Abstract
Parasitized red blood cells (RBCs) from children suffering from severe malaria often adhere to complement receptor 1 (CR1) on uninfected RBCs to form clumps of cells known as “rosettes.” Despite a well documented association between rosetting and severe malaria, it is controversial whether rosetting is a cause or a correlate of parasite virulence. CR1-deficient RBC show greatly reduced rosetting; therefore, we hypothesized that, if rosetting is a direct cause of malaria pathology, CR1-deficient individuals should be protected against severe disease. In this study, we show that RBC CR1 deficiency occurs in up to 80% of healthy individuals from the malaria-endemic regions of Papua New Guinea. This RBC CR1 deficiency is associated with polymorphisms in the CR1 gene and, unexpectedly, with α-thalassemia, a common genetic disorder in Melanesian populations. Analysis of a case-control study demonstrated that the CR1 polymorphisms and α-thalassemia independently confer protection against severe malaria. We have therefore identified CR1 as a new malaria resistance gene and provided compelling evidence that rosetting is an important parasite virulence phenotype that should be a target for drug and vaccine development.
Severe malaria remains one of the largest causes of childhood mortality in the world. The number of deaths is high at ≈1–2 million per year; however, this number represents a small fraction of the total number of clinical malaria episodes that occur worldwide, estimated to be ≈400–900 million (1). Much work has been done to determine the factors that lead to the development of severe malaria, with parasite virulence phenotypes and host genetic factors being two major foci of research. One such Plasmodium falciparum virulence phenotype is rosetting, an adhesion property in which parasitized red blood cells (RBCs) bind to unparasitized RBCs to form clumps of cells (2). Rosetting is thought to contribute to malaria pathology by causing microvascular obstruction and impaired tissue perfusion (3, 4). Rosetting has been associated with severe malaria in many studies in Africa (e.g., refs. 5–7), although no association with severe disease was seen in Southeast Asia (8) or Papua New Guinea (PNG) (9). This inconsistency in the association of rosetting with severe disease has led some investigators to question the role of rosetting in malaria pathogenesis (10).
Rosetting is mediated by the parasite ligand PfEMP1 on the surface of infected RBCs (11, 12), binding to a variety of uninfected RBC receptors, including complement receptor 1 (CR1) (11, 13). CR1 is an immune-regulatory protein found on RBCs and a variety of leukocytes, and its functions include control of complement activation and the clearance of immune complexes (14). On RBCs, CR1 levels vary between individuals in the range of 50–1,200 molecules per cell (15–21). In Caucasians, RBC CR1 levels are genetically determined and are associated with at least three single-nucleotide polymorphisms (SNPs) in the CR1 gene (in exon 22, intron 27, and exon 33; ref. 22). These SNPs comprise high (H) and low (L) CR1 expression haplotypes that are codominant and are associated with CR1 level on RBC, but not on other cell types, such as B cells and macrophages (15). We have shown previously that CR1-deficient RBCs from LL homozygotes with <200 CR1 molecules per cell show greatly reduced rosetting with P. falciparum-infected RBCs (ref. 11 and J.A.R., unpublished data). We reasoned that if rosetting is important in malaria pathology, then CR1 deficiency should protect against severe disease by reducing rosettemediated microvascular obstruction. CR1 deficiency is thought to be rare in most populations; however, a review article (23) stated that 28 of 67 Melanesians had CR1-deficient RBCs. No experimental details were given, and no further information was published to validate this claim. This tantalizing suggestion that CR1 deficiency could be very common in Melanesians led us to examine the prevalence and genetic basis of CR1 deficiency in PNG and to determine whether RBC CR1 deficiency protects against severe malaria.
Methods
Study Sites and Sample Collection. We collected fresh blood samples into EDTA from healthy adult volunteers at two malaria-endemic sites within PNG (New Ireland and Madang) and from Edinburgh, United Kingdom. Informed consent was given by all donors after the aims of the project were explained, and the PNG Medical Research Advisory Committee approved all protocols. In Edinburgh and New Ireland the blood samples were analyzed fresh, whereas in Madang, buffy coats were removed for DNA extraction and the RBCs were fixed in 5% formaldehyde (24) before shipping to the United Kingdom. Buffy-coat samples from the Eastern Highlands Province (PNG) were kindly made available to us by the PNG Institute of Medical Research and came from a village 2,000 m above sea level, where no regular malaria transmission exists. New Ireland and Madang both have intense year-round malaria transmission with a spleen enlargement rate of >70% (25, 26).
