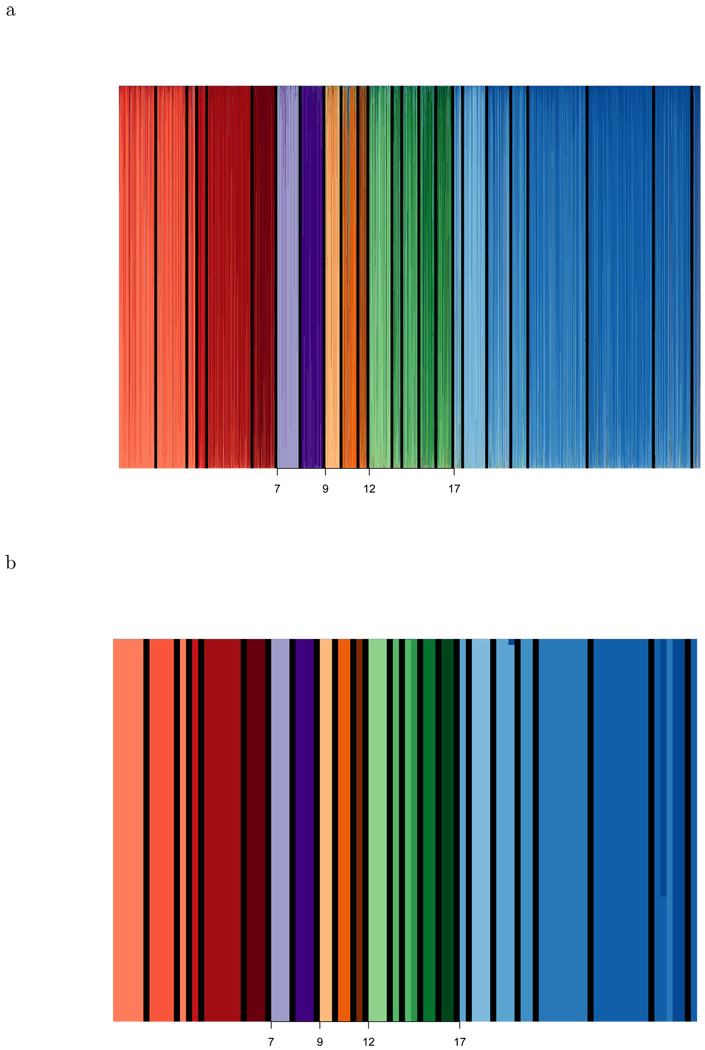
The error rate is still near 50% when Ω* includes 40 markers. However, a more detailed analysis of P(Y|G⃗(Ω), p̂ML, π̂) shows that the majority of errors involve classifying a subject from population k1 to population k2, where k1 and k2 are close to each other on the evolutionary tree. Figure 7, created by superStruct (available on author’s website), is similar to the output from STRUCTURE and shows that using 2000 markers reduces the error rate to near 0%. Each point on the axis corresponds to one of the subjects from one of the test data sets, and above that point, is a series of 24 stacked bars. Each bar has a unique color and represents a single population. The height of the colored bar corresponding to population k is proportional to the posterior probability, P(k|G⃗(Ω), p̂ML, π̂). Populations within the same continental region are different shades of the same color. The total number of subjects described by figure 7a is 3650 (= 73 subjects × 50 datasets). As for the overall potential for SNPs, we examined the predictive accuracy of all 2000 SNPs (40 SNPs × 50 datasets) and found near perfect identification (i.e. P(Ŷi = Yi|G⃗i(Ω), p̂ML, π̂) ≈ 1) for the majority of the 73 subjects (figure 7b). The 6 predictions that disagreed with the self-identification, (i.e. Ŷi ≠ Yi) were neighboring populations. This figure shows that we can do better than predicting continental origin.
Figure 7.
