Abstract
The 3D structure of the influenza virus polymerase complex was determined by electron microscopy and image processing of recombinant ribonucleoproteins (RNPs). The RNPs were generated by in vivo amplification using cDNAs of the three polymerase subunits, the nucleoprotein, and a model virus-associated RNA containing 248 nt. The polymerase structure obtained is very compact, with no apparent boundaries among subunits. The position of specific regions of the PB1, PB2, and PA subunits was determined by 3D reconstruction of either RNP–mAb complexes or tagged RNPs. This structural model is available for the polymerase of a negative-stranded RNA virus and provides a general delineation of the complex and its interaction with the template-associated nucleoprotein monomers in the RNP.
Many viruses contain RNA as genetic material and use RNA-dependent RNA polymerases as enzymes for replication and transcription. The atomic structures of several viral RNA (vRNA)-dependent RNA polymerases from positive- and double-stranded RNA viruses are available (1–6). These proteins show little sequence homology with other DNA-dependent polymerases but contain specific sequence motifs shared with other polymerases (7). They show a typical right-hand fold that includes the thumb, finger, and palm domains, the last of which contains the conserved sequence motifs involved in polymerase catalysis. These polymerases are medium-size proteins that show activity in vitro, although viral replication and transcription in vivo are carried out by polymerase-containing macrocomplexes. In these, other viral and cellular factors contribute to the efficiency and regulation of the polymerase or to the localization of the RNA synthetic machinery in the infected cell (reviewed in ref. 8).
The situation in negative-strand viruses (NSVs) is different. The templates for transcription and replication are ribonucleoproteins (RNPs) in which the negative-stranded RNA is complexed with the nucleoprotein (NP) and associated to the polymerase. In the NSVs, the polymerase is a very large protein, the L protein (≈ 250 kDa, except in the Orthomyxoviridae), which consists of a heterotrimer of similar aggregate size. The replication intermediates are RNPs similar to virion RNPs but containing positive-stranded RNA. Other viral proteins, such as the P protein for nonsegmented NSVs, and cellular factors are essential for transcription or replication of NSV RNPs. As a consequence of this complexity, very little structural information is available on NSV polymerases.
Within the NSVs, influenza A viruses are distinct in having a segmented genome and replicating in the cell nucleus (9). Their genome consists of eight RNPs that are transcribed and replicated by the viral polymerase, a heterotrimer composed by the PB1, PB2, and PA subunits (10, 11). Replication of vRNA involves de novo initiation of nascent RNA chains, whereas initiation of mRNA synthesis requires the generation of capped primers from cellular heterogeneous nuclear RNAs by a cap-stealing mechanism (12). Transcription termination needs the interaction of the polymerase with 5′ terminal sequences of the template (13) and occurs at the polyadenylation signal, which consists of an oligo-U sequence (14, 15).
Our knowledge about influenza polymerase has increased over the years. Regions of each subunit involved in complex formation have been defined (16–19), and the domains in the PB1 subunit that bind vRNA and cRNA have been determined (20–22). The PB1 subunit harbors the polymerase activity (7, 23), but the function of other subunits is becoming more diffuse. The PB2 subunit is a cap-binding protein (24), but the endonuclease activity has been located in the PB1 subunit (25). In addition, mutations in the N-terminal region of the PB2 subunit alter the viral replication capacity but not the transcription activity of RNPs (26). The PA subunit has been associated with the replication activity of the polymerase (27, 28). However, a PA mutation has been reported that abolishes cap-snatching but not RNA replication (29). On the other hand, our knowledge about the structure of the influenza virus RNP and its polymerase is very limited. Virion RNPs are supercoiled ribbon structures (30, 31) with the polymerase at one end of the supercoil (32), in association with both vRNA ends (33). vRNA–NP complexes or RNA-free NPs form structures similar to natural RNPs (34). More recently, we have reported the 3D reconstruction of recombinant RNPs containing short model vRNAs (35, 36). Here, we report the structure at a 23-Å resolution of the polymerase complex present in these RNPs, as determined by electron microscopy and image processing, and the localization of specific subunit domains by 3D reconstruction of tagged RNPs and RNP–mAb complexes.