Determination of RBC CR1 Level. The mean RBC CR1 level on both fresh and fixed blood samples was determined as described (24). In brief, the CR1 level was determined by flow cytometry using the anti-CR1 monoclonal antibody J3D3, with comparison to a standard curve derived from a set of reference RBCs with known CR1 levels. The reference RBCs used to establish the assay had their CR1 expression initially determined by the use of 125I-labeled antibody and Scatchard analysis to determine the number of antigenic sites per cell, controlling for the number of antibody-binding sites per CR1 molecule (24). We have shown that fresh and fixed RBC samples are similar and that CR1 level can be assessed accurately on fixed RBCs provided that the reference RBCs used to generate the standard curve are fixed in the same manner (24).
DNA Extraction and Genotyping. DNA was extracted from buffy coats by using the Nucleon BACC I kit (Amersham Pharmacia Life Science). Samples were genotyped for three SNPs in the CR1 gene at nucleotide 3650 in exon 22, a HindIII restriction fragment length polymorphism in intron 27, and at nucleotide 5507 in exon 33 by PCR and restriction digest as described (22). α-Thalassemia genotyping for the -α3.7 and -α4.2 deletions was done by multiplex PCR according to the method of Chong et al. (27). Southeast Asian ovalocytosis genotype was determined by PCR as described (28). DNA samples for the case-control study were as described (29, 30) and were amplified by primer-extension preamplification (31) before CR1 genotyping. α-Thalassemia genotyping for the case-control study was carried out by Southern blotting (29).
Determination of Glucose-6-phosphate Dehydrogenase (G6PD) Deficiency. Whole-blood samples from Madang and New Ireland were screened for G6PD deficiency by using Sigma Procedure no. 400 (Sigma).
Statistical Analysis. Statistical tests for differences in RBC CR1 levels between populations, between CR1 genotypes, and between α-thalassemia genotypes were performed by using multiway ANOVA and F tests in the statistical package SAS (SAS Institute, Cary, NC). Further analyses were performed to test for population differences in the frequencies of the CR1 alleles and α-thalassemia genotype by using the χ2 test or Fisher's exact test when the numbers of observations per cell were small. Odds ratios for the protective effects of genotypes in the case-control study were derived by logistic regression analysis in SAS. Data were analyzed by using both conditional and unconditional logistic regression; conditional logistic regression allows for matching of case-control pairs and maximizes power. However, because CR1 genotyping was not carried out in pairs, information was unnecessarily lost by using conditional logistic regression, and so results from unconditional analyses were used. This choice made no difference in the outcome of the analysis, nor did the fitting of ethnic groups in the model make a difference in the outcome.
Results
We measured the RBC CR1 levels of healthy adult volunteers from two highly malarious regions of PNG (Madang and New Ireland) and from a control Caucasian population from Edinburgh, United Kingdom. CR1 levels in Edinburgh varied between individuals in the range of 235–1,181 molecules per cell, with a mean of 786 (Fig. 1a). These results are similar to CR1 levels described in other Caucasian populations (Table 1), showing that our assay for CR1 (24) is similar to those used previously. In PNG, CR1 levels were significantly lower than in Edinburgh (Fig. 1a, P < 0.001), with 79% of individuals in Madang and 55% of individuals in New Ireland having fewer than 200 CR1 molecules per cell. The mean RBC CR1 levels in Madang and New Ireland are the lowest in the world (Table 1). These results indicate that, in malarious regions of PNG, CR1 deficiency is extremely common and could therefore play a major role in influencing susceptibility to severe malaria.
Fig. 1.