A graph to summarize the ancestry information for each individual. The x-axis indicates subject. For each subject, 24 bars, corresponding to the 24 populations, are stacked. The height of a bar k is the estimated probability that the individual is from that population. Each bar is a different color. Populations from one continent are varying shades of a single color: Red = Africa, Orange = America, Green = S.E. Asia, Blue = EuroAsia. Black lines separate populations. Top (a) and Bottom (b) images show the expected results using 40 and 2000 markers respectively. The total number of subjects described in the top half of the figure is 3650 (= 73 subjects × 50 datasets), whereas the bottom half of the figure describes only 73 subjects.
Data 2
We used to select an optimal set of 100 SNPs. Those SNPs are listed in the supplementary material.
Discussion
This article has introduced two ideas with an influence that should extend beyond SNP selection procedures. First, we offer an improved method for estimating the population specific allele frequencies. Second, we offer an improved method for estimating the error rate for prediction rules using genotypes. In fact, this latter method can be applied to any classification problem based on logistic regression. Focusing on the SNP selection procedures, we have demonstrated that selecting SNPs to minimize , instead of can lead to a group of SNPs that can predict ancestry with high accuracy.
The apparent error rate and maximum likelihood estimates have been successfully used in the past for selecting SNPs [23]. However, here the apparent error performed poorly. The main difference is that the number of populations has increased. With more populations, SNPs that truly separate a small group of populations no longer stand out. The population differences in p̂ML caused by sample selection are relatively small, but, in terms of the overall importance of a SNP, these differences are additive. Also, as the number of populations increases, we need more SNPs. As the number of needed SNPs increases, the improvement due to each additional SNP decreases, and it is more likely that a non-informative SNP can appear to be the best candidate. More general limitations of the apparent error rate are that it estimates the wrong quantity and cannot account for the fact that estimates of allele frequencies for some populations (i.e. those with more subjects in the training dataset) should be more accurate than others.
In this manuscript, we never actually considered using , and here we discuss one of its limitations and our reason for avoiding it. Although it has gone unstated in the literature, can be heavily biased because using p̂ML will exaggerate the true accuracy of the prediction rule. The following, simple, example illustrates that . Let there be one gene and two populations, where the allele frequencies in the populations are p1 and p2. As an extremely rough approximation, suitable only for illustration, consider the error to be a function of the difference log(p1) − log(p2) ≡ log(p1/p2). The true error generally increases as the distance between p1 and p2 decreases, with err attaining its maximum of 0.5 when log(p1/p2) = 0, or when the allele frequencies are the same in both populations. Now, assume we are unlucky, and the truth happens to be log(p1/p2) = 0. The estimate, is distributed around its true value, resulting in .
This bias discussed for is absent from , and therefore without the additional information from the evolutionary tree, would be the preferred method for estimating the error rate. However, we did find that performed favorably when compared to in the results section. Because of the nature of non-parametric estimates, it would be difficult to introduce the information from the evolutionary tree into .
Our study focused on individuals with only a single ancestry. However, our general conclusions about the SNP selection procedure and the selected SNPs will be valid when our goal is to identify the multiple ancestries of admixed individuals. Obviously the selected group of SNPs will need to be expanded to attain similar error rates. We suspect that the total number of SNPs needed to identify one of the admixed ancestries will be inversely proportional to the percentage of an individual’s genome originating with that ancestry. Instead of looking for ancestries of an individual, we would now be looking for ancestries of sections of the chromosomes. Admixture therefore requires a selection procedure that assumes only a random subset of the chosen SNPs will actually be available to identify a given ancestry. Therefore, the selected set should include some redundancy. This will also safeguard against genotyping error. We are currently exploring solutions for our two objectives in admixtures.
The next goal, already under examination, is how to incorporate the HGDP data and the knowledge of the optimal set of SNPs in identifying population substructure in Genome Wide Association Studies. First, most GWAS are large enough to contribute their own information about allele frequencies in populations. Second, GWAS are often more influenced by large population substructure, and may not need to identify populations that are not greatly present in the study. However, this focus is likely to change as we start searching for rare disease causing mutations.
