Abstract
As a result of technical advances in recombinant DNA technology and nucleotide sequencing, entire genome sequences have become available in the past decade and offer potential in understanding diseases. However, a central problem in the biochemical sciences is that the functions of only a fraction of the genes/proteins are known, and this is also an issue in pharmacology. This review is focused on issues related to the functions of cytochrome P450 (P450) enzymes. P450 functions can be categorized in several groups: 1) Some P450s have critical roles in the metabolism of endogenous substrates (e.g., sterols and fat-soluble vitamins). 2) Some P450s are not generally critical to normal physiology but function in relatively nonselective protection from the many xenobiotic chemicals to which mammals (including humans) are exposed in their diets [as well as more anthropomorphic chemicals (e.g., drugs, pesticides)]. 3) Some P450s have not been extensively studied and are termed “orphans” here. With regard to elucidation of any physiological functions of the orphan P450s, the major subject of this review, it is clear that simple trial-and-error approaches with individual substrate candidates will not be very productive in addressing questions about function. A series of liquid chromatography/mass spectrometry/informatics approaches are discussed, along with some successes with both human and bacterial P450s. Current information on what are still considered “orphan” P450s is presented. The potential for application of some of these approaches to other enzyme systems is also discussed.
I. Introduction
A. The General Problem in Biology Today
In many respects, our approach to biology has changed because of the rate at which genome sequence information is becoming available. Seldom do research problems still involve a historic approach of moving from an in vitro phenomenon to purification of an enzyme or receptor and then cDNA cloning (Fig. 1). A more common scenario involves trying to annotate genes based on their sequence similarities, heterologously expressing the proteins, and then attempting to find specific catalytic activities that might be associated with the protein. One bottleneck in this approach is heterologous expression, which overall is not as difficult as classic enzyme purifications from tissues, etc., but can be problematic. However, an even larger problem is selecting what chemicals (or macromolecules) should be chosen as candidates for assays. We still know the functions of only approximately half of the genes in the model organism Escherichia coli (Hanson et al., 2010) and other bacteria, and of course the fraction is less when we consider mammals and other higher eukaryotes.
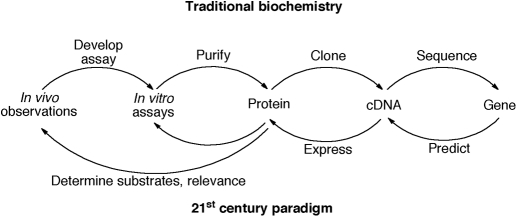
Fig. 1.
Classic (upper sequence) and “reversed” (lower sequence) approaches to understanding biochemical functions of enzymes (Guengerich et al., 2010, 2011).
B. The Problem Applied to Cytochrome P450s
P450 genes can be identified by their signature sequence element FXXGXXXCXG, where the underlined cysteine serves as an axial ligand to the heme iron. The Human Genome Project has set the number of human P4501 genes at 57 (Table 1), although we do not necessarily know that all are expressed. For reference, there are 103 mouse P450 genes and 89 rat P450 genes (http://drnelson.uthsc.edu/cytochromeP450.html).
TABLE 1.
Classification of human P450s based on major substrate class
This classification (Guengerich, 2005) is somewhat arbitrary (e.g., P450s 1B1 and 27A1 could be grouped in either of two different categories).
| Sterols | Xenobiotics | Fatty Acids | Eicosanoids | Vitamins | Unknown |
|---|---|---|---|---|---|
| 1B1 | 1A1 | 2J2 | 4F2 | 2R1 | 2A7 |
| 7A1 | 1A2 | 4A11 | 4F3 | 24A1 | 2S1 |
| 7B1 | 2A6 | 4B1 | 4F8 | 26A1 | 2U1 |
| 8B1 | 2A13 | 4F12 | 5A1 | 26B1 | 2W1 |
| 11A1 | 2B6 | 8A1 | 26C1 | 3A43 | |
| 11B1 | 2C8 | 27B1 | 4A22 | ||
| 11B2 | 2C9 | 4F11 | |||
| 17A1 | 2C18 | 4F22 | |||
| 19A1 | 2C19 | 4V2 | |||
| 21A2 | 2D6 | 4X1 | |||
| 27A1 | 2E1 | 4Z1 | |||
| 39A1 | 2F1 | 20A1 | |||
| 46A1 | 3A4 | 27C1 | |||
| 51A1 | 3A5 | ||||
| 3A7 |
On the basis of our general knowledge of biochemical pathways, results with transgenic mice, and information about inherited human diseases, we can say that fewer than half of P450s have critical physiological functions, including those involved in the metabolism of steroids, fat-soluble vitamins (A and D), and some eicosanoids (e.g., P450s 5A1, 8A1) (Table 1). The focus of this review is the P450s in the far right column of Table 1, those of unknown function. These will be referred to as “orphans,” a term adopted from descriptions of the steroid nuclear receptor superfamily (Mangelsdorf and Evans, 1995). However, the point should be made that the P450s in the xenobiotics category of Table 1 have established reactions with drugs, carcinogens, and so forth, but do not seem to be essential. This is based on knowledge of human polymorphisms and work with transgenic mice (Gonzalez and Yu, 2006). Even when these P450s have reasonable catalytic activities toward endogenous substrates, there is no strong evidence that the reaction has major physiological relevance (i.e., testosterone hydroxylation by P450 3A4).
The problem with the use of rodent models for discerning P450 function is apparent in comparing the numbers of genes in the species (see above). Some of the P450 subfamilies contain many more genes in rodents than humans, and the matches of function of the members of the large subfamilies are not particularly good (e.g., the 2C and 3A subfamilies). The complexity of these subfamilies is also problematic for transgenic studies.
II. Current Views of the Functions of P450s
Interest in the P450 field began with work in the fields of endocrinology (Ryan, 1958), chemical carcinogenesis (Mueller and Miller, 1948), and drug metabolism (Remmer, 1959). Largely because of the practical issues involved in these areas, resources have been available and fueled progress in P450 research for half a century. Today interest continues in the above three areas and has also expanded to diverse fields, including drug discovery, bioremediation, insect control, and crop science.
Some mention should be made of the variety of types of reactions that P450s can catalyze. Most reactions are mixed-function oxidations, which include not only production of alcohols and epoxides but also a great variety of other transformations (Guengerich, 2001; Isin and Guengerich, 2007; Guengerich and Isin, 2011). P450s can even catalyze reactions that do not involve net oxidation-reduction chemistry (Cheng et al., 2010). Much of the literature on seemingly unusual P450 reactions was developed from transformations observed with drugs (or drug candidates) and natural products.
A. General: Specific versus Multifunctional/Low-Specificity Cytochrome P450s
There are two major views of the functions of P450s. One is that these enzymes have specific and important physiological functions. The other view is that P450s have low specificity and, related to that, low catalytic activities; their function is to protect organisms (especially mammals, including humans) from natural products (Jakoby, 1980). Both views are correct, and P450s (at least mammalian P450s) fit into both categories (Table 1).
A number of heritable metabolic diseases are associated with loss of particular P450s, especially those involved in the metabolism of steroids and fat-soluble vitamins (Nebert and Russell, 2002). Studies with transgenic mice also support this view (Gonzalez and Yu, 2006). In addition, deletion of NADPH-P450 reductase is lethal to mouse embryos (Shen et al., 2002), although the exact reason is not clear [attenuation of the reductase in selected organs in mice has a relatively modest effect (Gu et al., 2003)]. Clinical issues have also been associated with polymorphisms in the reductase gene in humans (http://www.cypalleles.ki.se/por.htm). The P450s considered to be in this category generally do not vary considerably in levels of expression among individuals (Shinkyo and Guengerich, 2011) to the same extent as the “xenobiotic-metabolizing” P450s (Guengerich, 2005). Some of these P450s—but not all—have relatively high catalytic efficiencies (e.g., kcat/km 2 × 106 M−1 s−1 for purified cholesterol 7α-hydroxylase P450 7A1) (Shinkyo and Guengerich, 2011).
Mammals ingest considerable amounts of natural products (e.g., terpenes, alkaloids, flavonoids) in their food along with amino acids, carbohydrates, lipids, and so forth. Cells have two major means of protection: one is to transport the materials out (e.g., Mdr1 and related genes) and the other is metabolism. Thus, the presence of high levels of a set of enzymes (P450s) that are capable of removing a great variety of compounds from cells has a role in protecting cells from harmful chemicals. Drugs, pollutants, and so forth have structures that have some general resemblance to these natural products, at least in general terms of molecular weight, lipophilicity, polar surface area, and so forth. The information available to date indicates that even when these P450s (i.e., those using xenobiotics as substrates) do oxidize endogenous compounds (e.g., human P450 3A4 and testosterone hydroxylation), the reactions do not have important physiological consequences. For instance, many people (e.g., ∼ 7% of white persons) do not have P450 2D6 but seem to live normal lives. However, a caveat to this general view is that a small effect might have been missed in epidemiological studies, and a change of a few years of life expectancy or protection against some disease might have been missed.
Mammals have this less selective group of P450s to breakdown natural products, but the P450s in plants, which are much more numerous, seem to be devoted largely to anabolic reactions (i.e., generation of secondary metabolites that reduce infection, animal consumption, or enhance symbiotic interactions with other organisms for improved growth and reporduciton). For instance, several P450s have been identified in individual pathways to synthesize plant pigments and other secondary products (Humphreys and Chapple, 2000; Morant et al., 2003; Takei et al., 2004). Whether plants also have more general P450 systems for protection against xenobiotics is unknown; however, several cases of P450 involvement and adaptation to plant-insect “warfare” have been described previously (Gonzalez and Nebert, 1990; Schuler, 1996; Li et al., 2003). Common enteric bacteria do not have P450s (e.g., E. coli and Salmonella typhimurium). With more complex bacteria (e.g., Actinomyces spp.), the situation is less clear. Some functions of P450s have been described in the production of secondary products (Lamb et al., 2006), and there is also evidence that some can deal with a wide variety of xenobiotics (e.g., P450 105D1) (Taylor et al., 1999).
B. Classification by Substrates
Table 1 has already been discussed. However, the classification has some caveats. For instance, P450 1B1 oxidizes a wide variety of xenobiotics (Shimada et al., 1996) and also estrogens; this P450 is considered important in physiology in that polymorphisms have been shown to be associated with glaucoma (Stoilov et al., 1997; Vasiliou and Gonzalez, 2008). In addition, P450 27A1 oxidizes both cholesterol (a steroid) and vitamin D3 and could be placed in either category. Many P450s that oxidize xenobiotics can also oxidize fatty acids (Niwa et al., 2009), but only a few P450s are included in this category in Table 1 when it seems to be a primary function for them. Exactly how critical these fatty acid reactions occur has not been established.