Materials and Methods
Biological Materials. Plasmids pGPB1, pGPB2, pGPA, and pGNP(poly)A have been described (37). Plasmid pT7ΔNSRT clone 23, encoding a 248-nt model vRNA under a T7 promoter and the hepatitis δ ribozyme, has been described (35). Plasmid pBS1479 (38), containing the tandem affinity purification (TAP) tag, was provided by B. Séraphin (Centre National de la Recherche Scientifique, Gif-sur-Yvette, France). mAbs specific for PA and PB2 have been described (39).
Mutant Construction. For construction of the pGEMPB2-His plasmid, a PB2 ORF was amplified with a forward primer containing a NcoI site at the initiation codon. The PCR product was digested with NcoI and EcoRI and cloned into pTRC2B plasmid in-frame with the vector C-terminal His tag. The His-tagged PB2 gene was subcloned into pGPB2 by exchanging the PflmI–ScaI restriction fragment. The pGPB2-TAP plasmid was constructed by replacement of the BsaBI–XbaI fragment of pGPB2 (40 bp, containing the C terminus of the PB2 ORF) by a PCR product of the TAP tag. The forward primer contained the codons of PB2 that were eliminated by excision of the BsaBI–XbaI fragment, thereby restoring a complete TAP-tagged PB2 gene. To generate the pGPB1-TAP plasmid, the PB2 ORF present in pGPB2-TAP was eliminated by restriction with KpnI and BamHI and replaced by a PCR product of the PB1 ORF. The structure of the plasmids was tested by restriction analysis and sequencing. The expression of the encoded tagged protein was ascertained by infection/transfection (35) and Western blotting with anti-polymerase and anti-tag Abs. The functionality of the recombinant proteins was tested by reconstitution of RNPs as indicated below.
Reconstitution and Purification of Recombinant RNPs. Reconstitution and in vivo amplification of recombinant RNPs was carried out essentially as described (35, 36) by transfection of vaccinia vTF7-3-infected cells with plasmids pGPB1, pGPB2, pGPA, pGNP(poly)A, and pT7ΔNSRT clone 23. In the appropriate experiments, plasmids pGPB2-His or pGPB1-TAP were substituted for the corresponding WT plasmids. For purification of His-tagged RNPs, the clarified extract was centrifuged in a glycerol step gradient in TN buffer (150 mM NaCl/50 mM Tris·HCl, pH 7.8) for 17 h at 35,000 rpm and 4°C in a SW41 rotor. Occasionally, the centrifugation step was repeated under the same conditions. The transcriptionally active fractions were diluted to 5 ml in 50 mM Tris·HCl/100 mM NaCl/5 mM MgCl2/0.1% Nonidet P-40/20 mM imidazol (pH 8.0) and incubated with an Ni2+-NTA agarose resin for 12–14 h at 4°C, and the resin was washed 10 times with 10 column volumes of the same buffer. The bound His-tagged RNPs were eluted with 50 mM Tris·HCl/100 mM KCl/100 mM EDTA (pH 8.0). For purification of RNP–Ab complexes, the active fractions after the first glycerol step gradient were incubated with excess purified mAb, and the mixture was centrifuged again in a similar glycerol gradient. Finally, the RNP–Ab complexes were purified on an Ni2+-NTA agarose column as described above.
Protein Analyses. mAbs were purified from culture supernatants by chromatography on IgG purification resin (Stratagene). Western blotting was carried out as described (35), by using a mixture of anti-PA or anti-PB2 mAbs, anti-PB1, or anti-NP serum. The His tag was revealed with anti-His IgG coupled to peroxidase. The TAP tag was revealed with unrelated rabbit Ab. Rabbit IgGs were revealed with biotin-labeled goat anti-rabbit Ab. For silver staining, the gels were fixed for 30 min each in 25% ethanol/10% acetic acid and 25% ethanol/0.5% acetic acid, soaked twice for 20 min in silver nitrate (1.8 g/liter), and washed three times with H2O. Reduction was made in 10 mM NaBH4/0.75 M NaOH/0.75% formaldehyde, and the reaction was stopped in 10% acetic acid.