Supplementary Material
Acknowledgements
This work was supported, in part, by NIJ grants 2007-DN-BX-K197 and 2010-DN-BX-K225 to KKK awarded by the National Institute of Justice, Office of Justice Programs, US Department of Justice. Points of view in this document are those of the authors and do not necessarily represent the official position or policies of the US Department of Justice.
Appendix
5.1 Definition of FST and In
The fixation index (FST) is a measure of population differentiation, and is usually the correlation of randomly chosen alleles within the same sub-population relative to that found in the entire population. To define FST, let be the frequency of allele “A” in population k, k ∈ {1,…, ne}, where ne is the number of ancestries and . Then
| (23) |
| (24) |
| (25) |
where wk is the relative size of the kth subpopulation and ∑k wk = 1. In our definitions of these quantities, wk is the relative size of each subpopulation in some larger, natural, population, not among the individuals sampled.
Information, I, is based on the idea of statistical entropy as described in [24]. The information, or Informativeness of Assignment is
| (26) |
where and q* = 1 − p*.
5.2 Calculating pB
Recall, for each SNP j, we defined a set of indicator variables, S⃗j ≡ {S2j,…, Snnj}, representing bottleneck events. Formally, a bottleneck is an evolutionary event in which only a small subset of individuals survives to the next generation, leading to a dramatic change in population allele frequencies. We use the term more liberally to imply any event that allows two neighboring populations to have different allele frequencies. Recall that NSj is the number of unique allele frequencies, n(κ)j is the number of subjects with the κth unique allele frequency, and ne is the total number of ancestries. Central to this calculation is the fact that the density of the estimated parameters, p̂, conditional on the true parameters, p, can be defined by
| (27) |
where
| (28) |
Note that f(·|·) represents a generic conditional density function, with the exact form depending on the variables in that function. Clearly, as f(p|S) is a constant when non-zero,
| (29) |
Because we know that f(p|S, p̂) is a density function, we know that f(p|S, p̂) must have the form for all non-zero values, where fβ(·|α, β) is the β density.
We know E[p|p̂] = ∑S E[p|S, p̂]f(S|p̂). We start by calculating f(S|p̂),
| (30) |
where we calculate C2 by noting that is a multiple of f β(p(κ)|n(κ)1 + 1, n(κ)0 + 1),
| (31) |
where B is beta function. Because we know that f(S|p̂) is a density function, we know that we can define and conclude that
| (32) |
To calculate E[p|S, p̂], note that for each value of S, we now know
| (33) |
where 1(pj = p(κ)) indicates whether population j shares the κth unique allele frequency. Therefore, we have arrived at
| (34) |
5.3 Estimating Error
Here, we show that log(P̂(Yi = k| G⃗i(Ω), p̂ML)) is asymptotically normal with mean μki and variance defined in equation (10). Start by focusing on SNP j in population k. Recall, nk is the number of subjects in population k and ne is the total number of ancestries. Then, our estimate of pkj is
| (35) |
where by the central limit theorem we know,
| (36) |
Next, we want to define our estimate for
| (37) |
and
| (38) |
We approximate the distribution of m̂kj by a linear function of p̂kj, specifically,
| (39) |
where and
| (40) |
Then, we have the following approximation
| (41) |
where
| (42) |
5.4 Greedy Algorithm
If the total of number of SNPs available is around 1,000,000, the number of possible groups grows at a rate of 1, 000, 000NS. It is computationally infeasible to search such a large space, making solution 2a impractical. Therefore, we propose using a greedy algorithm with NS steps.
Step 1: Select the single SNP j that minimizes the expected error rate: j1 = argminj err({j}, p*, π⃗*, p*, π⃗*).
Step 2…Ns: Given a set of n − 1 SNPs, {j1, j2,…, jn − 1}, select the SNP that when added to that current set minimizes the error rate: jn = argminj err({j1, j2,…, jn−1, j}, p*, π⃗*, p*, π⃗*).
Although {j1,…, jNs}, the set chosen by the greedy algorithm, is not guaranteed to be the optimal set, the set should perform satisfactorily, in that the resulting error rate should be similar to the true minimum.