C. Major Cytochrome P450s Involved in Xenobiotic Metabolism
1. Drugs.
Estimates have been made that ∼75% of the enzymatic transformations of (“small molecule”) drugs are attributable to P450 reactions, with the UDP-glucuronosyl transferases and esterases constituting the next two most prominent subsets (Williams et al., 2004). Furthermore, five P450s—1A2, 2C9, 2C19, 2D6, and 3A4—have been estimated to account for ∼90% of the P450 oxidations of drugs in two studies (Williams et al., 2004; Wienkers and Heath, 2005). The caveats should be stated that not all drugs are extensively metabolized and that these estimates are based largely on the drugs (and drug candidates) of the pharmaceutical companies that employ the authors (i.e., Pfizer, Amgen) (Williams et al., 2004; Wienkers and Heath, 2005) and may not totally reflect the entire universe of all drug candidates. Nevertheless, these estimates are generally considered reliable in the field.
2. Carcinogens.
Analyses of the type reported above for drugs have not been made for carcinogen metabolism, although tables of the P450s and carcinogenic substrates have been published (Guengerich, 2005). The patterns have some overlap with those for drug metabolism, but not much. The main P450s involved are 1A1, 1A2, 1B1, 2A6, 2A13, 2E1, and 3A4. There is considerable overlap among the family 1 P450s because of their structural similarity (Sansen et al., 2007; Wang et al., 2011) yet some selectivity. In brief, the family 1 P450s are involved in the metabolism of polycyclic aromatic hydrocarbons, arylamines, and heterocyclic amines, P450s 2A6 and 2A13 are involved in nitrosamine oxidation, P450s 2A6 and 2E1 are involved in the oxidation of nitrosamines and low-molecular-weight halogenated hydrocarbons and vinyl monomers, and P450 3A4 is involved in the oxidation of a number of larger compounds (e.g., some mycotoxins such as aflatoxin B1). P450 2W1 can activate a variety of carcinogens (Wu et al., 2006b). P450 2D6 does not have identified roles in carcinogen metabolism.
III. Approaches to Deorphanization
In this context “deorphanization” simply means identification of the function of an enzyme, at least with regard to reactions it catalyzes (giving it a home, to be colloquial). The discussion in this review will be focused on enzymes that use small molecules as substrates, particularly the P450s.
Much of the field we call biochemistry has used the following paradigm (Fig. 1): phenomena were discovered in in vivo settings and the transformations were replicated in vitro, with an appropriate assay. A goal was to use the assay to purify the enzyme. As recombinant DNA technology developed, cDNAs could be identified for enzymes and then used to characterize genes. Today we have a situation in which we know the nucleotide sequences of entire genomes, including humans, and can predict open reading frames and protein sequences, but methods for rapidly identifying functions are missing.
For a number of reasons, the in silico prediction of gene function has its limits. This approach is often successful when 1) the function of a gene is known in one species and 2) there is a high degree of conservation (of the sequence). One problem is that of “protein promiscuity,” in that enzymes can use similar amino acid side-chain chemistry to perform different overall functions. For example, the enzymes in the enolase and vicinal oxygen chelate (VOC) families have some identity but catalyze a wide variety of enzymatic reactions (Gerlt and Babbitt, 2001). Another problem arises in predicting which molecules will fit into active sites. In many cases, including most P450s, even the crystal structure of a protein (determined in the absence of ligand) may be changed considerably upon binding (Poulos and Johnson, 2005). Thus, the quest for functions of orphan enzymes will continue to be one that involves experiments.
One approach is to hypothesize possible reactions for the enzyme and try these. This may be a reasonable approach if there is information about substrates for related genes. For example, P450s 27A1 and 27B1 are involved in the oxidation of vitamin D. However, in our work on the orphan P450 27C1, we detected no enzymatic activity toward vitamin D (Wu et al., 2006a). The quest is even more complicated when considering a problem de novo. Simply put, there are too many chemicals on the shelf (and in freezers, etc.) to try one at a time, especially if an assay needs to be developed for each. Moreover, we do not know what all the minor chemicals in the human body are, and the situation is even more complex when considering the possibilities in microorganisms and plants.
The main approaches involve what is generally referred to as metabolomics, and that field of literature should be followed for insights into new types of experiments. The general concept here, in contrast to some other aspects of metabolomics, is to compare two sets of experimental data and look for a change. In one set the enzyme is added, whether in purified state or in vivo. Obviously this is a challenge in complex systems and pushes the limits of both instrumental capabilities and information processing.
A. Instrumental Needs
The initial need is a system that can identify small differences between samples derived from two states (i.e., with and without active enzyme). The samples must be necessarily complex, either tissue extracts or body fluids. Therefore it is important to have a detection system that does not discriminate among molecules. Thus, UV and fluorescence are not really acceptable for initial screening.
1H-NMR analysis has been used extensively in metabolomics studies (Bollard et al., 2005). However, resolution is limited. and most of the applications published so far involve changes in the patterns of major metabolites in body fluids, (e.g., amino acids, major carbohydrates, carboxylic acids of low relative molecular mass).
The most useful instrument for the purpose of identifying the functions of orphan enzymes is a mass spectrometer, for a number of reasons, including the following: the ability to resolve complex mixtures of compounds (either by LC coupling or high-resolution methods) (Fig. 2), 2) the generality of the detection mode (but see caveats below), and 3) the provision for annotation of candidate signals (e.g., using high resolution and/or fragmentation). There are several considerations about choices of mass spectrometry systems that need to be made, however, before commencing with analysis. One of the most important things to remember is that the goal in such work is to identify small changes between two complex mixtures that can be attributed to the action of a single enzyme, and all planning for instruments and informational analysis should be focused on this goal.
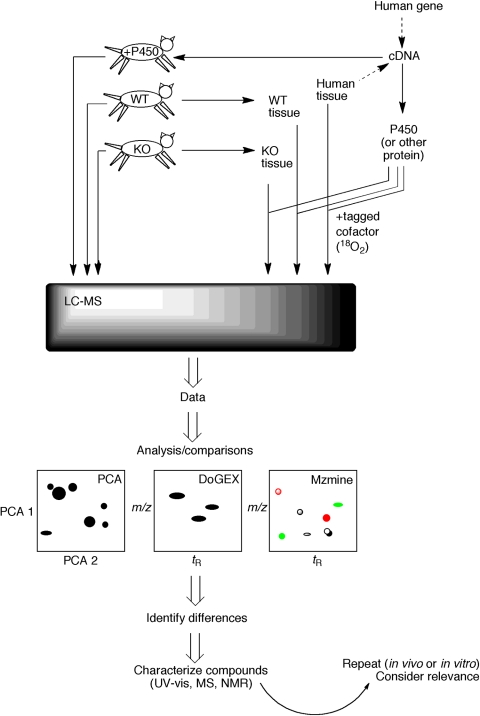
Fig. 2.
LC-MS based strategies for elucidating substrates of human enzymes (Guengerich et al., 2010, 2011). One approach (in vivo) involves either expressing the human protein or deleting its apparent ortholog in a transgenic animal model. LC-MS metabolomic analysis can be done with body fluids from the animals or by removing tissues and using the in vitro incubations. Alternatively, a total in vitro approach can be used in which the protein is heterologously expressed and used with extracts of human tissue (with or without isotopically labeled cofactor). Data from the LC-MS analysis are used in comparisons (e.g., wild-type versus transgenic mice, in vitro enzyme incubations versus controls) made with one or more programs (e.g., PCA, DoGEX, MZmine, XCMS). The goal is to identify differences. In some cases, the mass spectrum (i.e., molecular formula) may be sufficient to identify possible matches of candidates, and the authentic materials may be available for comparison. However, if unknown compounds are involved, then not only the high-resolution MS spectra but also UV visible and NMR spectra may be needed. Ultimately, more experiments will be needed to assess the biological relevance of any findings.
In principle, it is possible to separate a complex mixture of compounds by direct injection into a high-resolution mass spectrometer (i.e., quadrupole time-of-flight, Orbitrap, or Fourier transform-MS). A spectrum would reveal the elemental formulae of all compounds (or at least provide a list of the best choices for each peak). Such approaches have been reported for annotation of the peaks in complex mixtures. However, there are at least three deficiencies associated with such an approach. One is the existence of isomers (i.e., for these purposes, compounds with the same elemental formulae). A second is that the lack of separation (in the LC phase) would lead to undue ion suppression in the MS phase. The third (linked with the second) is that comparing the quantities of individual compounds in the two samples is difficult.
More commonly, separation is done by GC and LC before introduction into the mass spectrometer. Capillary GC has excellent resolving power, but relatively few biological compounds (at least mammalian) are amenable to direct separation by GC; thus, derivatization is needed and imposes a preselection on which compounds will be analyzed.
A more common approach involves LC. Modern ultra-high-performance liquid chromatography systems have high resolving power, and typical reversed-phase methods have the capability to resolve low-molecular-weight biological molecules, particularly relatively nonpolar ones of the type that are P450 substrates. Typical water-to-acetonitrile gradients, with ammonium acetate (or formate), can be used with octadecylsilane (C18)-based columns.
If compounds are not derivatized, then a choice must be made as to what ionization mode should be used for LC-MS work—positive ion electrospray ionization (ESI), negative ion ESI, or atmospheric pressure chemical ionization. Molecules bearing a positive charge (e.g., amines) ionize best with positive-ion ESI; molecules bearing a negative charge (i.e., carboxylic acids) ionize best with negative-ion ESI. Neutral molecules (e.g., steroids) are problematic and in our own experience work best with APCI. When one does not know what to expect in an LC-MS analysis, the most reliable approach is to do the analysis in all three modes. Obviously this increases the time and cost of analysis 3-fold.
Two approaches have been developed in our own laboratory to facilitate the analysis of P450 reaction products. Many of the products of P450 reactions are alcohols, although not all are (Guengerich, 2001; Isin and Guengerich, 2007; Guengerich and Isin, 2011). In the past, dansyl chloride has been used to convert amines and nucleophilic phenols to fluorescent derivatives. However, dansylated compounds also have excellent ionization properties in positive-ion ESI MS because of the positive charge on the amine moiety. We modified conditions (heat, use of 4,4-dimethylaminopyridine as a reagent) for derivatization of unactivated alcohols (Tang and Guengerich, 2010). The method was shown to be applicable to several alcohols that can be formed by the action of P450s on steroids, fatty acids, and vitamin D3. In some cases, the limit of detection was lowered by >103-fold (Tang and Guengerich, 2010). The products can be further defined by fragmentation, which removes the dansyl group.