Electron Microscopy and Image Processing. Samples were applied to carbon-coated copper grids previously glow-discharged at low air pressure, stained with 2% uranyl acetate, and visualized in a JEOL 1200 EXII microscope at a nominal magnification of ×40,000 and a defocus range of 3,000–5,000 Å. Photographic plates exposed under a low-dose protocol were digitized at 3.5 Å per pixel. Individual images were processed in 2D and aligned by cross-correlation free-pattern methods (40) by using the xmipp package (41). When appropriate, the images were subjected to Kohonen's self-organizing feature maps (42). Homogeneous populations were obtained and averaged. For 3D reconstruction, the sample stage was tilted 30°. The model was refined by using the angular refinement algorithms provided by spider (43), and the reconstruction was performed by using art (44). The handedness of the reconstructed volumes was chosen arbitrarily because of the intrinsic ambiguity generated by the electron microscopy reconstruction procedure. The resolution of the model was estimated by Fourier ring correlation of two independent reconstructions, and these values were used to low-pass filter the volume in the final model.
Results and Discussion
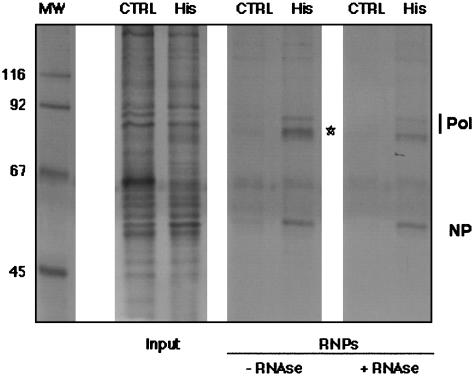
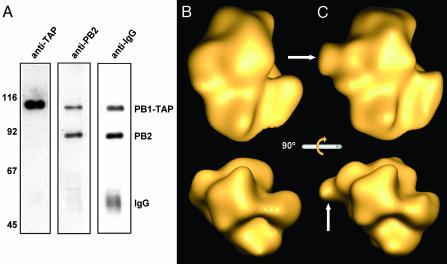
Purification of His-Tagged, Recombinant RNPs Generated by in Vivo Amplification. We have reported the structural characterization of influenza virus recombinant RNPs containing a 248-nt model vRNA (35, 36) that were generated by in vivo amplification and purified by successive glycerol gradient centrifugations. To increase the purity and yield of recombinant RNPs and to allow the structural analysis of the polymerase complex, we incorporated in the RNP tags for affinity purification. Because the RNPs were amplified by replication in transfected cells, the presence of the tag should not have impaired the ability of the polymerase to replicate. Addition of a C-terminal His tag at the PB2 subunit did not affect replication or transcription of His-tagged RNPs (data not shown). They were purified by gradient centrifugation to separate them from soluble proteins and by affinity chromatography on an Ni2+-NTA agarose resin. In the purified RNP preparations, the polymerase proteins, the NP, and the vRNA model were prominent after silver staining (Fig. 1, star; compare the samples with and without RNase treatment), as compared with control preparations in which the template RNA was omitted.
Fig. 1.
Purification of recombinant mini-RNPs. RNPs were reconstituted and amplified in vivo by using His-tagged PB2 (His). As a control, a reconstitution was performed in the absence of RNA (CTRL). RNP preparations after glycerol gradient centrifugation (Input) and after elution from Ni2+-NTA agarose (RNPs) were analyzed by silver staining. The positions of the RNP components are indicated to the right and that of vRNA is indicated by a star. The molecular mass markers (in kDa) are indicated to the left. MW, molecular weight.
