Abstract
4-Hydroxy-5-methoxy-2,3-dihydro-1H-[1,3]benzodioxolo[5,6-c]pyrrolo[1,2-f]-phenanthridium chloride (NK314) is a benzo[c] phenanthridine alkaloid that inhibits topoisomerase IIα, leading to the generation of DNA double-strand breaks (DSBs) and activating the G2 checkpoint pathway. The purpose of the present studies was to investigate the DNA intercalating properties of NK314, to evaluate the DNA repair mechanisms activated in cells that may lead to resistance to NK314, and to develop mechanism-based combination strategies to maximize the antitumor effect of the compound. A DNA unwinding assay indicated that NK314 intercalates in DNA, a property that likely cooperates with its ability to trap topoisomerase IIα in its cleavage complex form. The consequence of this is the formation of DNA DSBs, as demonstrated by pulsed-field gel electrophoresis and H2AX phosphorylation. Clonogenic assays demonstrated a significant sensitization in NK314-treated cells deficient in DNA-dependent protein kinase (DNA-PK) catalytic subunit, Ku80, ataxia telangiectasia mutated (ATM), BRCA2, or XRCC3 compared with wild-type cells, indicating that both nonhomologous end-joining and homologous recombination DNA repair pathways contribute to cell survival. Furthermore, both the DNA-PK inhibitor 8-(4-dibenzothienyl)-2-(4-morpholinyl)-4H-1-benzopyran-4-one (NU7441) and the ATM inhibitor 2-(4-morpholinyl)-6-(1-thianthrenyl)-4H-pyran-4-one (KU55933) significantly sensitized cells to NK314. We conclude that DNA-PK and ATM contribute to cell survival in response to NK314 and could be potential targets for abrogating resistance and maximizing the antitumor effect of NK314.
Introduction
4-Hydroxy-5-methoxy-2,3-dihydro-1H-[1,3]benzodioxolo[5,6-c]pyrrolo[1,2-f]-phenanthridium chloride (NK314) is a novel synthetic benzo[c]phenanthridine alkaloid that has been shown to inhibit topoisomerase IIα activity by stabilizing the topoisomerase IIα cleavage complex and generating DNA double-strand breaks (DSBs), which activate the G2 DNA damage checkpoint pathway and lead to cytotoxicity (Fig. 1A) (Guo et al., 2007; Toyoda et al., 2008). The specific topoisomerase IIα-targeting activity of NK314 and lack of activity against topoisomerase IIβ (Toyoda et al., 2008) distinguish its actions from other topoisomerase II inhibitors such as etoposide and doxorubicin, in that inhibition of topoisomerase IIβ is associated with secondary malignancies and toxicity (Azarova et al., 2007). NK314 also showed activity toward tumors resistant to other topoisomerase II inhibitors (Onda et al., 2008). Because NK314 has been demonstrated to induce rapid and extensive DNA DSBs (Guo et al., 2007; Onda et al., 2008), further investigation of the response of DNA repair pathways to NK314 is needed to understand the cell survival and resistance mechanisms and develop mechanism-based combination strategies.
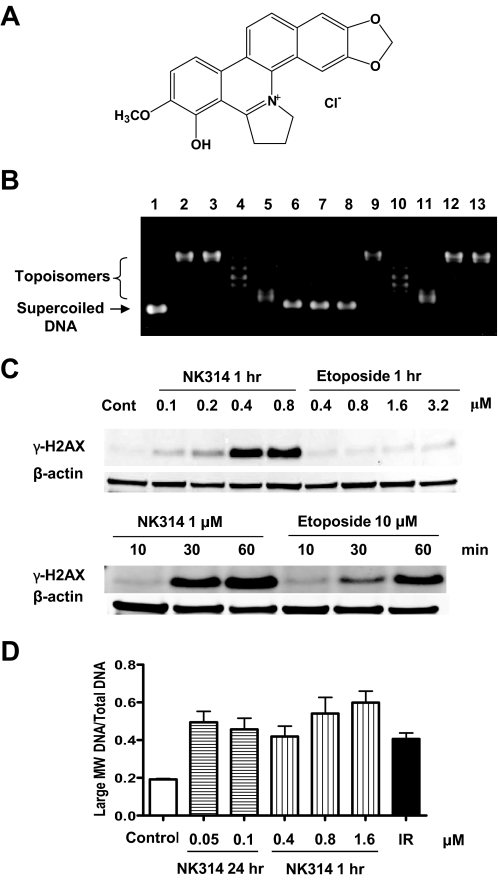
Fig. 1.