Another approach we have used involves mixtures of 16O2 and 18O2 cofactors (Sanchez-Ponce and Guengerich, 2007). In this approach, the atmosphere of a P450 reaction is replaced with a 1:1 mixture of 16O2/18O2, resulting in oxygenated products (e.g., alcohols, epoxides) with a 1:1 mixture of 16O and 18O atoms, which should be detected as M/M + 2 doublets, as in the case of natural abundance 79Br/81Br doublets. A MATLAB-based program (DoGEX) was developed to search for such doublets [alternatively, the program can be set to seek other isotopic products with other ratios (e.g., chlorine)] (Fig. 3) (Sanchez-Ponce and Guengerich, 2007). The program DoGEX also has features to allow baseline adjustment, noise removal, and calibration of retention times. With regard to technical aspects of handling 18O2 gas, the procedures are relatively straightforward in that pressurized vessels of 18O2 are now commercially available (e.g., Cambridge Isotope Laboratories, Andover, MA). Samples (of enzyme and a tissue extract) are placed in Thunberg tubes, and air is removed by alternate cycles of (mild) vacuum and exposure to argon gas. Isotopically labeled oxygen can be purchased as a 1:1 mixture of 16O2 and 18O2 (Cambridge Isotope Laboratories), although we have experienced unexplained problems with 16O-18O being present in some lots. A simpler system involves preparing separate/parallel samples with 16O2 and 18O2 and then mixing the contents after completion of the incubations.
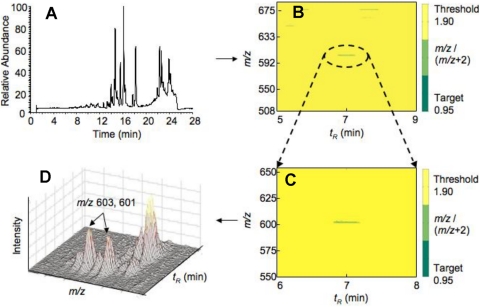
Fig. 3.
Use of the program DoGEX in screening for candidate substrates. Raw LC-MS data from an incubation of a P450, NADPH-P450 reductase, NADPH, a tissue extract, and a 1:1 mixture of 6O2/18O2 is shown in A. B shows the analysis of mass-to-charge ratio versus retention time, with a green positive spot on a yellow background for a ratio of ∼1.0 and Δ atomic mass units = 2.0, with the positive zone expanded in part C. The data from C are produced as a plot of relative ion intensity versus mass-to-charge ratio in part D. In this case the P450 uses P450 7A1, and the product was (the succinate ester of) 7α-hydroxycholesterol (Guengerich et al., 2010, 2011; Tang et al., 2010).
The approach has been validated using P450 3A4 and the substrate testosterone (Sanchez-Ponce and Guengerich, 2007). A more rigorous validation involved finding labeled 7α-hydroxycholesterol in an incubation of P450 7A1 and human liver extract (Fig. 3) (Tang et al., 2010). Fatty acid substrates were identified using the approach (with negative-ion ESI) with P450s 1A2, 2C8, and 2C9 (Tang et al., 2009) and 4F11 (Tang et al., 2010). Furthermore, the approach can be coupled with the dansylation procedure mentioned above; e.g., dansylated 7α-cholesterol could be identified in human liver extracts after incubation with P450 7A1 and 18O2 (Tang and Guengerich, 2010). In principle, capillary electrophoresis-MS could be used as an approach (Dunayevskiy et al., 1996) but would probably yield a more restricted set of resolved analytes into the mass spectrometer (compared with LC).
B. Informatics
The goal of data processing is to identify a few major changes among thousands of compounds that distinguish the two different states. In the MS modes used in this work (see section III.A), the full scan mode is used across a wide mass-to-charge ratio range, resulting in very large data sets. Therefore it is unrealistic to analyze the data by visual inspection and specialized software is needed for this kind of data analysis.
Principal component analysis (PCA) is a well established general approach for comparing two or more data sets. PCA methods have been used widely in NMR metabolomics comparisons (Bollard et al., 2005) but apparently not extensively for MS approaches to enzyme function.
A number of freely available software programs have been developed to identify the differences between metabolite profiles acquired by on-line chromatography mass spectrometry techniques: XCMS (Smith et al., 2006), MZmine (Katajamaa et al., 2006), DoGEX (Sanchez-Ponce and Guengerich, 2007), MetAlign (Tolstikov et al., 2003), and MSFACTs (Duran et al., 2003). All of these programs are capable of adjusting small but problematic differences in retention time, one of the chief problems when comparing two (or more) complex LC-MS/full-scan profiles. In our laboratory, XCMS and MZmine have been the most widely used programs for untargeted metabolic profiling. We successfully identified an unusual endogenous dipentaenone substrate of P450 154A1 using XCMS and MZmine by comparing the metabolic profiles of wild-type and P450 154A1-knockout Streptomyces coelicolor (Fig. 4) (Cheng et al., 2010). DoGEX has already been mentioned. This program has been used in analyses involving isotope tags, and several fatty acids were verified as substrates of four hepatic P450s (1A2, 2C8, 2C9, 4F11) by this approach (Fig. 3) (Tang et al., 2009, 2010). In addition, some commercially available software is applicable to studies of this type [e.g., ACD/IntelliXtract (ACD/Labs, Toronto, ON, Canada), MetaboLynx (Waters, Milford, MA), and Mass Frontier (HighChem, Bratislava, Slovakia)].
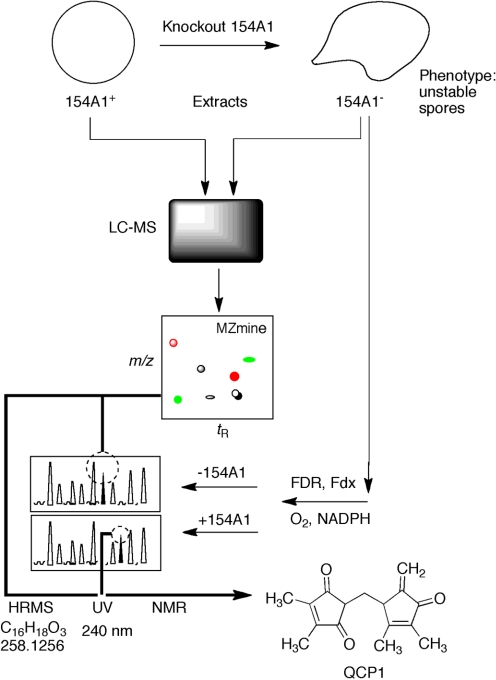
Fig. 4.
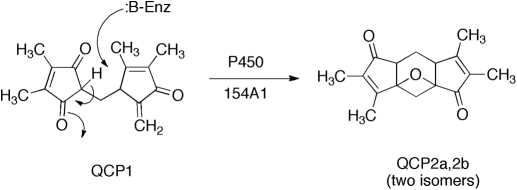
Use of an MZmine approach to identification of a substrate and product for S. coelicolor P450 154A1. LC-MS data were obtained for wild-type and P450 154AA1-knockout bacteria. Comparison of the LC-MS data by LC-MS using MZmine suggested a compound with MH+ 259. Extracts of the P450 154A1 bacteria (in which a substrate would be expected to accumulate) with P450 154A1 indicated that the candidate compound disappeared during incubation and a new peak appeared. Subsequent analytical work showed 1) the structures of the substrate QCP1 and product QCP2a,b (2 isomers) and 2) that no redox function was related to the change (Fig. 5) (Cheng et al., 2010).
C. In Vivo Approaches
There are inherently two approaches to experimental studies defining enzyme function, in vivo and in vitro. In vivo approaches will be discussed first, although this laboratory has only applied in vivo methods in the case of P450s in microorganisms. The discussion here will focus on mammals.
1. Transgenic Animals.
In principle, the function of an enzyme can be discerned by eliminating it from an animal model and observing phenotypes. In practice, this approach has several limitations: 1) If the gene product is essential, then lethality will result (embryonic or later), but the reason may not be clear, and studies with a knockout cell line may be in order. 2) The gene and protein may have functions but redundancy may obscure observation of a phenotype. 3) A human gene may not have a mouse counterpart (e.g., P450 27C1).
Another approach is to add a human gene to a mouse model and look for changes in the phenotype. One issue is achieving tissue-selective expression. Another issue is that the added gene may have several possibly orthologous counterparts in the host (mouse), (e.g., multiple P450 2d mouse genes in the case where analyzing human P450 2D6 is of interest).
These approaches have been applied more in the cases of bacteria and plants. For instance, phenotypes have been observed with a number of P450 knockout bacteria (Fig. 4) (McLean et al., 2007; Cheng et al., 2010; Lamb et al., 2011). In some cases, the organization of genes into operons is useful in identifying functions through the relationships. Characterizing the large number of P450 genes in plants (e.g., 273 in Arabidopsis thaliana, 412 in rice) is challenging, but many interesting phenotypes have been reported and have led to analysis of plant function.
In addition to actual transgenics, other short-term approaches can be considered in regard to analysis of function, in vivo and also in cell cultures. One approach is the use of siRNA down-regulation of individual genes, looking for a transient phenotype. Liver-specific expression of proteins is also possible with the use of intravenous hydrodynamic injection methods in rodents (Kameda et al., 2003).
2. Metabolomics.
One approach to discerning functions of proteins is through simple observations (e.g., is a knockout lethal, does the color of a plant change). However, this approach will probably not provide molecular details for an integral change. One approach to details of function is metabolomics, which compares the compounds present in biological sources differing only in the enzyme under consideration, in this application. For in vivo studies, a typical source could be urine or plasma. As pointed out earlier, the samples could be compared by any of several instrumental/bioinformatic approaches, LC-MS having several advantages. An example of this approach with P450 involves some work on P450 2D6 by the Gonzalez laboratory (Yu et al., 2003).
D. In Vitro Approaches
In vitro approaches have advantages in that the level of complexity can be reduced and, in principle, the mechanism underlying a phenotypic change may be seen directly.
1. Recombinant Enzymes.
One approach to elucidating functions of enzymes is to express and purify the proteins and use them as reagents. An advantage is that use of such a system can provide a very clean background for experiments, at least with regard to the protein component. As an alternative to a purified protein, an extract from a heterologous system can be used [e.g., bacterial membranes containing the (expressed) protein of interest].
2. Libraries versus Tissue Extracts.
With a recombinant enzyme of interest, one can begin searches for reaction in several ways.
One approach is to hypothesize substrates and try each individually. This approach is most likely to work if there is already information available about the catalytic activities of enzymes that have similar primary sequences. For example, P450s 27A1 and 27B1 both have catalytic activity with vitamin D substrates; therefore, testing P450 27C1 for such activity seemed reasonable (Wu et al., 2006a). However, this approach is less amenable with P450s that have little similarity to others (e.g., P450 20A1) (Stark et al., 2008b).