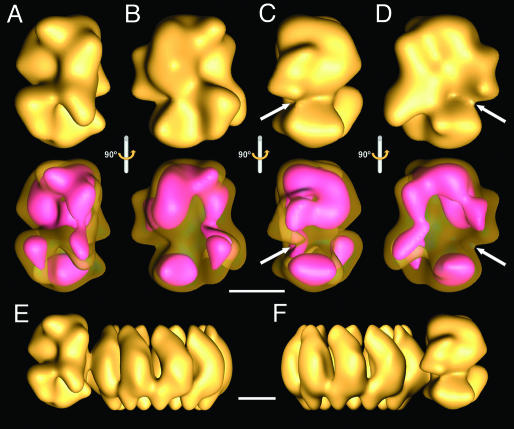
3D Reconstruction of the Viral Polymerase Complex. His-tagged RNPs were analyzed by electron microscopy of stained samples, and images from individual RNPs were collected from tilted samples. The polymerase and the adjacent NP monomers from each RNP image were extracted, and the images obtained were centered on the polymerase, aligned, and averaged. These images were used for 3D reconstruction, using as initial model the volume of the polymerase present in the 3D reconstruction of the RNP (36) and using a radius that would exclude the information derived from the NP monomers. The final model obtained (Fig. 2 A–D) was derived from 11,194 images and acquired from some 500 photographic plates, and the resolution obtained was 23 Å. The isosurface presented (Fig. 2 A–D Upper) corresponds to a volume of approximately the molar mass of the polymerase complex. The structure is very compact and does not reflect the N- to C-terminal arrangement of the subunits deduced from in vitro interaction studies (16–19). This observation is strengthened by the fact that a low-threshold illustration of the volume (Fig. 2 A–D Lower), which indicates areas of the highest density, does not reveal the boundaries among subunits. The polymerase complex shows an area of lower density (Fig. 2 C and D, arrows) in which a groove could be present, although at the resolution reached it is not possible to establish it unambiguously. The topology of the polymerase structure in the context of the complete RNP model is presented in Fig. 2 E and F. Two specific mass accumulations are in close contact with the adjacent NP monomers in the RNP. These regions of the complex may represent the polymerase–template NP interaction domains and show different contact sizes. At present, it is not possible to ascertain which polymerase subunits belong to these domains, although biochemical data indicate that PB1 and PB2, but not PA, proteins interact with soluble NP (45).
Fig. 2.
3D reconstruction of the influenza virus polymerase complex. The images of the polymerase complex from isolated RNPs were used for 3D reconstruction. (A–D) The isosurface representations at a threshold that corresponds to the mass of the polymerase (Upper) and a lower threshold representation (pink) under a transparent isosurface (Lower). They represent the side views (A and C), front view (B), and rear view (D) of the model. The arrows point to a possible groove in the complex. (E and F) Side views of the 3D model of a recombinant RNP (36) showing the relative topology of the new model of the polymerase with regard to the NP ring. (Scale bars, 50 Å.)
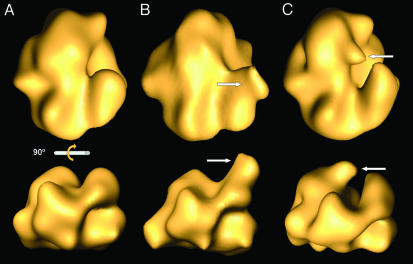
Localization of Specific Polymerase Domains by 3D Reconstruction of Polymerase–Ab Complexes. To delineate the polymerase subunits within the complex, we attempted the structural characterization of RNP–mAb complexes. The Abs chosen (PB2–25 and PA-2) recognize epitopes located within the N-terminal 113 aa in PB2 and the C-terminal region of PA (residues 400–716), respectively. Complexes of RNPs and mAb IgGs of either specificity contained polymerase, NP, and PB2- or PA-specific IgG, whereas parallel preparations in which unrelated mAbs were used did not show the presence of IgG (data not shown).
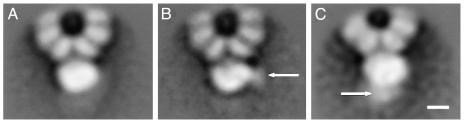
Each preparation of RNP–mAb complexes was processed independently, and the images were classified by using a self-organizing feature map (42). Fifteen, or 24%, of the images corresponded to RNP–mAb complexes in the different data sets. The average images from the complexes containing subunit-specific mAbs showed additional densities at sites specific for either PB2–25 IgG (Fig. 3B, arrow) or PA-2 IgG (Fig. 3C, arrow), as compared with the full set of images (Fig. 3A). In view of these results, we carried out 3D reconstructions of the polymerase–mAb complexes, using the sets of images whose 2D averages are shown in Fig. 3 B and C. As starting model in the refinement, we used the 3D volume of the polymerase obtained from a similar number of images of Ab-free RNPs; the results are presented in Fig. 4. Additional masses showing the contact sites of the monoclonal Fab fragments were detected in the 3D volumes of RNP–PB2–25 and RNP–PA-2 complexes (Fig. 4 B and C) as compared with the Ab-free polymerase (Fig. 4A). The 3D models presented indicate that the N-terminal region of PB2 is located in the bulge of the polymerase, on the right side of the front view (Fig. 4B Upper), whereas the C-terminal region of PA is situated at the center of the complex, as seen in its front view (Fig. 4C Upper); i.e., they are located opposite the polymerase–NP interaction side.