DNA intercalation and DSBs induced by NK314. A, structure of NK314. B, NK314 intercalates into DNA in vitro. Supercoiled plasmid DNA (pHOT1) was incubated with topoisomerase I (topo I) and various concentrations of NK314, etoposide, and m-AMSA. Lane 1, pHOT1 plasmid DNA; lane 2, pHOT1 and topo I (no drug control); lanes 3 to 7, pHOT1 and topo I in the presence of 0 (solvent control), 2, 10, 20, and 40 μM NK314; lane 8, pHOT1 and 40 μM NK314 (no enzyme control); lanes 9 to 11, pHOT1 and topo I in the presence of 0 (solvent control), 10, and 50 μM m-AMSA; lanes 12 and 13, pHOT1 and topo I in the presence of 0.1 and 1 mM etoposide. C, phosphorylation of H2AX in response to NK314. ML-1 cells were incubated with 0.1 to 0.8 μM NK314 or 0.4 to 3.2 μM etoposide for 24 h, and samples were collected to detect γ-H2AX by immunoblotting. ML-1 cells were also incubated with 1 μM NK314 or 10 μM etoposide, and samples were collected at 10, 30, and 60 min to detect γ-H2AX by immunoblotting. D, the fraction of the intensity of large molecular weight DNA fragment and total DNA was calculated by Kodak 1D Image Analysis Software 3.5. Each column is the mean of two independent experiments.
DNA DSBs are considered to be the most lethal DNA damage if they are not repaired properly, because they lead to the formation of chromosomal aberrations that then cause cell death from a loss of genetic material. DSBs are repaired either through homologous recombination (HR) or nonhomologous end-joining (NHEJ) (Khanna and Jackson, 2001). HR requires the presence of an identical sequence, often from a sister chromatid available in late S or G2 phase after DNA replication, as a template for repair of the break. Ataxia telangiectasia mutated (ATM) plays an important role in HR and is required in the activation of the HR repair pathway in response to ionizing radiation (Morrison et al., 2000). Other important proteins involved in HR include RAD51, BRCA2, and XRCC3 (San Filippo et al., 2008). In contrast, NHEJ does not need homologous sequences to repair damaged DNA. In NHEJ, DNA breaks are joined after resection to small regions of homology (Chen et al., 2000; Ahnesorg et al., 2006). Although NHEJ is error-prone, it is especially important before the cell has replicated its DNA because there is no template available for repair by homologous recombination. The DNA-dependent protein kinase (DNA-PK) is a key component of the NHEJ apparatus promoting the joining of broken DNA ends (Durocher and Jackson, 2001). DNA-PK comprises a catalytic subunit (DNA-PKcs) and regulatory subunits Ku70 and Ku80. Ku70 and Ku80 bind directly to free DNA termini and activate DNA-PKcs to form the DNA-PK complex (Collis et al., 2005).
DNA DSBs repair pathways are crucial for the maintenance of genome integrity and cell survival. Impairing DSB repair pathways using specific inhibitors of repair proteins might sensitize tumor cells to particular DNA-damaging agents, especially those that induce DSBs. Thus, inhibitors of ATM and DNA-PK are being developed as potential therapeutics for the treatment of cancer (Ding et al., 2006). Inhibition of ATM may disrupt the HR repair pathway and sensitize cells to DSB-inducing agents. DNA-PK, ATM, and ATM and Rad3-related (ATR) all belong to the phosphoinositide 3-kinase-related kinase family and are DNA damage sensors functioning in different pathways. A small-molecule inhibitor of ATM [2-(4-morpholinyl)-6-(1-thianthrenyl)-4H-pyran-4-one (KU55933)] has been shown to sensitize cells to ionizing radiation, etoposide, doxorubicin, and camptothecin (Hickson et al., 2004). Several small-molecule inhibitors of DNA-PKcs have also been developed, including 8-(4-dibenzothienyl)-2-(4-morpholinyl)-4H-1-benzopyran-4-one (NU7441), which sensitizes cells to ionizing radiation and etoposide, increases DSBs, and increases the antitumor activity of etoposide in vivo (Leahy et al., 2004; Zhao et al., 2006). Inhibitors of these repair pathways are being prepared to enter clinical trials, offering the promise of mechanism-based combinations with DNA damaging agents in cancer therapeutics.