RBC CR1 deficiency is common in Melanesians and is associated with SNPs in the CR1 gene. (a) RBC CR1 levels in Edinburgh (United Kingdom), Madang (PNG), and New Ireland (PNG). Each point represents the mean RBC CR1 level of a single individual in molecules per cell. SD, standard deviation. (b–d) RBC CR1 levels in relation to CR1 genotype based on typing at the n3650 polymorphism in exon 22. H, high-expression allele; L, low-expression allele; na, not applicable.
Table 1. RBC CR1 levels and frequency of the CR1 low-expression (L) allele in different populations.
| Population | No. tested | Mean RBC CR1 in molecules per cell (range) | Gene frequency of the CR1 L allele | Ref. |
|---|---|---|---|---|
| Edinburgh, U.K. | 31 | 786 (235—1181) | 0.23 | This study |
| New Ireland (PNG)* | 47 | 253 (40—839) | 0.73 | This study |
| Madang (PNG)* | 96 | 124 (0—439) | 0.60 | This study |
| Eastern Highlands Province (PNG)† | 17 | ND | 0.41 | This study |
| United Kingdom | 60 | 624 (119—1220) | 0.27 | 16 |
| France | 84 | 613 (161—712) | 0.21 | 17 |
| African Americans | 54 | 635 (158—1023) | 0.25 | 19 |
| Caucasian Americans | 53 | 547 (89—1166) | 0.25 | 19 |
| India | 48 | 648 (140—1294) | 0.23 | 18 |
| Mali | 149 | 415 (58—1032) | 0.14 | 21 |
| China | 100 | 446 (27—1039) | 0.28 | 20 |
| Thailand | 30 | ND | 0.52 | 36 |
To ascertain whether RBC CR1 levels in Melanesians are genetically determined, we studied the three SNPs in the CR1 gene that have been associated with CR1 expression levels in Caucasians (22). The CR1 exon 22 SNP (A/G at nucleotide 3650) showed the strongest association with RBC CR1 level in all three populations; therefore, only exon 22 data are shown. At all three study sites the exon 22 genotype had a highly significant effect on RBC CR1 level, with carriers of the G3650 low (L) expression allele having significantly lower CR1 levels than HH individuals (Fig. 1 b–d, F2,96 = 50, P < 0.001). The RBC CR1 levels of HL individuals were intermediate between those of HH and LL individuals, as expected for codominant alleles. The frequency of the CR1 L allele in the malarious regions of PNG is the highest described in the world to date (Table 1, significantly different to Edinburgh, χ2 >6, P < 0.01). DNA samples were also studied from the nonmalarious Eastern Highlands Province of PNG, and the frequency of the L allele was found to be significantly lower than in the malarious regions (Table 1, χ2 = 8.1, P < 0.01). This may indicate selection for the L allele in areas with high malaria mortality, although other explanations cannot be excluded. The data above indicate that RBC CR1 levels in Melanesians are associated with polymorphisms in the CR1 gene. However, even when matched for exon 22 genotype, CR1 expression levels in Edinburgh, Madang, and New Ireland remained significantly different from each other (F2,96 = 66.0, P < 0.001). For example, the CR1 levels of HH individuals from PNG are lower than those of HH individuals from Edinburgh (Fig. 1 b–d), suggesting that additional factors influence CR1 levels in Melanesians.
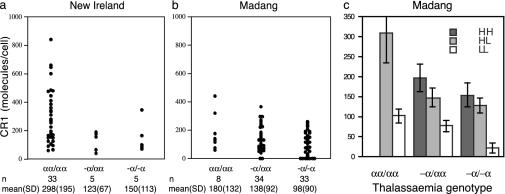
We investigated whether other RBC polymorphisms that occur commonly in PNG such as ovalocytosis (30), G6PD deficiency (32), and α-thalassemia (32) could affect RBC CR1 levels. α-Thalassemia occurred in 89% of individuals in Madang and in 23% of individuals in New Ireland. At both sites, individuals with one or more α-thalassemia mutations had RBC CR1 levels significantly lower than those of nonthalassemic individuals (Fig. 2 a and b, F1,111 = 12.4, P < 0.001). The effect of α-thalassemia genotype on RBC CR1 level was independent of the CR1 exon 22 polymorphism, because both genotypes were statistically significant when simultaneously included in the analysis (F2,110 = 10, P < 0.001 for α-thalassemia genotype; F2,110 = 44, P < 0.001 for exon 22 genotype). Thus, an individual's RBC CR1 level depends on both their CR1 exon 22 genotype and their α-globin genotype (Fig. 2c). Significant population differences still remained after including α-thalassemia and CR1 exon 22 genotype in the statistical model, suggesting that additional factors that influence RBC CR1 levels in Melanesians are undetected. Ovalocytosis and G6PD deficiency occurred in up to 15% of individuals in Madang and New Ireland and were not associated with RBC CR1 level (ovalocytosis, F1,115 = 0.2, P > 0.10; G6PD deficiency, F1,115 = 1.8, P > 0.10).