Fig. 3.
2D average images of recombinant polymerase–mAb complexes. Images obtained from negative-stained preparations of RNP–PB2–25 and RNP–PA-2 complexes were centered and aligned. The average image obtained is presented in A. Sorting by a self-organizing feature map led to data collections lacking or containing bound IgG. The average images obtained for the PB2–25- and PA-2-containing collections are presented in B and C, respectively. The arrows indicate the additional masses observed. (Scale bar, 50 Å.)
Fig. 4.
3D reconstruction of polymerase–anti-PB2 and polymerase–anti-PA monoclonal complexes. The images at the bottom are the result of a 90° rotation of those at the top. The images whose 2D averages are presented in B and C were used for 3D reconstruction. (A) The isosurface representation of the WT polymerase is shown. (B and C) The corresponding models for polymerase–anti-PB2 and polymerase–anti-PA complexes, respectively, are shown. The arrows point to the additional masses representing the domains of interaction of the mAb IgGs.
Mapping PB1 Subunit Domains by 3D Reconstruction of Tagged Polymerase Complexes. To localize the PB1 subunit in the polymerase, we prepared recombinant RNPs that contained a TAP tag (38) of ≈170 aa at the C terminus of PB1. Recombinant RNPs containing TAP-tagged PB1 and His-tagged PB2 were purified as indicated above. The TAP tag was exposed at the surface of the polymerase, because these double-tagged RNPs could be efficiently retained on an IgG Sepharose column (data not shown). Therefore, we prepared complexes of double-tagged recombinant RNPs and purified unrelated rabbit IgG. These IgG molecules recognize the protein A domain in the TAP tag by their Fc regions and should provide a mass recognizable by electron microscopy.
The purification of these RNP–TAP–IgG complexes is presented in Fig. 5A. The presence of PB1-TAP is apparent by the increase in size of this subunit (from 757 to 927 aa) and its reactivity in a Western blot with unrelated rabbit serum. In addition, the presence of IgG molecules bound to the TAP-tagged RNPs is apparent (Fig. 5A). Such IgG molecules were not present in control untagged RNPs (data not shown). Images from these complexes were obtained and processed for 3D reconstruction as indicated above for RNP–mAb complexes, and the results are shown in Fig. 5 B and C. An additional mass became apparent in the 3D model (Fig. 5C, arrows) as compared with the WT polymerase (Fig. 5B), which is located opposite the site where the N-terminal region of PB2 was mapped (Fig. 4B). These results indicate that the C-terminal region of PB1 is situated on the left of the polymerase complex as seen in its front view.
Fig. 5.
3D reconstruction of PB1-tagged influenza virus polymerase. (A) Purification of RNP–TAP–IgG complexes. RNPs were amplified by using TAP-tagged PB1 and His-tagged PB2, purified, and incubated with an excess of purified, unrelated rabbit IgGs. The RNP–TAP and RNP–TAP–IgG complexes were purified by affinity chromatography on Ni2+-NTA agarose. The successive Western blotting of the affinity-purified material is shown; unrelated rabbit serum was used to reveal the TAP tag (anti-TAP), anti-PB2 rabbit serum (anti-PB2), and anti-rabbit IgG biotin (anti-IgG). Molecular mass markers (in kDa) are indicated to the left. (B and C) The images obtained for RNP–TAP–IgG complexes were used for 3D reconstruction. Relevant views of the tagged polymerase (C) are presented in comparison with WT polymerase (B). The arrows point to additional masses contributed by the TAP tag and the bound IgG. The lower images are the result of a 90° rotation of the upper images.