Testing chemicals as substrates one by one is a laborious process, in that the probability of success is low with each compound. Moreover, each assay may require optimization of conditions. One way to approach the problem is to react a mixture of chemicals with the enzyme in a single exposure, with analysis by LC-MS (or another method) to detect which of the compounds, if any, was transformed. A synthetic library can contain representatives of a number of groups (e.g., some steroids, some fatty acids, some drugs), with the proviso that the appearance of any activity toward one of these could be followed by analysis with another library based on compounds related to the one yielding the positive result.
In the last case, the library is an extract of a tissue in which the enzyme has been shown to be expressed. [In our experience it is far easier to identify sites of expression at the RNA level than protein, although less reliably (Wu et al., 2006a).] The working assumption here is that the enzyme has a substrate in this tissue. That assumption may not be useful, however, in that in an endocrine model the substrate could be outside the tissue or, as seen with some of the “xenobiotic-metabolizing” P450s, there is not an apparent endogenous substrate (i.e., some P450s; Table 1), and the function of an enzyme may be protection from xenobiotic chemicals.
The demands on the analytical chemistry increase considerably in moving from trials with individual chemicals to tissue extracts. Thus, the need to separate compounds and compare complex sets of data cannot be overemphasized.
This laboratory has used the above approach with tissue extracts to identify some P450 substrates in human liver. A first approach was made with human liver and human P450s 1A2, 2C8, and 2C9 with the use of LC-MS in the ESI− mode (Tang et al., 2009). The approach generated a set of positives, which were found to be several fatty acids. Studies with the individual substrates confirmed the reactions and yielded values for catalytic efficiencies. Similar approaches with orphan human P450 4F11 also yielded several fatty acid substrates (Tang et al., 2010).
One of the issues in work with tissue extracts, or any large unbiased libraries, is the variation in both sensitivity and abundance of various compounds that could be substrates and products. Relatively little can be done about the abundance of individual compounds [with the exception of knocking out the gene for the enzyme to facilitate accumulation of the substrate, in the case of mice or certain model organisms (Cheng et al., 2010)[. The problem with limited sensitivity is that those molecules that ionize well in LC-MS will be favored, and those that do not will be invisible. Thus, some of our searches have yielded fatty acids as substrates (Tang et al., 2009, 2010), probably because of the propensity of these carboxylic acids to ionize in negative ion MS.
One approach to improving and, to a large extent, equalizing responses of different reaction products is derivatization with a chemical entity that produces a strong response in the instrument, and such an approach has been used in this laboratory. Many of the products of P450 reactions are unactivated alcohols, and the generation of derivatives is often difficult. In contrast, derivatives of phenols can be more easily generated. For instance, dansyl chloride is well known as a fluorescent reagent that reacts with amines and phenols. However, this moiety also has excellent LC-MS properties because of the basic tertiary amine, which is positively charged in the solvents typically used in ESI LC-MS. Modified derivatization conditions were developed that produce dansylated products in high yield. The method is particularly effective for uncharged molecules such as sterols and, in the best cases, increases the sensitivity several thousandfold (Tang and Guengerich, 2010). The method need not be quantitative for this work, only relatively high yield and general. The “equalization” of the response is an important factor. Furthermore, the method can be coupled with isotopic labeling approaches (e.g., DoGEX) (Tang and Guengerich, 2010).
E. In Vivo/In Vivo Combinations
Preliminary assays can be done in vitro or in vivo. However, combination approaches can also be used to advantage. In addition, it is important to emphasize that ultimately both in vitro and in vivo approaches are needed to understand mechanisms and to put the results into a physiological perspective.
1. Transgenic Animals and Tissue Extracts as Enzyme Sources.
One example comes from the work of Yu et al. (2003, 2004). Comparisons were made with liver microsomes derived from control mice and transgenic mice expressing human P450 2D6. Comparisons were done with a set of selected phenethylamines and indolethylamines. A number of these were shown not to be substrates, but two indolethylamines were—5-methoxy-N,N-dimethyltryptamine and picoline (6-methoxy-1,2,3,4-tetrahydro-β-carboline) (Yu et al., 2003, 2004). Furthermore, the catalytic efficiencies (kcat/km) for P450 2D6 were 9 × 104 and 2.4 × 105 M−1 s−1, which can be considered respectable. However, in this approach, the human P450 2D6 is overlaid onto an endogenous background (of mouse 2d enzymes), so the somewhat orthologous mouse P450s must catalyze these reactions only at very low rates. The exact significance of these reactions in humans is unknown.
Another example comes from our work with Streptomyces spp. P450s (Cheng et al., 2010). A P450 154A knockout strain of S. coelicolor was compared with wild-type: extracts of each were compared by using LC-MS and the program MZmine (Fig. 4) (Katajamaa and Oresic, 2005; Katajamaa et al., 2006). This approach yielded a candidate, QCP1 (i.e., a putative substrate that accumulated in the knockout strain). An extract of this strain was also incubated with purified P450 154A1, which led to the disappearance of the peak of the substrate (QCP1) and the appearance of two new peaks as products (QCP2a,b) (Figs. 4 and 5) (Cheng et al., 2010). Thus, the in vitro approach was used to confirm the initial (in vivo) result. In this case, the knockout strain also had a discernable phenotype, the instability of spores in long-term storage. However, experiments have not been done to add the isolated substrate (or product) to the knockout strain in attempts to recover the wild-type phenotype. Nevertheless, this approach to combining in vitro and in vivo analysis should be applicable in any case in which a knock-out experimental animal model and a heterologously expressed protein are both available.
Fig. 5.
Conversion of an endogeneous compund found in S. coelicolor to products without net oxidation-reduction (Cheng et al., 2010).
2. Importance of Extending In Vitro Studies In Vivo and Vice Versa.
In vitro studies can establish that a particular enzyme can catalyze a reaction. Furthermore, the catalytic efficiency can be measured, and, assuming that no factors are missing, this information can provide some insight into how important the reaction might be. However, it is possible that other enzymes might be present in the organism, doing the same thing, and that the enzyme under consideration may contribute very little.
Caveats are also associated with results that are only demonstrated in vivo. If the only result is a difference between two strains—e.g., wild type and knockout—then the enzyme reaction has not been demonstrated and alternatives may be possible. Another point is that an in vivo result might be related to the food or medium and not inherent to the organism. Thus, it is necessary to address this issue by assaying food or media. As in traditional biochemistry, the most solid results come from applying a mixture of both in vitro and in vivo approaches to questions about the physiological or other function of a protein.
IV. Current Status of Cytochrome P450 Orphans
See Table 2 for summary.
TABLE 2.
Human P450 Orphans
| P450 | Location | Sites of Expression (mRNA) | Heterologous Expression | Possible Substrates |
|---|---|---|---|---|
| 2A7 | 19q13.2 | Liver (Ding et al., 1995; Koskela et al., 1999) | HepG2 (Yamano et al., 1990), COS-7 (Ding et al., 1995), COS-1 (Oscarson et al., 2002)a | |
| 2S1 | 19q13.1 | Skin, liver (also trachea, lung, stomach, small intestine, spleen, colon, urinary tract, nasal cavity, bronchi, bronchioli, tumors) (Rylander et al., 2001; Smith et al., 2003; Saarikoski et al., 2005a,b) | S. cerevisiae (Smith et al., 2003), E. coli (Bui and Hankinson, 2009; Wu et al., 2006b; Yoshioka et al., 2006) | 4QN (Nishida et al., 2010), several eicosanoid peroxides (Bui et al., 2010) (retinoic acid, carcinogens?) (see also Wu et al., 2006b; Bui et al., 2009)b |
| 2U1 | 4q25 | Thymus, brain (Chuang et al., 2004; Karlgren et al., 2004) (also keratinocytes, kidney, heart, ovarian cancer) (Downie et al., 2005; Du et al., 2006) | Sf9 insect cells (Chuang et al., 2004) | Fatty acids (ω, ω-1) (Chuang et al., 2004) |
| 2W1 | 7q22.3 | Tumors (Karlgren et al., 2005, 2006) | HEK293 cells (Karlgren et al., 2005), E. coli (Wu et al., 2006b) | Several carcinogens (Wu et al., 2006b), 4QN (Nishida et al., 2010) |
| 3A43 | 7q21 | Liver, brain (also kidney, pancreas, prostate) (Domanski et al., 2001; Westlind et al., 2001) | E. coli (Domanski et al., 2001), COS-1 cells (Agarwal et al., 2008) | Alprazolam (Agarwal et al., 2008), testosterone (Domanski et al., 2001) |
| 4A22 | 1q33 | Liver (Savas et al., 2003) | ||
| 4F11 | 19q13.1-2 | Liver (Kalsotra et al., 2004), also kidney, heart, skeletal muscle (Cui et al., 2000) | S. cerevisiae (Kalsotra et al., 2004), insect cells (Dhar et al., 2008), E. coli (Tang et al., 2010) | Fatty acids (ω-1, ω-2, ω-3) (Kalsotra et al., 2004; Tang et al., 2010), β-hydroxy fatty acids (Dhar et al., 2008; Tang et al., 2010), benzphetamine, ethylmorphine, erythromycin (Tang et al., 2010)3 |
| 4F22 | 19q13.1-2 | Skin, testis (also bone marrow, liver, small intestine, skeletal muscle, brain) (Lefevre et al., 2006) | S. cerevisiae (Nilsson et al., 2010) | Arachidonic acid (Nilsson et al., 2010) |
| 4V2 | 4q35 | Eye | Sf9 insect cells (Nakano et al., 2009) | Fatty acids (Nakano et al., 2009) |
| 4X1 | 1q33 | Brain (amygdala), liver (esp. fetal liver), skin, prostate, heart, aorta, breast, colon, trachea, aorta kidney (Savas et al., 2005; Stark et al., 2008a) | E. coli (Stark et al., 2008a) | Anandamide (Stark et al., 2008a) |
| 4Z1 | 1p33 | Breast cancer (Rieger et al., 2004) | S. pombe (Zöllner et al., 2009) | Fatty acids (Zöllner et al., 2009) |
| 20A1 | 2q33.2 | Liver, brain (esp. hippocampus and substantia nigra) (Stark et al., 2008b) | E. coli (Stark et al., 2008b) | |
| 27C1 | 2q14.3 | Liver, kidney, others (Wu et al., 2006a) | E. coli (Wu et al., 2006a) |
A. Cytochrome P450 2A7
mRNA transcripts have been reported in human liver samples (Ding et al., 1995; Koskela et al., 1999) and possibly esophagus (Godoy et al., 2002). Alternative splicing is apparently involved in producing P450 2A6/2A7 hybrid alleles (Koskela et al., 1999; Oscarson et al., 2002). The P450 2A6/2A7 “hybrid” allele has lower lower enzymatic activity than P450 2A6 (Oscarson et al., 2002). To date, attempts at heterologous expression of P450 2A7 protein in several systems have not produced active enzyme (Yamano et al., 1990; Ding et al., 1995), and it is not clear that an active form of P450 2A7 exists.