Fig. 2.
RBC CR1 deficiency in Melanesians is associated with α-thalassemia. (a and b) RBC CR1 level in relation to α-thalassemia genotype. Thalassemia genotypes are the following: -α/αα, heterozygotes for the -α4.2 deletion (32); -α/-α, homozygotes for the -α4.2 deletion; and αα/αα, normal α-globin gene structure. Only one individual carried the -α3.7 deletion (32); therefore, this case was excluded from the analysis. (c) The effect of α-thalassemia and CR1 exon 22 genotype on RBC CR1 level are independent of each other. The mean CR1 level for the Madang population subdivided by CR1 genotype (n3650 polymorphism) and α-thalassemia (-α4.2 deletion) is shown, and standard errors are indicated. The HH/normal α-globin genotype was not represented.
To determine whether the low CR1 levels in PNG influence susceptibility to severe malaria, we studied DNA samples from a case-control study carried out in Madang. These samples have been used previously to show that α-thalassemia and ovalocytosis confer protection against severe malaria (29, 30). One hundred eighty severe malaria cases and 179 community controls (matched for age, sex, ethnicity, season, and residential location) were genotyped for the CR1 exon 22 polymorphism. The data were analyzed by logistic regression, incorporating the previously identified protective factors of ovalocytosis (30) and α-thalassemia (29) into the model. We found that HL individuals for the CR1 exon 22 polymorphism were significantly protected against severe malaria (odds ratio, 0.33; P = 0.01; Table 2). The protective effect of the HL genotype was most prominent when present on a homozygous α-thalassemia background (odds ratio, 0.13; 95% confidence intervals, 0.02–0.67; P = 0.005). In contrast, HH/normal α-globin individuals, who would be expected to have the highest CR1 levels (Figs. 1 and 2), were most at risk of severe malaria, with this genotype being found in seven severe cases but no controls (P = 0.015 by Fisher's exact test). When the cases were subdivided into different severe malaria syndromes, the odds ratios for the HL genotype were reduced for severe anemia and cerebral malaria but not for other forms of severe malaria (a pool of metabolic complications, Table 2). The odds ratios for the LL genotype were also reduced for all severe malaria cases, severe anemia, and cerebral malaria but did not reach statistical significance (Table 2).
Table 2. The effect of the L allele on susceptibility to severe malaria.