5.5 Population Names
(Continental Region 1) 1: Yoruba (25), 2: Mandeka (20), 3: Bantu (8), 4: San (7), 5: Biaka Pygmy (32), 6: Mbuti Pygmy (15) (Continental Region 2) 7: Papuan (16), 8: Melanesian (17), 9: Pima (11), 10: Maya (13), 11: Columbian (7), 12: Yakut (15), 13: Mongola (9), 14: Daur (10), 15: Cambodian (10), 16: Yi (10) (Continental Region 3) 17: Burusho (7), 18:Kalash (18), 19: Balochi (15), 20: Russian (13), 21: Druze (43), 22: Beduin (47), 23: Palestinian (26), 24: Mozabite 96) Population Number: Population Name (Number of subjects in population)
References
- 1.Inferring ancestral origin using a single multiplex assay of ancestry-informative marker snps. Forensic Science International: Genetics. 2007;1(3–4):273–280. doi: 10.1016/j.fsigen.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Bamshad MJ, Wooding S, Watkins WS, Ostler CT, Batzer MA, Jorde LB. Human Population Genetic Structure and Inference of Group Membership. Nature Genetics. 2003;72(3):578–589. doi: 10.1086/368061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnholtz-Sloan JS, McEvoy B, Shriver MD, Rebbeck TR. Ancestry Estimation and Correction for Population Stratification in Molecular Epidemiologic Association Studies. Cancer Epidemiology Biomarkers and Prevention. 2008;17(3):471–477. doi: 10.1158/1055-9965.EPI-07-0491. [DOI] [PubMed] [Google Scholar]
- 4.Budowle B, van Daal A. BioTechniques. 2008;44:603–610. doi: 10.2144/000112806. [DOI] [PubMed] [Google Scholar]
- 5.Claeskens G, Croux C, Kerckhoven JV. Variable selection for logistic regression using a prediction-focused information criterion. Biometrics. 2006 December;62:972–979. doi: 10.1111/j.1541-0420.2006.00567.x. [DOI] [PubMed] [Google Scholar]
- 6.Daniel R, Walsh SJ, Piper A. Investigation of single-nucleotide polymorphisms associated with ethnicity. International Congress Series; Progress in Forensic Genetics 11 - Proceedings of the 21st International ISFG Congress held in Ponta Delgada, The Azores, Portugal between 13 and 16 September 2005; 2006. pp. 79–81. [Google Scholar]
- 7.Efron B. Estimating the error rate of a prediction rule: Improvement on cross-validation. Journal of the American Statistical Association. 1983;78(382):316–331. [Google Scholar]
- 8.Efron B. How biased is the apparent error rate of a prediction rule? Journal of the American Statistical Association. 1986;81(394):461–470. [Google Scholar]
- 9.Efron B, Tibshirani R. Improvements on cross-validation: The .632+ bootstrap method. Journal of the American Statistical Association. 1997;92(438):548–560. [Google Scholar]
- 10.Farris JS. Estimating phylogenetic trees from distance matrices. The American Naturalist. 1972;106(951):645–668. [Google Scholar]
- 11.Freedman ML, Reich D, Penney KL, McDonald GJ, Mignault AA, Patterson N, Gabriel SB, Topol EJ, Smoller JW, Pato CN, Pato MT, Petryshen TL, Kolonel LN, Lander ES, Sklar P, Henderson B, Hirschhorn JN, D A. Assessing the impact of population stratification on genetic association studies. Nature Genetics. 2004;36:388–393. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- 12.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning - Data Mining, Inference and Prediction. 2001 Springer [Google Scholar]
- 13.Hemminger BM, Saelim B, Sullivan PF. TAMAL: an integrated approach to choosing SNPs for genetic studies of human complex traits. Bioinformatics. 22(5):626–627. doi: 10.1093/bioinformatics/btk025. [DOI] [PubMed] [Google Scholar]
- 14.Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung H-C, Szpiech ZA, Degnan JH, Wang K, Guerreiro R, Bras JM, Schymick JC, Hernandez DG, Traynor BJ, Simon-Sanchez J, Matarin M, Britton A, van de Leemput J, Rafferty I, Bucan M, Cann HM, Hardy JA, Rosenberg N, Singleton AB. Genotype, haplotype, and copy number variation in worldwide human populations. Nature. 2008 February;451:998–10003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 15.Jorde LB, Wooding SP. Genetic variation, classification and ‘race’. Nat Genet. 2004;36:s28–s33. doi: 10.1038/ng1435. [DOI] [PubMed] [Google Scholar]