Materials and Methods
Cell Culture.
ML-1, a human acute myeloid leukemia cell line, was a gift from Dr. Michael B. Kastan (St. Jude Children's Research Hospital, Memphis, TN) (Kastan et al., 1991). OCI-AML3 was kindly provided by Dr. Michael Andreeff (The University of Texas, MD Anderson Cancer Center, Houston, TX) (Carter et al., 2001). ML-1 and OCI-AML3 cells were maintained in exponential growth phase in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) at 37°C in a humidified atmosphere with 5% CO2. AT22IJE-T (AT-C), a fibroblast cell line derived from a patient with ataxia telangiectasia, and lines stably transfected with full-length ATM cDNA (AT22IJE-TpEBS7-YZ5, AT-AT) were gifts from Dr. Yosef Shiloh (Tel Aviv University, Tel Aviv, Israel) (Ziv et al., 1997), and were cultured in Dulbecco's modified Eagle's medium with high glucose and 20% FBS. Glioma-derived cell lines M059-K (wild-type) and M059-J (DNA-PKcs-deficient) were obtained from Dr. M. J. Allalunis-Turner (Brookhaven National Laboratory, Upton, NY) (Anderson et al., 2001) and cultured in α-minimum essential medium (MEM) supplemented with 20% FBS. F02-98hTERT, an ATR-deficient Seckel fibroblast cell line, and 1BRhTERT ATR wild-type cells, gifts from Dr. Penny A. Jeggo (University of Sussex, Sussex, UK),were cultured in α-MEM supplemented with 20% FBS. ATR protein was not detectable in F02-98hTERT cells by immunoblotting (Fig. 4A), consistent with that reported in the original article (O'Driscoll et al., 2003). Colon carcinoma cell line HCT116 with wild-type p53 was provided by Dr. Bert Vogelstein (The Johns Hopkins University School of Medicine, Baltimore, MD), and cultured in McCoy's 5A with 10% FBS. Cervical cancer cell line HeLa CCL2 was purchased from the American Type Culture Collection (Manassas, VA) and cultured in MEM with nonessential amino acids, sodium pyruvate, and 10% fetal bovine serum. Chinese hamster ovary line AA8 was purchased from the American Type Culture Collection. AA8-derived mutant irs1SF (Xrcc3) and Chinese hamster lung cell line V79 and its mutant V-C8 (Brca2) were generously provided by Dr. R. Legerski (MD Anderson Cancer Center, Houston, TX). xrs6 (Ku80) and xrs6-hamKu80 (Ku80-repleted) Chinese hamster ovary cells (Ross et al., 1995) were obtained from European Collection of Cell Cultures (Salisbury, Wiltshire, UK). All hamster cells were maintained in α-MEM supplemented with 10% FBS. All human cell lines were authenticated by short tandem repeat analysis at an MD Anderson core facility.
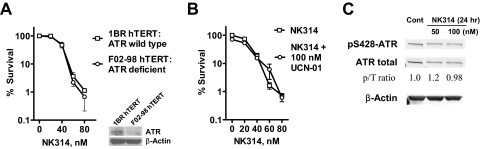
Fig. 4.
ATR does not play a role in survival in response to NK314. A, ATR wild-type 1BRhTERT and ATR-deficient F02-98hTERT Seckel cells were incubated with various concentrations of NK314 for 24 h. The insert shows immunoblotting of ATR and β-actin (loading control) in these two cell lines. B, ML-1 cells were incubated with various concentrations of NK314 in the presence or absence of 100 nM UCN-01 for 24 h. For experiments in A and B, colonies were counted after eight doubling times. Each data point represents the mean ± S.E.M. of triplicate samples. C, ATR phosphorylation level does not change in response to NK314. HeLa cells were exposed to 50 and 100 nM NK314 for 24 h, and lysates were subjected to SDS-polyacrylamide gel electrophoresis and then immunoblotting with indicated antibodies.
Chemicals and Antibodies.