| Genotype | Severe malaria cases, n (%) | Community controls, n (%) | Adjusted odds ratio (95% CI)* | P value |
|---|---|---|---|---|
| All severe cases | ||||
| n = 180 | n = 178 | |||
| HH | 21 (12) | 9 (5) | 1 | |
| HL | 57 (32) | 81 (46) | 0.33 (0.14—0.77) | 0.01 |
| LL | 102 (56) | 88 (49) | 0.55 (0.24—1.28) | 0.16 |
| Severe anemia | ||||
| n = 113 | n = 109 | |||
| HH | 15 (13) | 5 (5) | 1 | |
| HL | 34 (30) | 45 (41) | 0.30 (0.10—0.93) | 0.04 |
| LL | 64 (57) | 59 (54) | 0.46 (0.15—1.40) | 0.17 |
| Cerebral malaria | ||||
| n = 45 | n = 46 | |||
| HH | 4 (9) | 2 (4) | 1 | |
| HL | 15 (33) | 21 (46) | 0.40 (0.06—2.50) | 0.33 |
| LL | 26 (58) | 23 (50) | 0.60 (0.10—3.69) | 0.55 |
| Other severe malaria† | ||||
| n = 58 | n = 58 | |||
| HH | 5 (9) | 6 (10) | 1 | |
| HL | 18 (31) | 25 (43) | 1.18 (0.29—4.79) | 0.82 |
| LL | 35 (60) | 27 (47) | 2.23 (0.56—8.87) | 0.26 |
Odds ratios for the HL and LL genotypes are calculated in relation to the HH genotype after adjusting for the potential confounding factors ovalocytosis and α-thalassemia. CI, confidence intervals
Hypoglycemia, acidosis (low plasma bicarbonate), and hyperlactatemia; the odds ratios were similar for all three syndromes when analyzed separately
Discussion
This study shows that RBC CR1 deficiency is extremely common in malaria-endemic regions of PNG and that polymorphisms associated with CR1 deficiency confer protection against severe malaria. As parasites invade and grow normally in both CR1-deficient normal (33) and thalassemic RBCs (34), the simplest interpretation of our findings is that these polymorphisms protect against severe malaria through CR1 deficiency bringing about reduced rosetting (11, 35). Positive selection of rosette-reducing polymorphisms in a human population with high malaria mortality is strong evidence that CR1-mediated rosetting plays a causal role in the pathogenesis of severe malaria.
The case-control study results shown here indicate that heterozygotes for the CR1 low-expression allele (HL) are significantly protected from severe malaria, whereas homozygotes (LL) have a reduced odds ratio, but this reduction does not reach statistical significance. It is unclear whether the difference between the levels of protection provided by the HL and LL genotypes is genuine, and a larger sample size would be required to resolve this issue. The only previous small study of CR1 polymorphisms and malaria suggested that the LL genotype is a risk factor for severe malaria in Thai adults (36). In Southeast Asia, severe malaria in characterized mainly by metabolic disturbances and multiorgan failure (37) rather than by severe anemia and cerebral malaria, which occur commonly in children in PNG (38). Our data for the small subgroup of children with metabolic forms of severe malaria indicated a trend toward increased risk for LL individuals. It is possible that the L allele protects against the forms of severe malaria caused by microvascular obstruction and RBC damage (by means of reduced rosetting), but it increases risk for other syndromes with different pathological mechanisms, such as multiorgan failure and metabolic disturbances (39).
The results shown here linking RBC CR1 expression to α-thalassemia are striking. Although the protective effect of this disorder is well known (25, 29), the mechanism is debated (40). Parasite growth is unimpaired in thalassemic cells (34), so investigators have sought explanations based on reduced cytoadherence (41) or enhanced immune recognition (42). Carlson et al. (35) showed a large reduction in rosetting with thalassemic RBCs. They argued that this could be a result of microcytosis, because thalassemic cells have ≈15% less surface area than normal RBCs (42). However, thalassemic RBCs formed rosettes less well than microcytic RBCs with normal hemoglobin (35). Microcytosis is insufficient to explain the reduction in RBC CR1, which is ≈50% lower in thalassemic individuals than in those with normal α-globin (Fig. 2c). The mechanism responsible for low CR1 on thalassemic RBCs is unclear. CR1 is preferentially lost in vesicles from the cell surface as RBCs age or become ATP-depleted (43, 44). It is possible that biochemical abnormalities in thalassemic RBCs promote a similar process that would reduce their CR1 level.
The high prevalence of CR1 deficiency in Southeast Asia and PNG could explain why rosetting is not associated with malaria severity in this region (8, 9). Rosetting is greatly reduced in CR1-deficient RBCs, but some small rosettes still form in in vitro cultures, presumably by using other RBC receptors such as the A or B blood group sugars (45). These small and weak rosettes are unlikely to withstand the sheer forces encountered in the circulation in vivo. Therefore, many of the rosettes observed in vitro in Southeast Asia and PNG may not be physiologically relevant. In Africa, on the other hand, RBC CR1 deficiency does not appear to have been selected to high frequencies (19, 21). Instead, African populations possess other CR1 polymorphisms, such as the Knops blood group antigens that reach high frequencies in East and West Africa (ref. 46 and J.A.R. and J.M.M., unpublished data). Our initial DNA-based studies of these African CR1 polymorphisms have been complicated by a high level of genotype/phenotype mismatch (47); therefore, a prospective case-control study is underway to examine the effect of RBC CR1 phenotype on severe malaria in Africans. Clearly, the selective pressure of malaria has acted in different ways on the CR1 gene in diverse populations.