B. Cytochrome P450 2S1
mRNA expressed has been detected in human skin and liver, and there are also reports of expression in trachea, lung, stomach, small intestine, and spleen (Table 2). The gene is reported to be regulated by the aryl hydrocarbon receptor (Rivera et al., 2002, 2007).
Heterologous expression has been achieved in both yeast and E. coli systems, with relatively high levels in the latter case. The catalytic specificity of P450 2S1 has been controversial. Smith et al. (2003) reported that retinoic acid is a substrate for P450 2S1 but did not quantify a rate. Subsequently, the oxidation of retinoic acid has not been repeated in other attempts (Wu et al., 2006b; Nishida et al., 2010). Although P450 2S1 has been reported to be inducible via the aryl hydrocarbon receptor system, no carcinogens have been found to be substrates (Wu et al., 2006b). Bui and Hankinson (2009) reported that P450 2S1 could not be reduced by NADPH-P450 reductase and that several catalytic activities of P450 2S1 could be observed if reactions were supported by oxygen surrogates, e.g., hydroperoxides (Bui and Hankinson, 2009; Bui et al., 2009, 2010). Nishida et al. (2010) reported that P450 2S1 is capable of catalyzing the reduction of the substrate AQ4N, the di-N-oxide derivative of AQ4 [1,4-bis([2-(dimethylamino)ethyl]amino)-5,8-dihydroxyanthracene-9,10-dione], a topoisomerase-inhibiting drug. The rate was 12 min−1, and a ferrous-carbon monoxide complex could be produced with the reductase (rate not measured). Therefore P450 2S1 seems capable of being reduced via electron flow from the reductase. A serious caveat in the work of Bui and Hankinson (2009) is that the reduction work was done aerobically, under conditions known to allow ferrous P450s to reoxidize rapidly (Guengerich et al., 1976). As a result of the delivery of oxygen from the capillaries, parts of organs are anoxic (e.g., reduction of CCl4 by P450), and some tumors are rather anaerobic. Nishida et al. (2010) also point out that the conclusion of Bui et al. (Bui and Hankinson, 2009; Bui et al., 2009, 2010) that P450 2S1 normally uses lipid peroxides for its catalytic function is inconclusive, in that many P450s can react with lipid hydroperoxides through a shunt reaction that generates lipid alkoxy and peroxy radicals. These radicals can enter cooxidation reactions outside of P450 active sites (Mansuy et al., 1982; Ortiz de Montellano, 1995).
In conclusion, the only demonstrated reaction of P450 2S1 is reduction of AQ4N, and no oxidation reactions have been clearly established at this time. The rate of reduction of P450 2S1 by the reductase has recently been measured at 40 s−1, and the AQ4N enhancement of NADPH oxidation has been demonstrated, clearly indicating the reduction of P450 2S1 (Xiao et al., 2011).
C. Cytochrome P450 2U1
mRNA has been detected in brain and thymus (Table 2) (Chuang et al., 2004; Karlgren et al., 2004). Heterologous expression of P450 2U1 in insect cells has been reported. The only reported reactions are hydroxylations of fatty acids, both ω- and ω-1 (Chuang et al., 2004).
D. Cytochrome P450 2W1
Attempts to find mRNA expression of P450 2W1 in human tissues were unsuccessful (Karlgren et al., 2005; Wu et al., 2006b), and expression seems to occur only in tumorous cells, particularly colorectal cancer (Gomez et al., 2010). Gomez et al. (2010) have also reported that a fraction of P450 2W1 expressed in human embryonic kidney 293 cells is glycosylated and also on the lumenal site of the endoplasmic reticulum, suggesting that it might not interact well with NADPH-P450 reductase.
P450 2W1 has been demonstrated to catalyze benzphetamine N-demethylation at a rate of ∼3 min−1 (Wu et al., 2006b) and AQ4N reduction at a rate of 12 min−1 (Nishida et al., 2010), when coupled with NADPH-P450 reductase. Very low rates of fatty acid oxidation have also been reported (Karlgren et al., 2006; Wu et al., 2006b). Some activity toward indole and indole derivatives has been observed (Wu et al., 2006b; Yoshioka et al., 2006; Gomez et al., 2010). Of particular significance is the ability of P450 2W1 to activate a variety of chemical carcinogens to genotoxic forms (Wu et al., 2006b), which may be of particular relevance in light of the localization of this P450 in cancers.
E. Cytochrome P450 3A43
This P450 is one of the four P450s in the 3A subfamily. Some controversy exists as to its significance, particularly as regards its importance relative to the other Subfamily 3A P450s, particularly the more dominant P450 3A4 (Guengerich, 1999). Nevertheless, 3A7 is generally expressed only in fetal tissue, many people do not express P450 3A5 because of polymorphisms (html://www.cypalleles.ki.se/), and some people do not appear to express active P450 3A4 (Guengerich and Turvy, 1991; Yang et al., 2010). mRNA levels of P450 3A43 were ∼0.1% of the mean levels of P450 3A4 and 2% the mean levels of P450 3A5 (Westlind et al., 2001). The localization of some of the mRNA in brain (Table 2) may be important, as well as a reported selectivity for oxidation of the drug alprazolam (Agarwal et al., 2008). Heterologous expression of active protein has been reported in E. coli (Domanski et al., 2001) and COS-1 cells (Agarwal et al., 2008) but not in yeast or mouse hepatic H2.35 cells (Westlind et al., 2001). Another substrate (with very low activity) is testosterone (Domanski et al., 2001).
F. Cytochrome P450 4A22
This gene does not seem to be expressed very extensively, with mRNA levels much lower than for P450 4A11 (Savas et al., 2003). This gene was originally named 4A11, but what was 4A22 (95% sequence identity) has now been renamed 4A11 and shown to be a dominant fatty acid ω-hydroxylase in human liver (Guengerich, 2005).
G. Cytochrome P450 4F11
The mRNA is expressed in liver (Table 2). Heterologous expression has been achieved in yeast, insect cells, and E. coli (Table 2). P450 4F11 has catalytic activity toward fatty acids and also β-keto fatty acids (Kalsotra et al., 2004; Dhar et al., 2008; Tang et al., 2010). No bioactivation was observed when a number of procarcinogens were tested (Tang et al., 2010). In recent studies, some activity in oxidation of vitamin K1 has been detected (A. E. Rettie, Y. Xiao, and F. P. Guengerich, unpublished results).
H. Cytochrome P450 4F22
Haplotype and gene mapping for nonsyndromic autosomal recessive congenital ichthyosis revealed a new gene (FLJ39501) on chromosome 19p12-q12. Seven mutations were identified in the gene, later identified as CYP4F22 (http://drnelson.uthsc.edu/CytochromeP450.html). mRNA expression was detected in skin and testis, with minor levels found in bone marrow, liver, small intestine, skeletal muscle, and brain Lefèvre et al. (2006) proposed that P450 4F22 is involved in the bioactivation of products derived from 12R-hydroperoxyeicosatetraenoic acid and 8R,11R,12R-epoxy-hydroxyeicosatrienoic acid. Nilsson et al. (2010) expressed P450 4F22 in Saccharomyces cerevisiae and reported that it oxidizes arachidonic acid to a mixture of hydroxyeicosatetraenoic acids.
I. Cytochrome P450 4V2
Mutations in the CYP4V2 gene (chromosome 4q35.1) have been found to be related to a rare eye disorder, Bietti crystalline corneoretinal dystrophy (BCD). This is an autosomal recessive chorioretinal dystrophy characterized by progressive night blindness (Li et al., 2004). Approximately 20 single-nucleotide polymorphisms have been reported, most of which have been discovered in Japanese or other Asian populations. The CYP4V2 gene consists of 11 exons, and mRNA has been found in human retina. Biochemical studies of patients with BCD indicate impaired fatty acid metabolism, which, along with homology to other P450 proteins, suggests that P450 4V2 may play a role in fatty acid and steroid metabolism (Li et al., 2004).
P450 4V2 has been expressed in insect cells and shown to catalyze ω-hydroxylation of medium-chain fatty acids (Nakano et al., 2009). However, the function is still unclear in that serum fatty acids and desaturase activites in BCD were not affected (in their subgroup analysis) by the different genotypes found in BCD patients (Lai et al., 2010), even though ocular crystal deposits were (Yokoi et al., 2010).
J. Cytochrome P450 4X1
An orthologous cDNA was cloned from rat brain (Bylund et al., 2002). Northern blotting indicated a specific expression to the brain, primarily in neurons of the hippocampus, cerebellum, and cortex. Full-length human P450 4X1 cDNA was cloned using human genome and expressed sequence tag databases, and the mRNA was found to be expressed in the trachea and aorta (Savas et al., 2005). The peroxisome proliferator activator receptor α has been reported to be involved in the activation of CYP4X1 gene transcription in a HepG2 cell line (Savas et al., 2005). In human brain, the distribution pattern shows a striking pattern with the highest levels in amygdala (Stark et al., 2008a). Human P450 4X1 could be expressed in E. coli and purified (Stark et al., 2008a). It is of interest that this P450 could catalyze a selective epoxidation of anandamide, a fatty acid amide, but not arachidonic acid (Stark et al., 2008a). However, the catalytic efficiency of the reaction was low, and the physiological significance of the product is unknown.
K. Cytochrome P450 4Z1
Transcriptome database screening for ESTs revealed the presence of P450 4Z1, a new P450, in a breast cancer cell line (SK-BR-3) (Rieger et al., 2004). P450 4Z1 mRNA expression is almost undetectable or at a low level in most human tissues, whereas abundant expression is noted in mammary tissue and breast carcinomas. It is still not known which cells express this P450 within breast tissue (Rieger et al., 2004; Savas et al., 2005). Expression has been confirmed in at least two known breast cancer cell lines, T47-D and MCF-7. In the progesterone-positive cell line T47-D, 15-fold induction of P450 4Z1 mRNA levels was found after treatment with progesterone, and a 10-fold induction after treatment with the glucocorticoid analog dexamethasone. This induction was suppressed by coadministration of the antagonist mifepristone (Savas et al., 2005).