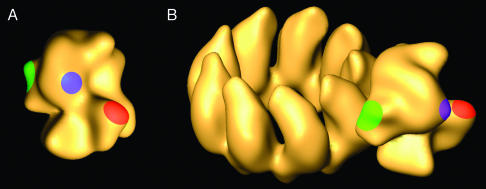
Biological Implications of the Polymerase Structure. The influenza virus polymerase is a heterotrimer with an aggregate molecular mass of ≈250 kDa. The complex is quite stable and can be purified by affinity chromatography on coexpression from cDNAs with a variety of tags at different locations (E.T. and E.A., unpublished results). These biochemical properties correlate with a very compact 3D structure, as described here (Figs. 2 and 6). The N-terminal region of PB2 and the C-terminal region of PA are exposed at the surface of the polymerase present in the RNP, as indicated by the possibility of forming complexes with purified specific mAbs. The structural information obtained from these complexes (Fig. 4) allowed the localization of these regions of PB2 and PA subunits at specific sites of the complex (Fig. 6A), opposite the domains of the polymerase that interact with the adjacent NP monomers in the RNP (Fig. 2 E and F). The localization of the C terminus of PB1 has been accomplished by inserting a TAP tag downstream of the gene. Because this sequence contributes some additional 170 aa and can be labeled specifically with IgG, its position in the complex could be determined (Fig. 5 B and C). A similar strategy was attempted to localize the C terminus of PA. However, the low capacity of PA-TAP and PB2-His double-tagged RNPs to replicate in vivo precluded their 3D study.
Fig. 6.
3D model for the influenza virus polymerase. The colored areas indicate the location of specific subunit domains: red, N-terminal region of PB2; blue, C-terminal region of PA; green, C-terminal region of PB1. (A) The model obtained by 3D reconstruction from electron micrograph images. (B) The position of the polymerase subunits in the context of the structure of the RNP.
The 3D structure determined suggests the existence of a low-density area in the center of the polymerase complex (see Fig. 2 C and D, arrows) that may represent a groove for interaction with the template or for the exit of the newly synthesized RNA. The available evidence indicates that the NP monomers recognize the template RNA by contacting the sugar–phosphate backbone, leaving the bases accessible to chemical modification (46). This accessibility would allow the polymerase to use the NP–RNA complex as template without needing to dissociate their interaction. Under these circumstances, the polymerase would move along the successive NP monomers, using the exposed RNA sequence as a template and extruding the mRNA transcript through the potential groove proposed in Fig. 2. The structure of the RNP (36) and the data presented here indicate that two separate polymerase–NP interactions exist that show different contact surfaces (Figs. 2 E and F and 5A). It is tempting to speculate that one of them is repeatedly broken and reformed as the polymerase proceeds along the RNP for transcription while the other is stably maintained. Such a possibility would imply that the 5′ terminus of vRNA is located close to the more stable polymerase–NP interaction. In this way, the polymerase could read the template until the polyadenylation signal (14, 47), which is associated with the last NP monomer, is reached. The conformational stress determined by the proximity of the polyadenylation signal and the 5′ terminal sequence, still bound to the polymerase, would induce the enzyme to stutter and synthesize the (poly)A tail.
The 3D structure described corresponds to an inactive form of the polymerase bound to the vRNA promoter. Previous information indicates that the purified RNPs used for structure analysis contain mainly vRNA (35), and therefore the 3D model of the polymerase presented here would correspond to the complex poised for transcription but not yet activated by binding to the cap primer. It is conceivable that the polymerase would present a different structure in the absence of vRNA, as cap-binding only occurs on recognition of the promoter 5′ sequences (48). Likewise, binding to the cRNA promoter may imply structural alterations, because vRNA and cRNA panhandles are recognized in different ways by the polymerase (22). On the other hand, the replicating polymerase could present a number of additional structural alterations as a consequence of its interaction with soluble NP or with cellular factors like hCLE (49) or Hsp90 (50).
In conclusion, the 3D model for the influenza polymerase complex presented in this report provides structural information about the viral enzyme, including the localization of specific regions of their subunits.
Acknowledgments
We thank A. Nieto for critical comments on the manuscript; B. Moss, A. Portela, and B. Séraphin for providing biological materials; and Y. Fernández and J. Fernández for technical assistance. E.A. was a Comunidad de Madrid Fellow; P.G. was a Gobierno Vasco Fellow; and E.T. was a Fondo de Investigaciones Sanitarias Fellow. This work was supported by Programa Sectorial de Promoción General del Conocimiento Grants PB97-1160, BMC01-1223, and BMC02-00996 and Ministerio de Ciencia y Tecnología Grant PTR1995-0564-OP.