The results of this study have therapeutic implications. Case fatality rates for severe malaria are high, even in areas with intensive care facilities, so new approaches to treatment are urgently needed (48). Our findings indicate that therapies aimed at inhibiting CR1-mediated rosetting, such as soluble recombinant CR1 (49), could be of benefit in treating severe malaria. Alternatively, development of a vaccine against the parasite rosette-mediating ligand PfEMP1 (11) may prevent some cases of severe disease. The selective pressure of malaria on a major rosetting receptor indicated by our results strongly supports a direct role for CR1-mediated rosetting in the pathogenesis of severe disease and emphasizes that further research to develop rosette-inhibiting interventions should be a high priority.
Acknowledgments
We thank all the volunteers for donating blood for the study, Dr. E. Mackinnon for the collection of blood samples from New Ireland, and Professor David Weatherall and Professor John Clegg for permission to use the case-control samples. This work was funded by Wellcome Trust Grants 055167 and 067431 and by the United Kingdom Medical Research Council.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PNG, Papua New Guinea; G6PD, glucose-6-phosphate dehydrogenase; CR1, complement receptor 1; SNP, single-nucleotide polymorphism.
References
- 1.Breman, J. G. (2001) Am. J. Trop. Med. Hyg. 64, 1-11. [DOI] [PubMed] [Google Scholar]
- 2.David, P. H., Handunnetti, S. M., Leech, J. H., Gamage, P. & Mendis, K. N. (1988) Am. J. Trop. Med. Hyg. 38, 289-297. [DOI] [PubMed] [Google Scholar]
- 3.Kaul, D. K., Roth, E. F. J., Nagel, R. L., Howard, R. J. & Handunnetti, S. M. (1991) Blood 78, 812-819. [PubMed] [Google Scholar]
- 4.Nash, G. B., Cooke, B. M., Carlson, J. & Wahlgren, M. (1992) Br. J. Haematol. 82, 757-763. [DOI] [PubMed] [Google Scholar]
- 5.Carlson, J., Helmby, H., Hill, A. V., Brewster, D., Greenwood, B. M. & Wahlgren, M. (1990) Lancet 336, 1457-1460. [DOI] [PubMed] [Google Scholar]
- 6.Rowe, A., Obeiro, J., Newbold, C. I. & Marsh, K. (1995) Infect. Immun. 63, 2323-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ringwald, P., Peyron, F., Lepers, J. P., Rabarison, P., Rakotomalala, C., Razanamparany, M., Rabodonirina, M., Roux, J. & Le Bras, J. (1993) Infect. Immun. 61, 5198-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho, M., Davis, T. M., Silamut, K., Bunnag, D. & White, N. J. (1991) Infect. Immun. 59, 2135-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.al-Yaman, F., Genton, B., Mokela, D., Raiko, A., Kati, S., Rogerson, S., Reeder, J. & Alpers, M. (1995) Trans. R. Soc. Trop. Med. Hyg. 89, 55-58. [DOI] [PubMed] [Google Scholar]
- 10.Miller, L. H., Baruch, D. I., Marsh, K. & Doumbo, O. K. (2002) Nature 415, 673-679. [DOI] [PubMed] [Google Scholar]
- 11.Rowe, J. A., Moulds, J. M., Newbold, C. I. & Miller, L. H. (1997) Nature 388, 292-295. [DOI] [PubMed] [Google Scholar]