NK314 was provided by Nippon Kayaku Co. Ltd (Tokyo, Japan). A stock solution (20 mM) was prepared in 5% glucose solution, sterilized by filtration, stored at −70°C, and diluted in sterile 5% glucose solution just before use. The DNA-PK inhibitor NU7441 was kindly provided by Dr. Graeme C. M. Smith (KuDOS Pharmaceuticals, Cambridge, UK) (Zhao et al., 2006), and the ATM inhibitor KU55933 was purchased from EMD Chemicals (Gibbstown, NJ) (Hickson et al., 2004; Zhao et al., 2006). All other chemicals were of reagent grade. Sources of antibodies are as follows: phospho-Ser139 of H2AX (Millipore, Billerica, MA), mouse monoclonal antibodies against β-actin (Sigma-Aldrich, St. Louis, MO), IRDye 680 goat anti-mouse, or IRDye 800CW goat anti-rabbit IgG (Li-Cor Inc., Lincoln, NE).
DNA Unwinding Assay.
Supercoiled pHOT1 plasmid DNA (0.25 μg; TopoGEN, Inc., Port Orange, FL) was incubated with 1 μl of topoisomerase I in reaction buffer composed of 10 mM Tris-HCl, pH 7.9, 1 mM EDTA, 0.15 M NaCl, 0.1% bovine serum albumin, 0.1 mM spermidine, and 5% glycerol for 30 min at 37°C. Various concentrations of NK314, etoposide and m-AMSA were added, and the mixture was incubated for another 30 min. The reaction was terminated by the addition of SDS to 1% and digested by 50 μg/ml proteinase-K for 15 min at 56°C. The reaction products were separated on a 1% agarose gel, then visualized with 0.5 μg/ml ethidium bromide, and photographed under UV light (Bauer, 1978; Okuhara et al., 1999). m-AMSA was used as a positive control, and etoposide was used as a negative control.
Pulsed-Field Gel Electrophoresis.
Drug-treated cells (106) were washed in PBS, resuspended in 0.5 ml of PBS, and mixed with 0.5 ml of 1% agarose. Plugs were cast and cooled at 4°C for 20 min before incubation in 1.5 ml of lysis buffer (1% sarkosyl, 50 mM EDTA, 50 mM Tris-HCl, pH 7.8, and 1 mg/ml proteinase K) overnight at 45°C. The plugs were loaded into the wells of an 11 × 11-cm 0.5% agarose gel and sealed in the well with molten agarose. Electrophoresis was performed in a contour-clamped homogeneous field DR II unit (Bio-Rad Laboratories, Hercules, CA) at 40 V with a 75-min switch time for 16 h in 0.5× Tris-borate-EDTA buffer. Gels were stained with ethidium bromide (1 μg/ml) and photographed under UV light (Schwartz and Cantor, 1984; Story et al., 1994). Quantitation was done using Kodak 1D 3.5 software (Eastman Kodak Co., Rochester, NY).
Clonogenic Assays.
Cells were incubated with different concentrations of drug for a population doubling time, washed in fresh medium, and dilutions of 0.5 to 10 × 103 suspension cells were plated in 35-mm plastic dishes containing MethoCult H4230 methylcellulose medium (Stem Cell Technologies, Vancouver, BC, Canada). Adherent cells were plated in 60-mm Petri dishes containing fresh medium. Colonies of 100 to 200 cells were counted using a dissecting microscope after eight doubling times. The plating efficiencies of untreated controls were 30 to 50%. Results are reported as the mean ± S.D. of at least triplicate plates.
Immunoblotting.
Cell lysates were subjected to isolation by SDS-polyacrylamide gel electrophoresis and immunoblotting as described previously (Guo et al., 2007).
Cell Cycle Analysis.
Cells were washed with ice-cold PBS, pH 7.4, and fixed in 70% ethanol. Fixed cells were washed with PBS before incubation with 50 μg/ml propidium iodide (Sigma-Aldrich) and 2.5 μg/ml DNase-free RNase A (Roche Diagnostics, Indianapolis, IN). Fluorescence was measured using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA). At least 2 × 104 cells were measured for each sample.
Statistical Analysis.
Statistical analyses were performed using the Prism 5 software (GraphPad Software, Inc., San Diego, CA). The significance of results was determined using Student's paired t test. Results were considered significant when P < 0.05. Inhibitory concentration (IC50 and IC90) values were calculated by nonlinear regression (sigmoidal dose-response curve).
Results
NK314 Is a DNA Intercalator.