The protein has been expressed in Schizosaccharomyces pombe (Zöllner et al., 2009) and in insect cells (Y. Xiao and F. P. Guengerich, unpublished observations). The only reported substrates are the fatty acids lauric acid and myristic acid (Zöllner et al., 2009), but the relevance of these reactions to any phenotype is unknown.
L. Cytochrome P450 20A1
mRNA expression has been detected in liver and brain, especially the hippocampus and substantia nigra (Stark et al., 2008b). It is noteworthy that a similar pattern is seen in rats. The significance of this localization is not known at this time. No substrates have yet been identified. It is of interest that related forms of this P450 appear in lower animals [e.g., as low as Xenopus tropicalis, fish, worms, sea anemone, and sponges (http://drnelson.uthsc.edu/cytochromeP450.html)].
M. Cytochrome P450 27C1
P450 27C1 mRNA is expressed in a number of tissues, including liver, kidney, and spleen (Wu et al., 2006a). The protein is in the P450 27 family, and the other two family members (P450s 27A1 and 27B1) are involved in vitamin D hydroxylation (Guengerich, 2005). P450 27A1 also hydroxylates cholesterol. P450 27A1 and 27B1 are mitochondrial enzymes and receive electrons from adrenodoxin reductase and adrenodoxin, and presumably P450 27C1 does as well. P450 27C1 did not oxidize vitamin D, 25-hydroxyvitamin D, or cholesterol (Wu et al., 2006a). No substrates have yet been identified. Recently a tricistronic approach has been developed for expressing P450 27A1 or 27C1, adrenodoxin reductase, and adrenodoxin together in E. coli (Salamanca-Pinzón and Guengerich, 2011) and may facilitate further searches for P450 27C1 reactions. It is noteworthy that mice do not have a P450 27C1 gene (but do require 27A1 and 27B1 for vitamin D homeostasis).
V. Extensions of Approaches to Other Enzymes
Many of the approaches presented here are applicable to enzymes other than P450s. However, most would be restricted to those enzymes that have low relative molecular mass substrates. Many of the LC-MS approaches can be applied to any transformations that change the relative molecular mass or the LC-MS retention time of a substrate. The isotopic labeling approaches [e.g., the M/M + 2 doublet used for DoGEX (Sanchez-Ponce and Guengerich, 2007)] could be used in other cases in which a cofactor is incorporated into a substrate (e.g., addition of sulfate or methyl groups; M + 1 isotopic studies are also possible although more problematic because of the natural abundance of the 13C component). These approaches would usually be described in the context of metabolomics but not proteomics. In principle, it might be possible to use some of these approaches to characterize enzymes with protein substrates (e.g., kinases), but in many cases, the complexity of the separations would be too problematic.
One approach we have considered but not yet attempted involves binding screens. That is, a recombinant enzyme is produced with a tag (for recovery) and mixed with a cell extract to identify a substrate. The literature contains numerous examples of such pull-down approaches for protein-protein interactions, which tend to be stronger than for small molecules. However, Yuan et al. (2009) have employed such systems to search for ligands for orphan nuclear steroid receptors and identified fatty acids.
VI. Conclusions and Perspectives
In classic biochemistry, in vivo observations led to development of in vitro assays and the characterization of enzymes, the regulation of which could be studied at the gene level (Fig. 1). Today we have sequences of all of the genes in many organisms, including humans. The challenge is to develop ways of working in the opposite direction (i.e., to define functions for these genes). This is a problem not only in biochemistry but also in pharmacology, in that validation of targets is one of the most serious issues in drug discovery.
The number of approaches is still limited. What is clear is that two of the approaches provide only limited insight: 1) reliance only on in silico predictions and 2) trial and error, one hypothesis followed by another screening. New approaches, some of which have been presented here, are needed. These have been made possible by the rapid development of progress in two areas. One is MS, where increased sensitivity and resolution have been critical. The other is informatics approaches, which are absolutely essential. Several algorithms for the identification of differences in metabolite profiles are now available, most (except PCA) within the last few years. Even more powerful methods would be welcome.
Finally, the prospect of characterizing the function of the remaining “orphan” enzymes is exciting. Surely some enzyme functions will be found to overlap already known ones, but the prospect for new reactions in the human body provides excitement in biochemistry and pharmacology. With regard to the microbial and plant enzymes, there is considerable potential for practical application in pharmacology and agriculture.
Acknowledgments
This work was supported in part by the National Institutes of Health National Cancer Institute [Grant R37-CA090426] and the National Institutes of Health National Institute of Environmental Health Sciences [Grant P30-ES000267]. Thanks are extended to K. Trisler for assistance in preparation of the manuscript and to Y. Xiao for suggestions.
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Guengerich and Cheng.
Other: Guengerich acquired funding for the research.
This article is available online at http://pharmrev.aspetjournals.org.
doi:10.1124/pr.110.003525.
- APCI
- atmospheric pressure chemical ionization
- AQ4
- 1,4-bis([2-(dimethylamino)ethyl]amino)-5,8-dihydroxyanthracene-9,10-dione
- BCD
- Bietti crystalline corneoretinal dystrophy
- DoGEX
- Discovery of General Endo- and Xenobiotics
- ESI
- electrospray ionization
- GC
- gas chromatography
- LC
- liquid chromatography
- MS
- mass spectrometry
- P450
- cytochrome P450
- PCA
- principal component analysis.
References
- Agarwal V, Kommaddi RP, Valli K, Ryder D, Hyde TM, Kleinman JE, Strobel HW, Ravindranath V. (2008) Drug metabolism in human brain: high levels of cytochrome P4503A43 in brain and metabolism of anti-anxiety drug alprazolam to its active metabolite. PLos One 3:e2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollard ME, Stanley EG, Lindon JC, Nicholson JK, Holmes E. (2005) NMR-based metabonomic approaches for evaluating physiological influences on biofluid composition. NMR Biomed 18:143–162 [DOI] [PubMed] [Google Scholar]
- Bui PH, Hankinson O. (2009) Functional characterization of human cytochrome P450 2S1 using a synthetic gene-expressed protein in Escherichia coli. Mol Pharmacol 76:1031–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui PH, Hsu EL, Hankinson O. (2009) Fatty acid hydroperoxides support cytochrome P450 2S1-mediated bioactivation of benzo[a]pyrene-7,8-dihydrodiol. Mol Pharmacol 76:1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui P, Imaizumi S, Beedanagari SR, Reddy ST, Hankinson O. (2010) Human CYP2S1 metabolizes cyclooxygenase- and lipoxygenase-derived eicosanoids. Drug Metab Dispos 39:180–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund J, Zhang C, Harder DR. (2002) Identification of a novel cytochrome P450, CYP4X1, with unique localization specific to the brain. Biochem Biophys Res Commun 296:677–684 [DOI] [PubMed] [Google Scholar]
- Cheng Q, Lamb DC, Kelly SL, Lei L, Guengerich FP. (2010) Cyclization of a cellular dipentaenone by Streptomyces coelicolor cytochrome P450 154A1 without oxidation/reduction. J Am Chem Soc 132:15173–15175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang SS, Helvig C, Taimi M, Ramshaw HA, Collop AH, Amad M, White JA, Petkovich M, Jones G, Korczak B. (2004) CYP2U1, a novel human thymus- and brain-specific cytochrome P450, catalyzes ω- and (ω-1)-hydroxylation of fatty acids. J Biol Chem 279:6305–6314 [DOI] [PubMed] [Google Scholar]
- Cui X, Nelson DR, Strobel HW. (2000) A novel human cytochrome P450 4F isoform (CYP4F11): cDNA cloning, expression, and genomic structural characterization. Genomics 68:161–166 [DOI] [PubMed] [Google Scholar]
- Dhar M, Sepkovic DW, Hirani V, Magnusson RP, Lasker JM. (2008) Omega oxidation of 3-hydroxy fatty acids by the human CYP4F gene subfamily enzyme CYP4F11. J Lipid Res 49:612–624 [DOI] [PubMed] [Google Scholar]
- Ding S, Lake BG, Friedberg T, Wolf CR. (1995) Expression and alternative splicing of the cytochrome P-450 CYP2A7. Biochem J 306:161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanski TL, Finta C, Halpert JR, Zaphiropoulos PG. (2001) cDNA cloning and initial characterization of CYP3A43, a novel human cytochrome P450. Mol Pharmacol 59:386–392 [DOI] [PubMed] [Google Scholar]
- Downie D, McFadyen MC, Rooney PH, Cruickshank ME, Parkin DE, Miller ID, Telfer C, Melvin WT, Murray GI. (2005) Profiling cytochrome P450 expression in ovarian cancer: identification of prognostic markers. Clin Cancer Res 11:7369–7375 [DOI] [PubMed] [Google Scholar]
- Du L, Neis MM, Ladd PA, Lanza DL, Yost GS, Keeney DS. (2006) Effects of the differentiated keratinocyte phenotype on expression levels of CYP1–4 family genes in human skin cells. Toxicol Appl Pharmacol 213:135–144 [DOI] [PubMed] [Google Scholar]
- Dunayevskiy YM, Vouros P, Wintner EA, Shipps GW, Carell T, Rebek J., Jr (1996) Application of capillary electrophoresis-electrospray ionization mass spectrometry in the determination of molecular diversity. Proc Natl Acad Sci USA 93:6152–6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran AL, Yang J, Wang L, Sumner LW. (2003) Metabolomics spectral formatting, alignment and conversion tools (MSFACTs). Bioinformatics 19:2283–2293 [DOI] [PubMed] [Google Scholar]