- 12.Chen, Q., Barragan, A., Fernandez, V., Sundstrom, A., Schlichtherle, M., Sahlen, A., Carlson, J., Datta, S. & Wahlgren, M. (1998) J. Exp. Med. 187, 15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rowe, J. A., Rogerson, S. J., Raza, A., Moulds, J. M., Kazatchkine, M. D., Marsh, K., Newbold, C. I., Atkinson, J. P. & Miller, L. H. (2000) J. Immunol. 165, 6341-6346. [DOI] [PubMed] [Google Scholar]
- 14.Ahearn, J. M. & Fearon, D. T. (1989) Adv. Immunol. 46, 183-219. [DOI] [PubMed] [Google Scholar]
- 15.Wilson, J. G., Murphy, E. E., Wong, W. W., Klickstein, L. B., Weis, J. H. & Feardon, D. T. (1986) J. Exp. Med. 164, 50-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moldenhauer, F., David, J., Fielder, A. H., Lachmann, P. J. & Walport, M. J. (1987) Arthritis Rheum. 30, 961-966. [DOI] [PubMed] [Google Scholar]
- 17.Cohen, J. H., Caudwell, V., Levi-Strauss, M., Bourgeois, P. & Kazatchkine, M. D. (1989) Arthritis Rheum. 32, 393-397. [DOI] [PubMed] [Google Scholar]
- 18.Kumar, A., Kumar, A., Sinha, S., Khandekar, P. S., Banerjee, K. & Srivastava, L. M. (1995) Immunol. Cell Biol. 73, 457-462. [DOI] [PubMed] [Google Scholar]
- 19.Herrera, A. H., Xiang, L., Martin, S. G., Lewis, J. & Wilson, J. G. (1998) Clin. Immunol. Immunopathol. 87, 176-183. [DOI] [PubMed] [Google Scholar]
- 20.Moulds, J. M., Brai, M., Cohen, J., Cortelazzo, A., Cuccia, M., Lin, M., Sadallah, S., Schifferli, J., Bala Subramanian, V., Truedsson, L., et al. (1998) Exp. Clin. Immunogenet. 15, 291-294. [DOI] [PubMed] [Google Scholar]
- 21.Rowe, J. A., Raza, A., Diallo, D. A., Baby, M., Poudiougo, B., Coulibaly, D., Cockburn, I. A., Middleton, J., Lyke, K. E., Plowe, C. V., et al. (2002) Genes Immun. 3, 497-500. [DOI] [PubMed] [Google Scholar]
- 22.Xiang, L., Rundles, J. R., Hamilton, D. R. & Wilson, J. G. (1999) J. Immunol. 163, 4939-4945. [PubMed] [Google Scholar]
- 23.Molthan, L. (1983) Med. Lab. Sci. 40, 59-63. [PubMed] [Google Scholar]
- 24.Cockburn, I. A., Donvito, B., Cohen, J. H. & Rowe, J. A. (2002) J. Immunol. Methods 271, 59-64. [DOI] [PubMed] [Google Scholar]
- 25.Flint, J., Hill, A. V., Bowden, D. K., Oppenheimer, S. J., Sill, P. R., Serjeantson, S. W., Bana, K. J., Bhatia, K., Alpers, M. P., Boyce, A. J., et al. (1986) Nature 321, 744-750. [DOI] [PubMed] [Google Scholar]
- 26.Cattani, J. A., Tulloch, J. L., Vrbova, H., Jolley, D., Gibson, F. D., Moir, J. S., Heywood, P. F., Alpers, M. P., Stevenson, A. & Clancy, R. (1986) Am. J. Trop. Med. Hyg. 35, 3-15. [DOI] [PubMed] [Google Scholar]
- 27.Chong, S. S., Boehm, C. D., Higgs, D. R. & Cutting, G. R. (2000) Blood 95, 360-362. [PubMed] [Google Scholar]