Unwinding of the double strands of the DNA helix is a hallmark feature of intercalating agents, such as chloroquine, ethidium bromide, and m-AMSA. DNA unwinding assays, originally developed by Pommier et al. (1987), are used to identify intercalating agents. We applied a modified unwinding assay using the pHOT1 plasmid as a supercoiled DNA substrate to detect changes in the linking number of topoisomers, which are generated by relaxation of closed circular DNA with excess topoisomerase I. The presence of a DNA intercalator alters the distribution of topoisomers. NK314 started decreasing the linking number of topoisomers at a concentration as low as 2 μM, equivalent to the effect of 10 μM m-AMSA (Fig. 1B). This demonstrates that NK314 is a rather potent intercalator, much more effective than m-AMSA. Etoposide, which does not intercalate into DNA, did not have any effect. Our previous study demonstrated that NK314 stabilized topoisomerase IIα cleavage complex and inhibited its activity. Taken together, NK314 seems to act as a DNA intercalator that stabilizes topoisomerase IIα cleavage complex and inhibits its activity.
NK314 Induces DNA DSBs.
The observation that NK314 induces γ-H2AX formation suggested that NK314 may induce DSBs (Guo et al., 2007; Onda et al., 2008). However, H2AX may also be phosphorylated in response to replication stress or stalled replication forks (Ward and Chen, 2001; Ewald et al., 2007). To investigate whether NK314 induces DNA DSBs, ML-1 cells were treated with 50 and 100 nM NK314 for 24 h and with 0.4, 0.8, or 1.6 μM NK314 for 1 h before pulsed-field gel electrophoresis (PFGE) analysis (Fig. 1C). Ionizing radiation (10 Gy) was used as a positive control. Large molecular weight DNA fragments indicating DNA DSBs were observed after a 24-h treatment with 50 and 100 nM NK314 (data not shown). Concentrations of NK314 greater than 0.4 μM also induced DNA DSBs after 1 h, which was concomitant with the phosphorylation of H2AX (Fig. 1C). This was not seen in cells incubated with cytotoxic concentrations of etoposide. Quantitation of fluorescent intensities of DNA indicated that the fraction of large molecular weight DNA fragments in total DNA increased in a concentration-dependent manner after 1 h of treatment with NK314 (Fig. 1D). The DNA DSBs demonstrated by PFGE results were consistent with the phosphorylation of H2AX, supporting the conclusion that the H2AX phosphorylation caused by NK314 is attributable to DNA DSBs and not to another cause such as induction of apoptosis.
DNA-PK Contributes to Cell Survival in Response to NK314.
To study the function of DNA-PK in cell survival in response to NK314, the clonogenic survival of the DNA-PKcs wild-type M059K cell line was compared with that of the DNA-PKcs mutant M059J cells. A significant decrease in colony formation was observed in M059J cells compared with M059K cells (P = 0.02, paired t test) (Fig. 2A). In response to 200 nM NK314, M059J cells were approximately 10 times more sensitive than M059K cells were (0.5% colony formation compared with 4.4%). This indicates that DNA-PKcs contributes to the survival of the cells in response to NK314. These results provided a rationale for using a DNA-PKcs inhibitor to increase the cytotoxicity of NK314.
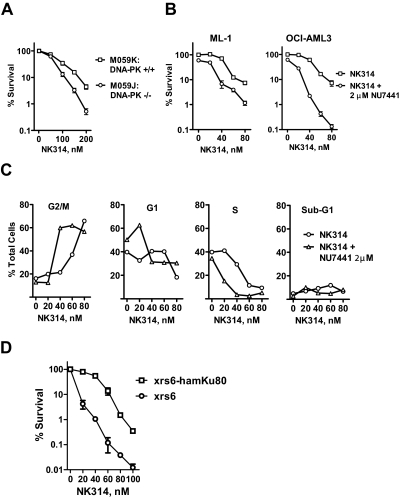
Fig. 2.