Abbreviations: vRNA, viral RNA; NSV, negative-strand virus; RNP, ribonucleoprotein; NP, nucleoprotein; TAP, tandem affinity purification.
References
- 1.Hansen, J. L., Long, A. M. & Schultz, S. C. (1997) Structure (London) 5, 1109-1122. [DOI] [PubMed] [Google Scholar]
- 2.Ago, H., Adachi, T., Yoshida, A., Yamamoto, M., Habuka, N., Yatsunami, K. & Miyano, M. (1999) Structure (London) 7, 1417-1426. [DOI] [PubMed] [Google Scholar]
- 3.Bressanelli, S., Tomei, L., Roussel, A., Incitti, I., Vitale, R. L., Mathieu, M., De Francesco, R. & Rey, F. A. (1999) Proc. Natl. Acad. Sci. USA 96, 13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lesburg, C. A., Cable, M. B., Ferrari, E., Hong, Z., Mannarino, A. F. & Weber, P. C. (1999) Nat. Struct. Biol. 6, 937-943. [DOI] [PubMed] [Google Scholar]
- 5.Butcher, S. J., Grimes, J. M., Makeyev, E. V., Bamford, D. H. & Stuart, D. I. (2001) Nature 410, 235-240. [DOI] [PubMed] [Google Scholar]
- 6.Ng, K. K., Cherney, M. M., Vazquez, A. L., Machin, A., Alonso, J. M., Parra, F. & James, M. N. (2002) J. Biol. Chem. 277, 1381-1387. [DOI] [PubMed] [Google Scholar]
- 7.Poch, O., Sauvaget, I., Delarue, M. & Tordo, N. (1990) EMBO J. 8, 3867-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai, M. M. (1998) Virology 244, 1-12. [DOI] [PubMed] [Google Scholar]
- 9.Lamb, R. A. & Krug, R. M. (1996) in Virology, eds. Fields, B. N., Knipe, D. M. & Howley, P. M. (Lippincott-Raven, Philadelphia), pp. 1353-1395.
- 10.Detjen, B. M., St. Angelo, C., Katze, M. G. & Krug, R. M. (1987) J. Virol. 61, 16-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Digard, P., Blok, V. C. & Inglis, S. C. (1989) Virology 171, 162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krug, R. M., Broni, B. A. & Bouloy, M. (1979) Cell 18, 329-334. [DOI] [PubMed] [Google Scholar]
- 13.Pritlove, D. C., Poon, L. L. M., Devenish, L. J., Leahy, M. B. & Brownlee, G. G. (1999) J. Virol. 73, 2109-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson, J. S., Schubert, M. & Lazzarini, R. A. (1981) J. Virol. 38, 157-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poon, L. L., Fodor, E. & Brownlee, G. G. (2000) J. Virol. 74, 418-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pérez, D. R. & Donis, R. O. (1995) J. Virol. 69, 6932-6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.González, S., Zürcher, T. & Ortìn, J. (1996) Nucleic Acids Res. 24, 4456-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toyoda, T., Adyshev, D. M., Kobayashi, M., Iwata, A. & Ishihama, A. (1996) J. Gen. Virol. 77, 2149-2157. [DOI] [PubMed] [Google Scholar]
- 19.Zürcher, T., de la Luna, S., Sanz-Ezquerro, J. J., Nieto, A. & Ortìn, J. (1996) J. Gen. Virol. 77, 1745-1749. [DOI] [PubMed] [Google Scholar]
- 20.Li, M. L., Ramìrez, B. C. & Krug, R. M. (1998) EMBO J. 17, 5844-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.González, S. & Ortìn, J. (1999) J. Virol. 73, 631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.González, S. & Ortìn, J. (1999) EMBO J. 18, 3767-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biswas, S. K. & Nayak, D. P. (1994) J. Virol. 68, 1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ulmanen, I., Broni, B. A. & Krug, R. M. (1981) Proc. Natl. Acad. Sci. USA 78, 7355-7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, M. L., Rao, P. & Krug, R. M. (2001) EMBO J. 20, 2078-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gastaminza, P., Perales, B., Falcón, A. M. & Ortìn, J. (2003) J. Virol. 76, 5098-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahy, B. W. J. (1983) in Genetics of Influenza Viruses, eds. Palese, P. & Kingsbury, D. W. (Springer, Vienna), pp. 192-253.