- 28.Jarolim, P., Palek, J., Amato, D., Hassan, K., Sapak, P., Nurse, G. T., Rubin, H. L., Zhai, S., Sahr, K. E. & Liu, S. C. (1991) Proc. Natl. Acad. Sci. USA 88, 11022-11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allen, S. J., O'Donnell, A., Alexander, N. D., Alpers, M. P., Peto, T. E., Clegg, J. B. & Weatherall, D. J. (1997) Proc. Natl. Acad. Sci. USA 94, 14736-14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen, S. J., O'Donnell, A., Alexander, N. D., Mgone, C. S., Peto, T. E., Clegg, J. B., Alpers, M. P. & Weatherall, D. J. (1999) Am. J. Trop. Med. Hyg. 60, 1056-1060. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, L., Cui, X., Schmitt, K., Hubert, R., Navidi, W. & Arnheim, N. (1992) Proc. Natl. Acad. Sci. USA 89, 5847-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yenchitsomanus, P., Summers, K. M., Chockkalingam, C. & Board, P. G. (1986) PNG Med. J. 29, 53-58. [PubMed] [Google Scholar]
- 33.Soubes, S. C., Reid, M. E., Kaneko, O. & Miller, L. H. (1999) Vox Sang 76, 107-114. [PubMed] [Google Scholar]
- 34.Luzzi, G. A., Torii, M., Aikawa, M. & Pasvol, G. (1990) Br. J. Haematol. 74, 519-524. [DOI] [PubMed] [Google Scholar]
- 35.Carlson, J., Nash, G. B., Gabutti, V., al-Yaman, F. & Wahlgren, M. (1994) Blood 84, 3909-3914. [PubMed] [Google Scholar]
- 36.Nagayasu, E., Ito, M., Akaki, M., Nakano, Y., Kimura, M., Looareesuwan, S. & Aikawa, M. (2001) Am. J. Trop. Med. Hyg. 64, 1-5. [DOI] [PubMed] [Google Scholar]
- 37.White, N. J. (1987) Acta Leidensia 56, 27-47. [PubMed] [Google Scholar]
- 38.Allen, S. J., O'Donnell, A., Alexander, N. D. & Clegg, J. B., (1996) QJM 89, 779-788. [DOI] [PubMed] [Google Scholar]
- 39.Warrell, D. A. (1987) Parasitology 94, S53-S76. [DOI] [PubMed] [Google Scholar]
- 40.Weatherall, D. J., Miller, L. H., Baruch, D. I., Marsh, K., Doumbo, O. K., Casals-Pascual, C. & Roberts, D. J. (2002) Hematology (Am. Soc. Hematol. Educ. Program) 1, 35-57. [DOI] [PubMed] [Google Scholar]
- 41.Udomsangpetch, R., Sueblinvong, T., Pattanapanyasat, K., Dharmkrong-at, A., Kittikalayawong, A. & Webster, H. K. (1993) Blood 82, 3752-3759. [PubMed] [Google Scholar]
- 42.Luzzi, G. A., Merry, A. H., Newbold, C. I., Marsh, K., Pasvol, G. & Weatherall, D. J. (1991) J. Exp. Med. 173, 785-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pascual, M., Lutz, H. U., Steiger, G., Stammler, P. & Schifferli, J. A. (1993) J. Immunol. 151, 397-404. [PubMed] [Google Scholar]
- 44.Dervillez, X., Oudin, S., Libyh, M. T., Tabary, T., Reveil, B., Philbert, F., Bougy, F., Pluot, M. & Cohen, J. H. (1997) Immunopharmacology 38, 129-140. [DOI] [PubMed] [Google Scholar]
- 45.Carlson, J. & Wahlgren, M. (1992) J. Exp. Med. 176, 1311-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moulds, J. M., Kassambara, L., Middleton, J. J., Baby, M., Sagara, I., Guindo, A., Coulibaly, S., Yalcouye, D., Diallo, D. A., Miller, L. & Doumbo, O. (2000) Genes Immun. 1, 325-329. [DOI] [PubMed] [Google Scholar]
- 47.Zimmerman, P. A., Fitness, J., Moulds, J. M., McNamara, D. T., Kasehagen, L. J., Rowe, J. A. & Hill, A. V. S. (2003) Genes Immun. 4, 368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warrell, D., A. (1999) Parassitologia (Rome) 41, 287-294. [PubMed] [Google Scholar]
- 49.Weisman, H. F., Bartow, T., Leppo, M. K., Marsh, H. C., Carson, G. R., Concino, M. F., Boyle, M. P., Roux, K. H., Weisfeldt, M. L. & Fearon, D. T. (1990) Science 249, 146-151. [DOI] [PubMed] [Google Scholar]