DNA-PK complex contributes to cell survival in response to NK314. A, M059J (DNA-PKcs mutant) and M059K (DNA-PKcs wild type) cells were incubated with various concentrations of NK314 for 24 h. Each data point represents the mean ± S.E.M. of triplicate samples. B, ML-1 and OCI-AML3 cells were treated with NK314 in the absence or presence of NU7441, a specific DNA-PK inhibitor. In both experiments, after 24 h, cells were washed and fresh medium was added. Colonies were counted after eight doubling times. Each data point represents the mean ± S.E.M. of triplicate samples. C, ML-1 cells were treated with NK314 in the absence or presence of 2 μM NU7441. Samples were collected at 24 h, stained with propidium iodide, and analyzed by flow cytometry. The data are representative of two independent experiments. D, xrs6 (Ku80-deficient) and xrs6-hamKu80 (Ku80-repleted) cells were treated with 0 to 100 nM NK314 for 24 h. Colonies were counted after 5 days. Each data point represents the mean ± S.E.M. of triplicate samples.
NU7441 is a potent and specific DNA-PK inhibitor that has been reported to potentiate the cytotoxicity of ionizing radiation and of etoposide (Leahy et al., 2004; Zhao et al., 2006). Treatment of ML-1 and OCI-AML3 cells with NU7441 abrogated the increase in phosphorylation of XRCC4, a downstream target of DNA-PKcs, induced by γ-irradiation in a concentration-dependent manner (Supplemental Fig. S1). Phosphorylation of SMC1 and Nbs1, targets of the ATM kinase, was not altered by NU7441, indicating its specificity for DNA-PKcs. Clonogenic assays in both ML-1 and OCI-AML3 cells demonstrated that 2 μM NU7441 increased the cytotoxicity of NK314 (Fig. 2B). NU7441 sensitized cells to 80 nM NK314 by approximately 6 times (1.2% colony formation compared with 7.4%) in ML-1 cells and approximately 60 times in OCI-AML3 cells (7.2 versus 0.13%). NU7441 increased the proportion of ML-1 cells arrested in G2 in response to 40 nM NK314 from 20 to 60%, possibly as a result of inhibition of repair of DNA damage (Fig. 2C). In contrast, NU7441 alone did not significantly diminish clonogenic survival or affect cell cycle distribution. Furthermore, NU7441 decreased the survival of M059K (P = 0.02, paired t test) but not M059J cells (P = 0.13, paired t test) treated with NK314 (Supplemental Fig. S2). These results indicate that DNA-PK is the target of NU7441 in these cells and that it is an important survival factor in response to NK314.
Ku80 is an important component of the NHEJ pathway, which binds and activates DNA-PKcs. Thus, Ku80-deficient xrs6 and Ku80-repleted xrs6-hamKu80 cells were used to study the function of Ku80 subunit in DNA-PK complex in response to NK314. A significant decrease in colony formation was observed in xrs6 cells compared with xrs6-hamKu80 cells (P = 0.003, paired t test) (Fig. 2D). In response to 60 nM NK314, xrs6 cells were approximately 100 times more sensitive than xrs6-hamKu80 cells were (0.12% colony formation compared with 14%). These results demonstrate that both DNA-PKcs and Ku80 contribute to the survival of the cells in response to NK314 and are consistent with the conclusion that NHEJ is probably the major repair pathway of the NK314-induced DNA damage.
Lack of ATM, BRCA2, or XRCC3 Sensitizes Cells to NK314.
ATM is a key protein involved in the homologous recombination repair of DNA DSBs; it phosphorylates BRCA1 (Cortez et al., 1999) and is required for efficient Rad51 focus formation (Jazayeri et al., 2006). ATM-deficient and -repleted cells were used in clonogenic assays to study the function of ATM in cell survival in response to NK314. A significant decrease in colony formation was observed in AT-C cells compared with that in AT-AT cells (P = 0.01, paired t test) (Fig. 3A), indicating that ATM and likely homologous recombination also contribute to the survival of the cells in response to NK314. On exposure to 160 nM NK314, AT-C cells (0.12% colony formation) were 70 times more sensitive than were AT-AT cells (8.7%). This provided a rationale for using an ATM-specific inhibitor to sensitize cells to NK314. KU55933 is a highly potent and specific ATM inhibitor that has been reported to increase the cytotoxicity of ionizing radiation (Hickson et al., 2004; Cowell et al., 2005). Preincubation of HCT116 and OCI-AML3 cells with KU55933 abolished phosphorylation of SMC1 and Nbs1 induced by NK314 (50 and 100 nM, respectively; Supplemental Fig. S3A) or γ-irradiation (Supplemental Fig. S3B), demonstrating specificity of KU55933 for ATM. Consistent with results with the mutant cells, clonogenic assays in HCT116 cells demonstrated that KU55933 increased the cytotoxicity of NK314 significantly (P = 0.003, paired t test) (Fig. 3B). For instance, KU55933 increased the sensitivity of HCT116 cells to 80 nM NK314 by approximately 14-fold.