- Gerlt JA, Babbitt PC. (2001) Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu Rev Biochem 70:209–246 [DOI] [PubMed] [Google Scholar]
- Godoy W, Albano RM, Moraes EG, Pinho PR, Nunes RA, Saito EH, Higa C, Filho IM, Kruel CD, Schirmer CC, et al. (2002) CYP2A6/2A7 and CYP2E1 expression in human oesophageal mucosa: regional and inter-individual variation in expression and relevance to nitrosamine metabolism. Carcinogenesis 23:611–616 [DOI] [PubMed] [Google Scholar]
- Gomez A, Nekvindova J, Travica S, Lee MY, Johansson I, Edler D, Mkrtchian S, Ingelman-Sundberg M. (2010) Colorectal cancer-specific cytochrome P450 2W1: intracellular localization, glycosylation, and catalytic activity. Mol Pharmacol 78:1004–1011 [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Nebert DW. (1990) Evolution of the P450 gene superfamily: animal-plant ‘warfare’, molecular drive and human genetic differences in drug oxidation. Trends Genet 6:182–186 [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Yu AM. (2006) Cytochrome P450 and xenobiotic receptor humanized mice. Annu Rev Pharmacol Toxicol 46:41–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Weng Y, Zhang QY, Cui H, Behr M, Wu L, Yang W, Zhang L, Ding X. (2003) Liver-specific deletion of the NADPH-cytochrome P450 reductase gene: impact on plasma cholesterol homeostasis and the function and regulation of microsomal cytochrome P450 and heme oxygenase. J Biol Chem 278:25895–25901 [DOI] [PubMed] [Google Scholar]
- Guengerich FP. (1999) Human cytochrome P-450 3A4: regulation and role in drug metabolism. Annu Rev Pharmacol Toxicol 39:1–17 [DOI] [PubMed] [Google Scholar]
- Guengerich FP. (2001) Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol 14:611–650 [DOI] [PubMed] [Google Scholar]
- Guengerich FP. (2005) Human cytochrome P450 enzymes, in Cytochrome P450: Structure Mechanism, and Biochemistry, 3rd ed (Ortiz de Montellano PR. ed) pp 377–530, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- Guengerich FP, Ballou DP, Coon MJ. (1976) Spectral intermediates in the reaction of oxygen with purified liver microsomal cytochrome P-450. Biochem Biophys Res Commun 70:951–956 [DOI] [PubMed] [Google Scholar]
- Guengerich FP, Isin EM. (2011) Unusual metabolic reactions and pathways, in Handbook of Metabolic Pathways of Xenobiotics (Prakash C, Gau L, Zhong D, Aizawa H, Lee P. eds), vol. 1, John Wiley & Sons, New York: In press [Google Scholar]
- Guengerich FP, Tang Z, Cheng Q, Salamanca-Pinzón SG. (2011) Approaches to deorphanization of human and microbial cytochrome P450 enzymes. Biochem Biophys Acta 1814:139–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich FP, Tang Z, Salamanca-Pinzón SG, Cheng Q. (2010) Characterizing proteins of unknown function: orphan cytochrome P450 enzymes as a paradigm. Mol Interv 10:153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich FP, Turvy CG. (1991) Comparison of levels of several human microsomal cytochrome P-450 enzymes and epoxide hydrolase in normal and disease states using immunochemical analysis of surgical liver samples. J Pharmacol Exp Ther 256:1189–1194 [PubMed] [Google Scholar]
- Hanson AD, Pribat A, Waller JC, de Crécy-Lagard V. (2010) ‘Unknown’ proteins and ‘orphan’ enzymes: the missing half of the engineering parts list–and how to find it. Biochem J 425:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys JM, Chapple C. (2000) Molecular ‘pharming’ with plant P450s. Trends Plant Sci 5:271–272 [DOI] [PubMed] [Google Scholar]
- Isin EM, Guengerich FP. (2007) Complex reactions catalyzed by cytochrome P450 enzymes. Biochim Biophys Acta 1770:314–329 [DOI] [PubMed] [Google Scholar]
- Jakoby WB. (1980) Detoxication enzymes, in Enzymatic Basis of Detoxication (Jakoby WB. ed), vol. 1, pp 1–6, Academic Press, New York [Google Scholar]
- Kalsotra A, Turman CM, Kikuta Y, Strobel HW. (2004) Expression and characterization of human cytochrome P450 4F11: putative role in the metabolism of therapeutic drugs and eicosanoids. Toxicol Appl Pharmacol 199:295–304 [DOI] [PubMed] [Google Scholar]
- Kameda S, Maruyama H, Higuchi N, Nakamura G, Iino N, Nishikawa Y, Miyazaki J, Gejyo F. (2003) Hydrodynamics-based transfer of PCR-amplified DNA fragments into rat liver. Biochem Biophys Res Commun 309:929–936 [DOI] [PubMed] [Google Scholar]
- Karlgren M, Backlund M, Johansson I, Oscarson M, Ingelman-Sundberg M. (2004) Characterization and tissue distribution of a novel human cytochrome P450-CYP2U1. Biochem Biophys Res Commun 315:679–685 [DOI] [PubMed] [Google Scholar]
- Karlgren M, Gomez A, Stark K, Svärd J, Rodriguez-Antona C, Oliw E, Bernal ML, Ramón y Cajal S, Johansson I, Ingelman-Sundberg M. (2006) Tumor-specific expression of the novel cytochrome P450 enzyme, CYP2W1. Biochem Biophys Res Commun 341:451–458 [DOI] [PubMed] [Google Scholar]
- Karlgren M, Stark K, Oliw E, Ingelman-Sundberg M. (2005) A novel human cytochrome P450 enzyme, CYP2W1. Analysis of function and distribution. Drug Metab Rev 37:51 [Google Scholar]
- Katajamaa M, Miettinen J, Oresic M. (2006) MZmine: toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics 22:634–636 [DOI] [PubMed] [Google Scholar]
- Katajamaa M, Oresic M. (2005) Processing methods for differential analysis of LC/MS profile data. BMC Bioinformatics 6:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskela S, Hakkola J, Hukkanen J, Pelkonen O, Sorri M, Saranen A, Anttila S, Fernandez-Salguero P, Gonzalez F, Raunio H. (1999) Expression of CYP2A genes in human liver and extrahepatic tissues. Biochem Pharmacol 57:1407–1413 [DOI] [PubMed] [Google Scholar]
- Lai TY, Chu KO, Chan KP, Ng TK, Yam GH, Lam DS, Pang CP. (2010) Alterations in serum fatty acid concentrations and desaturase activities in Bietti crystalline dystrophy unaffected by CYP4V2 genotypes. Invest Ophthalmol Vis Sci 51:1092–1097 [DOI] [PubMed] [Google Scholar]
- Lamb DC, Guengerich FP, Kelly SL, Waterman MR. (2006) Exploiting Streptomyces coelicolor A3(2) P450s as a model for application in drug discovery. Expert Opin Drug Metab Toxicol 2:27–40 [DOI] [PubMed] [Google Scholar]
- Lamb DC, Zhao B, Guengerich FP, Kelly SL, Waterman MR. (2011) Genomics of streptomyces cytochrome P450, in Streptomyces: Molecular Biology and Biotechnology, Horizon Press, Hethersett, UK: In press [Google Scholar]
- Lefèvre C, Bouadjar B, Ferrand V, Tadini G, Mégarbané A, Lathrop M, Prud'homme JF, Fischer J. (2006) Mutations in a new cytochrome P450 gene in lamellar ichthyosis type 3. Hum Mol Genet 15:767–776 [DOI] [PubMed] [Google Scholar]
- Li A, Jiao X, Munier FL, Schorderet DF, Yao W, Iwata F, Hayakawa M, Kanai A, Shy Chen M, Alan Lewis R, et al. (2004) Bietti crystalline corneoretinal dystrophy is caused by mutations in the novel gene CYP4V2. Am J Hum Genet 74:817–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Schuler MA, Berenbaum MR. (2003) Diversification of furanocoumarin-metabolizing cytochrome P450 monooxygenases in two papilionids: specificity and substrate encounter rate. Proc Natl Acad Sci USA 100 (Suppl 2):14593–14598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Evans RM. (1995) The RXR heterodimers and orphan receptors. Cell 83:841–850 [DOI] [PubMed] [Google Scholar]
- Mansuy D, Bartoli JF, Momenteau M. (1982) Alkane hydroxylation catalyzed by metalloporphyrins: evidence for different active oxygen species with alkylhydroperoxides and iodosobenzene as oxidants. Tetrahedron Lett 23:2781–2784 [Google Scholar]
- McLean KJ, Dunford AJ, Neeli R, Driscoll MD, Munro AW. (2007) Structure, function and drug targeting in Mycobacterium tuberculosis cytochrome P450 systems. Arch Biochem Biophys 464:228–240 [DOI] [PubMed] [Google Scholar]
- Morant M, Bak S, Møller BL, Werck-Reichhart D. (2003) Plant cytochromes P450: tools for pharmacology, plant protection and phytoremediation. Curr Opin Biotechnol 14:151–162 [DOI] [PubMed] [Google Scholar]
- Mueller GC, Miller JA. (1948) The metabolism of 4-dimethylaminoazobenzene by rat liver homogenates. J Biol Chem 176:535–544 [PubMed] [Google Scholar]
- Nakano M, Kelly EJ, Rettie AE. (2009) Expression and characterization of CYP4V2 as a fatty acid ω-hydroxylase. Drug Metab Dispos 37:2119–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert DW, Russell DW. (2002) Clinical importance of the cytochromes P450. Lancet 360:1155–1162 [DOI] [PubMed] [Google Scholar]
- Nilsson T, Ivanov IV, Oliw EH. (2010) LC-MS/MS analysis of epoxyalcohols and epoxides of arachidonic acid and their oxygenation by recombinant CYP4F8 and CYP4F22. Arch Biochem Biophys 494:64–71 [DOI] [PubMed] [Google Scholar]
- Nishida CR, Lee M, de Montellano PR. (2010) Efficient hypoxic activation of the anticancer agent AQ4N by CYP2S1 and CYP2W1. Mol Pharmacol 78:497–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa T, Murayama N, Yamazaki H. (2009) Oxidation of endobiotics mediated by xenobiotic-metabolizing forms of human cytochrome P450. Curr Drug Metab 10:700–712 [DOI] [PubMed] [Google Scholar]
- Ortiz de Montellano PR. (1995) Oxygen activation and reactivity, in Cytochrome P450: Structure, Mechanism, and Biochemistry, 2nd ed (Ortiz de Montellano PR. ed) pp 245–303, Plenum Press, New York [Google Scholar]
- Oscarson M, McLellan RA, Asp V, Ledesma M, Bernal Ruiz ML, Sinues B, Rautio A, Ingelman-Sundberg M. (2002) Characterization of a novel CYP2A7/CYP2A6 hybrid allele (CYP2A6*12) that causes reduced CYP2A6 activity. Hum Mutat 20:275–283 [DOI] [PubMed] [Google Scholar]
- Poulos TL, Johnson EF. (2005) Structures of cytochrome P450 enzymes, in Cytochrome P450: Structure Mechanism, and Biochemistry, 3rd Ed (Ortiz de Montellano PR. ed) pp 87–114, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- Remmer H. (1959) [The acceleration of evipan oxidation and the demethylation of methylaminopyrine by barbiturates.] Naunyn Schmiedebergs Arch Exp Pathol Pharmakol 237:296–307 [PubMed] [Google Scholar]