- 28.Perales, B., Sanz-Ezquerro, J. J., Gastaminza, P., Ortega, J., Fernández-Santarén, J., Ortìn, J. & Nieto, A. (2000) J. Virol. 74, 1307-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fodor, E., Crow, M., Mingay, L. J., Deng, T., Sharps, J., Fechter, P. & Brownlee, G. G. (2002) J. Virol. 76, 8989-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pons, M. W., Schulze, I. T. & Hirst, G. K. (1969) Virology 39, 250-259. [DOI] [PubMed] [Google Scholar]
- 31.Jennings, P. A., Finch, J. T., Winter, G. & Robertson, J. S. (1983) Cell 34, 619-627. [DOI] [PubMed] [Google Scholar]
- 32.Murti, K. G., Webster, R. G. & Jones, I. M. (1988) Virology 164, 562-566. [DOI] [PubMed] [Google Scholar]
- 33.Klumpp, K., Ruigrok, R. W. & Baudin, F. (1997) EMBO J. 16, 1248-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruigrok, R. W. & Baudin, F. (1995) J. Gen. Virol. 76, 1009-1014. [DOI] [PubMed] [Google Scholar]
- 35.Ortega, J., Martìn-Benito, J., Zürcher, T., Valpuesta, J. M., Carrascosa, J. L. & Ortìn, J. (2000) J. Virol. 74, 156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martìn-Benito, J., Area, E., Ortega, J., Llorca, O., Valpuesta, J. M., Carrascosa, J. L. & Ortìn, J. (2001) EMBO Rep. 2, 313-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perales, B. & Ortìn, J. (1997) J. Virol. 71, 1381-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M. & Séraphin, B. (1999) Nat. Biotechnol. 17, 1030-1032. [DOI] [PubMed] [Google Scholar]
- 39.Bárcena, J., Ochoa, M., de la Luna, S., Melero, J. A., Nieto, A., Ortìn, J. & Portela, A. (1994) J. Virol. 68, 6900-6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marco, S., Chagoyen, M., de la Fraga, L. G., Carazo, J. M. & Carrascosa, J. L. (1996) Ultramicroscopy 66, 5-10. [Google Scholar]
- 41.Marabini, R., Masegosa, I. M., San Martìn, C., Marco, S., Fernández, J. J., de la Fragra, L. G., Vaquerizo, C. & Carazo, J. M. (1996) J. Struct. Biol. 116, 237-240. [DOI] [PubMed] [Google Scholar]
- 42.Marabini, R. & Carazo, J. M. (1994) Biophys. J. 66, 1804-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frank, J., Radermacher, M., Penczek, P., Zhu, J., Li, Y., Ladjadj, M. & Leith, A. (1996) J. Struct. Biol. 116, 190-199. [DOI] [PubMed] [Google Scholar]
- 44.Marabini, R., Herman, G. T. & Carazo, J. M. (1998) Ultramicroscopy 72, 53-65. [DOI] [PubMed] [Google Scholar]
- 45.Biswas, S. K., Boutz, P. L. & Nayak, D. P. (1998) J. Virol. 72, 5493-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baudin, F., Bach, C., Cusack, S. & Ruigrok, R. W. (1994) EMBO J. 13, 3158-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poon, L. L. M., Pritlove, D. C., Fodor, E. & Brownlee, G. G. (1999) J. Virol. 73, 3473-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cianci, C., Tiley, L. & Krystal, M. (1995) J. Virol. 69, 3995-3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huarte, M., Sanz-Ezquerro, J. J., Roncal, F., Ortìn, J. & Nieto, A. (2001) J. Virol. 75, 8597-8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Momose, F., Naito, T., Yano, K., Sugimoto, S., Morikawa, Y. & Nagata, K. (2002) J. Biol. Chem. 277, 45306-45314. [DOI] [PubMed] [Google Scholar]