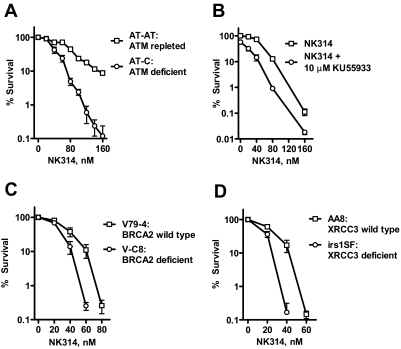
Fig. 3.
ATM, XRCC3, and BRCA2 are involved in DNA repair in response to NK314. A, AT-C (ATM-deficient) and AT-AT (ATM-repleted) cells were incubated with various concentrations of NK314 for 24 h. B, HCT116 cells were incubated with various concentrations of NK314 in the presence or absence of 10 μM KU55933 for 24 h. At 24 h, cells were washed and plated in the presence of 10 μM KU55933 in the medium. V79-4 (BRCA2 wild-type) and V-C8 (BRCA2-deficient) cells (C) and AA8 (XRCC3 wild-type) and irs1SF (XRCC3-deficient) cells (D) were incubated with various concentrations of NK314 for 24 h. At 24 h, cells were washed, and fresh medium was added. Colonies were counted after eight doubling times. Each data point represents the mean ± S.E.M. of triplicate samples.
BRCA2 and XRCC3 are crucial proteins in the HR pathway. To study their roles in cell survival in response to NK314, BRCA2-deficient V-C8 and wild-type V79-4 cells, and XRCC3-deficient irs1SF and wild-type AA8 cells were treated with NK314, and their viability was compared in clonogenic assays. A significant decrease in colony formation was observed in the cell lines deficient in these HR proteins compared with colony formation in wild-type cell lines (P = 0.01, paired t test) (Fig. 3, C and D). This sensitization of approximately 100-fold provides additional evidence that homologous recombination contributes to the survival of the cells in response to NK314.
ATR Does Not Contribute to Cell Survival in Response to NK314.
ATR is another important DNA-damage sensor that initiates S-phase cell cycle arrest in response to replication stress (Shiloh, 2001). To study the role of ATR in the survival of cells in response to NK314, ATR wild-type 1BRhTERT, and ATR-deficient F02-98hTERT Seckel fibroblasts were used in clonogenic assays. There was no significant difference in colony formation between these two cell lines in response to NK314 (P > 0.05, paired t test) (Fig. 4A), suggesting that ATR may not contribute to cell survival in response to NK314. Our earlier studies demonstrated that UCN-01 abrogates NK314-activated G2 checkpoint through inhibition of the Chk1 kinase (Guo et al., 2007). However, UCN-01 did not alter the survival of ML-1 cells after NK314 treatment (Fig. 4B). Similar results were observed for OCI-AML3 cells after NK314 treatment in the presence or absence of UCN-01 (data not shown). Therefore, depletion of ATR has a minimal impact on cell survival after exposure to NK314. In addition, phosphorylation of ATR on Ser428 was not altered by NK314 at cell cycle-arresting concentrations (50 and 100 nM) in HeLa cells (Fig. 4C), suggesting no further activation of ATR in response to NK314. Thus, ATR does not play a significant role in repairing NK314-induced DNA lesions.