- Rieger MA, Ebner R, Bell DR, Kiessling A, Rohayem J, Schmitz M, Temme A, Rieber EP, Weigle B. (2004) Identification of a novel mammary-restricted cytochrome P450, CYP4Z1, with overexpression in breast carcinoma. Cancer Res 64:2357–2364 [DOI] [PubMed] [Google Scholar]
- Rivera SP, Saarikoski ST, Hankinson O. (2002) Identification of a novel dioxin-inducible cytochrome P450. Mol Pharmacol 61:255–259 [DOI] [PubMed] [Google Scholar]
- Rivera SP, Wang F, Saarikoski ST, Taylor RT, Chapman B, Zhang R, Hankinson O. (2007) A novel promoter element containing multiple overlapping xenobiotic and hypoxia response elements mediates induction of cytochrome P4502S1 by both dioxin and hypoxia. J Biol Chem 282:10881–10893 [DOI] [PubMed] [Google Scholar]
- Ryan KJ. (1958) Conversion of androstenedione to estrone by placental microsomes. Biochim Biophys Acta 27:658–662 [DOI] [PubMed] [Google Scholar]
- Rylander T, Neve EP, Ingelman-Sundberg M, Oscarson M. (2001) Identification and tissue distribution of the novel human cytochrome P450 2S1 (CYP2S1). Biochem Biophys Res Commun 281:529–535 [DOI] [PubMed] [Google Scholar]
- Saarikoski ST, Rivera SP, Hankinson O, Husgafvel-Pursiainen K. (2005a) CYP2S1: a short review. Toxicol Appl Pharmacol 207:62–69 [DOI] [PubMed] [Google Scholar]
- Saarikoski ST, Wikman HA, Smith G, Wolff CH, Husgafvel-Pursiainen K. (2005b) Localization of cytochrome P450 CYP2S1 expression in human tissues by in situ hybridization and immunohistochemistry. J Histochem Cytochem 53:549–556 [DOI] [PubMed] [Google Scholar]
- Salamanca-Pinzón GS, Guengerich FP. (2011) A tricistronic human adrenodoxin reductase-adrenodoxin-cytrochrome P450 27A1 vector system for substrate hydroxylation in Escherichia coli. Protein Expr Purif doi:10.1016/j.pep.2011.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Ponce R, Guengerich FP. (2007) Untargeted analysis of mass spectrometry data for elucidation of metabolites and function of enzymes. Anal Chem 79:3355–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansen S, Yano JK, Reynald RL, Schoch GA, Griffin KJ, Stout CD, Johnson EF. (2007) Adaptations for the oxidation of polycyclic aromatic hydrocarbons exhibited by the structure of human P450 1A2. J Biol Chem 282:14348–14355 [DOI] [PubMed] [Google Scholar]
- Savas U, Hsu MH, Griffin KJ, Bell DR, Johnson EF. (2005) Conditional regulation of the human CYP4X1 and CYP4Z1 genes. Arch Biochem Biophys 436:377–385 [DOI] [PubMed] [Google Scholar]
- Savas U, Hsu MH, Johnson EF. (2003) Differential regulation of human CYP4A genes by peroxisome proliferators and dexamethasone. Arch Biochem Biophys 409:212–220 [DOI] [PubMed] [Google Scholar]
- Schuler MA. (1996) The role of cytochrome P450 monooxygenases in plant-insect interactions. Plant Physiol 112:1411–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen AL, O'Leary KA, Kasper CB. (2002) Association of multiple developmental defects and embryonic lethality with loss of microsomal NADPH-cytochrome P450 oxidoreductase. J Biol Chem 277:6536–6541 [DOI] [PubMed] [Google Scholar]
- Shimada T, Hayes CL, Yamazaki H, Amin S, Hecht SS, Guengerich FP, Sutter TR. (1996) Activation of chemically diverse procarcinogens by human cytochrome P450 1B1. Cancer Res 56:2979–2984 [PubMed] [Google Scholar]
- Shinkyo R, Guengerich FP. (2011) Cytochrome P450 7A1 cholesterol 7α-hydroxylation: individual reaction steps in the catalytic cycle and rate-limiting ferric iron reduction. J Biol Chem 286:4632–4643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. (2006) XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 78:779–787 [DOI] [PubMed] [Google Scholar]
- Smith G, Wolf CR, Deeni YY, Dawe RS, Evans AT, Comrie MM, Ferguson J, Ibbotson SH. (2003) Cutaneous expression of cytochrome P450 CYP2S1: individuality in regulation by therapeutic agents for psoriasis and other skin diseases. Lancet 361:1336–1343 [DOI] [PubMed] [Google Scholar]
- Stark K, Dostalek M, Guengerich FP. (2008a) Expression and purification of orphan cytochrome P450 4X1 and oxidation of anandamide. FEBS J 275:3706–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark K, Wu ZL, Bartleson CJ, Guengerich FP. (2008b) Heterologous expression and mRNA distribution of orphan cytochrome P450 20A1. Drug Metab Dispos 36:1930–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoilov I, Akarsu AN, Sarfarazi M. (1997) Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Hum Mol Genet 6:641–647 [DOI] [PubMed] [Google Scholar]
- Takei K, Yamaya T, Sakakibara H. (2004) Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-zeatin. J Biol Chem 279:41866–41872 [DOI] [PubMed] [Google Scholar]
- Tang Z, Guengerich FP. (2010) Dansylation of unactivated alcohols for improved mass spectral sensitivity and application to analysis of cytochrome P450 oxidation products in tissue extracts. Anal Chem 82:7706–7712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Martin MV, Guengerich FP. (2009) Elucidation of functions of human cytochrome P450 enzymes: identification of endogenous substrates in tissue extracts using metabolomic and isotopic labeling approaches. Anal Chem 81:3071–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Salamanca-Pinzón SG, Wu ZL, Xiao Y, Guengerich FP. (2010) Human cytochrome P450 4F11: heterologous expression in bacteria, purification, and characterization of catalytic function. Arch Biochem Biophys 494:86–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M, Lamb DC, Cannell R, Dawson M, Kelly SL. (1999) Cytochrome P450105D1 (CYP105D1) from Streptomyces griseus: heterologous expression, activity, and activation effects of multiple xenobiotics. Biochem Biophys Res Commun 263:838–842 [DOI] [PubMed] [Google Scholar]
- Tolstikov VV, Lommen A, Nakanishi K, Tanaka N, Fiehn O. (2003) Monolithic silica-based capillary reversed-phase liquid chromatography/electrospray mass spectrometry for plant metabolomics. Anal Chem 75:6737–6740 [DOI] [PubMed] [Google Scholar]
- Vasiliou V, Gonzalez FJ. (2008) Role of CYP1B1 in glaucoma. Annu Rev Pharmacol Toxicol 48:333–358 [DOI] [PubMed] [Google Scholar]
- Wang A, Savas U, Stout CD, Johnson EF. (2011) Structural characterization of the complex between α-naphthoflavone and human cytochrome P450 1B1. J Biol Chem 286:5736–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlind A, Malmebo S, Johansson I, Otter C, Andersson TB, Ingelman-Sundberg M, Oscarson M. (2001) Cloning and tissue distribution of a novel human cytochrome P450 of the CYP3A subfamily, CYP3A43. Biochem Biophys Res Commun 281:1349–1355 [DOI] [PubMed] [Google Scholar]
- Wienkers LC, Heath TG. (2005) Predicting in vivo drug interactions from in vitro drug discovery data. Nat Rev Drug Discov 4:825–833 [DOI] [PubMed] [Google Scholar]
- Williams JA, Hyland R, Jones BC, Smith DA, Hurst S, Goosen TC, Peterkin V, Koup JR, Ball SE. (2004) Drug-drug interactions for UDP-glucuronosyltransferase substrates: a pharmacokinetic explanation for typically observed low exposure (AUCi/AUC) ratios. Drug Metab Dispos 32:1201–1208 [DOI] [PubMed] [Google Scholar]
- Wu ZL, Bartleson CJ, Ham AJ, Guengerich FP. (2006a) Heterologous expression, purification, and properties of human cytochrome P450 27C1. Arch Biochem Biophys 445:138–146 [DOI] [PubMed] [Google Scholar]
- Wu ZL, Sohl CD, Shimada T, Guengerich FP. (2006b) Recombinant enzymes over-expressed in bacteria show broad catalytic specificity of human cytochrome P450 2W1 and limited activity of human cytochrome P450 2S1. Mol Pharmacol 69:2007–2014 [DOI] [PubMed] [Google Scholar]
- Xiao Y, Shinkyo R, Guengerich FP. (2011) Cytochrome P450 2S1 is reduced by NADPH-cytochrome P450 reductase. Drug Metab Dispos 39:944–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano S, Tatsuno J, Gonzalez FJ. (1990) The CYP2A3 gene product catalyzes coumarin 7-hydroxylation in human liver microsomes. Biochemistry 29:1322–1329 [DOI] [PubMed] [Google Scholar]
- Yang X, Zhang B, Molony C, Chudin E, Hao K, Zhu J, Gaedigk A, Suver C, Zhong H, Leeder JS, et al. (2010) Genetic and genomic analysis of cytochrome P450 enzyme activities in human liver. Genome Res 20:1020–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H, Kasai N, Ikushiro S, Shinkyo R, Kamakura M, Ohta M, Inouye K, Sakaki T. (2006) Enzymatic properties of human CYP2W1 expressed in Escherichia coli. Biochem Biophys Res Commun 345:169–174 [DOI] [PubMed] [Google Scholar]
- Yu AM, Idle JR, Gonzalez FJ. (2004) Polymorphic cytochrome P450 2D6: humanized mouse model and endogenous substrates. Drug Metab Rev 36:243–277 [DOI] [PubMed] [Google Scholar]
- Yu AM, Idle JR, Herraiz T, Küpfer A, Gonzalez FJ. (2003) Screening for endogenous substrates reveals that CYP2D6 is a 5-methoxyindolethylamine O-demethylase. Pharmacogenetics 13:307–319 [DOI] [PubMed] [Google Scholar]
- Yokoi Y, Nakazawa M, Mizukoshi S, Sato K, Usui T, Takeuchi K. (2010) Crystal deposits on the lens capsules in Bietti crystalline corneoretinal dystrophy associated with a mutation in the CYP4V2 gene. Acta Ophthalmol 88:607–609 [DOI] [PubMed] [Google Scholar]
- Yuan X, Ta TC, Lin M, Evans JR, Dong Y, Bolotin E, Sherman MA, Forman BM, Sladek FM. (2009) Identification of an endogenous ligand bound to a native orphan nuclear receptor. PloS one 4:e5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zöllner A, Dragan CA, Pistorius D, Müller R, Bode HB, Peters FT, Maurer HH, Bureik M. (2009) Human CYP4Z1 catalyzes the in-chain hydroxylation of lauric acid and myristic acid. Biol Chem 390:313–317 [DOI] [PubMed] [Google Scholar]