- 16.Lao O, Duijn KV, Kersbergen P, Knijff Pd, Kayser M. Proportioning whole genome single nucleotid polymorphism diversity for the identification of geographic population structure and genetic ancestry. American Journal of Human Genetics. 2006 April;78(4):680–690. doi: 10.1086/501531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM. Worldwide Human Relationships Inferred from Genome-Wide Patterns of Variation. Science. 2008;319(5866):1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 18.Lowe AL, Urquhart A, Foreman LA, Evett IW. Inferring ethnic origin by means of an str profile. Forensic Science International. 2001;119(1):17–22. doi: 10.1016/s0379-0738(00)00387-x. [DOI] [PubMed] [Google Scholar]
- 19.Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nature Genetics. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- 20.Michie D, Spiegelhalter DJ. Machine learning, neural and statistical classification. 1994 [Google Scholar]
- 21.Nassir R, Kosoy R, Tian C, White P, Butler L, Silva G, Kittles R, Alarcon-Riquelme M, Gregersen P, Belmont J, De La Vega F, Seldin M. An ancestry informative marker set for determining continental origin: validation and extension using human genome diversity panels. BMC Genetics. 2009;10(39) doi: 10.1186/1471-2156-10-39. 07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paschou P, Ziv E, Burchard EG, Choudhry S, Rodriguez-Cintron W, Mahoney MW, Drineas P. Pca-correlated snps for structure identification in worldwide human populations. PLoS Genet. 2007 Sep;3(9):e160. doi: 10.1371/journal.pgen.0030160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenberg NA. Algorithms for selecting informative marker panels for population assignment. Journal of Computational Biology. 2005 November;12(9):1183–1201. doi: 10.1089/cmb.2005.12.1183. [DOI] [PubMed] [Google Scholar]
- 24.Rosenberg NA, Li LM, Wark R, Pritchard JK. Information on genetic markers for inference of ancestry. Am J Hum Genet. 2003 December;73(6):1402–1422. doi: 10.1086/380416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic Structure of Human Populations. Science. 2002;298(5602):2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 26.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 27.Seldin MF, Price AL. Application of ancestry informative markers to association studies in european americans. PLoS Genet. 2008;4(1):e5. doi: 10.1371/journal.pgen.0040005. 01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shriver MD, Smith MW, Jin L, Marcini A, Akey JM, Deka R, Ferrell RE. Ethnic-affiliation estimation by use of population-specific dna markers. American Journal of Human Genetics. 1997;60 [PMC free article] [PubMed] [Google Scholar]
- 29.Weir BS, Cardon LR, Anderson AD, Nielsen DM, Hill WG. Measures of human population structure show heterogeneity among genomic regions. Genome Research. 2005;15(11):1468–1476. doi: 10.1101/gr.4398405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu H, Gregory SG, Hauser ER, Stenger JE, Pericak-Vance MA, Vance JM, Zuchner S, Hauser MA. SNPselector: a web tool for selecting SNPs for genetic association studies. Bioinformatics. 2005;21(22):4181–4186. doi: 10.1093/bioinformatics/bti682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaguchi-Kabata Y, Nakazono1 K, Takahashi A, Saito S, Hosono N, Kubo M, Nakamura Y, Kamatani N. Japanese Population Structure, Based on SNP Genotypes from 7003 Individuals Compared to Other Ethnic Groups: Effects on Population-Based Association Studies. American Journal of Human Genetics. 2008;83(4):445–456. doi: 10.1016/j.ajhg.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.