Discussion
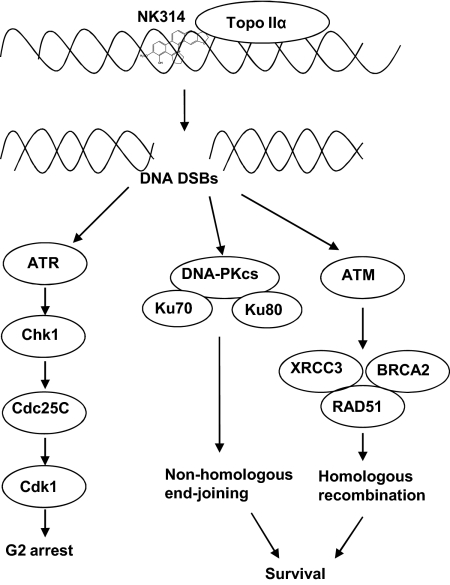
The initial studies of the actions of NK314 demonstrated that it was a potent inhibitor of topoisomerase IIα (Guo et al., 2007; Onda et al., 2008; Toyoda et al., 2008). The present investigation extended this work to develop an understanding that NK314 intercalates into DNA, an action that may contribute to its inhibition of topoisomerase IIα. This earlier work also demonstrated that cells respond to the DNA damage caused by NK314 by activating the G2 cell cycle checkpoint (Guo et al., 2007). This arrest in progression of cells through the cell cycle to mitosis could be a potential resistance mechanism by facilitating the repair of damaged DNA. However, present studies demonstrate that abrogation of the checkpoint with the Chk1 inhibitor, UCN-01, added little to the cytotoxicity of NK314. It is noteworthy that evaluation of the apical sensors of DNA damage and repair responses demonstrated that cells lacking the double-strand break repair pathways were substantially sensitized to the toxicity of NK314. Thus, we conclude that the major survival mechanisms in response to NK314-induced DNA damage are the DNA-PK–mediated NHEJ and ATM-mediated HR pathways to facilitate DNA repair (Fig. 5). Finally, inhibitors of DNA-PK and of ATM each sensitized cells to NK314, indicating a rationale for combination strategies.
Fig. 5.
The model of mechanism of action of NK314 and cellular responses. NK314 induces DNA DSBs, which are sensed by ATR, ATM, and DNA-PK. ATM and DNA-PK are important to cell survival. They may mediate the HR and NHEJ DNA repair pathways, respectively. ATR may lead to cell cycle checkpoint activation by activating Chk1.
A DNA unwinding assay demonstrated that NK314 at concentrations greater than 10 μM induced forced supercoiling of relaxed DNA (Fig. 1A). This concentration is consistent with that which also stabilized cleavage complexes in vitro and inhibited the activity of topoisomerase IIα in a previous study (Guo et al., 2007), indicating that NK314 may be a DNA intercalator. Other DNA intercalators such as anthracyclines, mitomycin, and amsacrine are potent topoisomerase II inhibitors and DNA DSBs inducers (Tewey et al., 1984; Liu, 1989) that have a wide spectrum of activity against solid tumors and hematological malignancies. However, some of these agents are known to generate free radicals that contribute to their cardiotoxicity and limit their usage (Singal and Iliskovic, 1998). This would seem less likely for NK314, a benzo[c]phenanthridine alkaloid that lacks the quinone structure responsible for free radical production.
Inhibition of topoisomerase II arrests the enzyme in the cleavable complex intermediate, preventing the religation of DNA cleavage product resulting in frank DNA DSBs (Long et al., 1985; Osheroff, 1989). Consistent with this mechanism of action, results of PFGE assays demonstrated that NK314 induced DNA DSBs both after 1 h at concentrations greater than 400 nM and in 24 h at concentrations less than 100 nM (Fig. 1B). This was associated with the phosphorylation of H2AX, a well documented marker of DSB (Rogakou et al., 1998). H2AX phosphorylation has been reported to occur as a result of cleavage stabilization caused by topoisomerase I inhibition in association with DNA replication (Furuta et al., 2003; Bonner et al., 2008). This alternative explanation must be considered, because the activity of NK314 against this enzyme has not been investigated. The fact that these events occurred within an hour makes it unlikely that the breaks were a result of either apoptosis or of replication over damaged DNA. NK314 is more potent than etoposide at inducing DNA breaks, possibly because of its DNA intercalation properties. NK314 also induced DNA DSBs at 24 h at concentrations less than 100 nM, conditions that induce G2 arrest in cells at 24 h (Guo et al., 2007), which is consistent with the conclusion that G2 arrest is a consequence of the DNA DSB formation.
Clonogenic studies demonstrated that cells lacking either DNA-PKcs or Ku80 were significantly more sensitive to NK314 than wild-type cells (Fig. 2, A and D), indicating that DNA-PK-mediated NHEJ is involved in the repair of NK314-induced DNA DSBs. These results are in accord with those of Toyoda et al. (2008), who reported sensitization in preB Nalm-6 cells lacking the NEHJ enzyme ligase 4. In addition, cells lacking ATM were significantly more sensitive to NK314 than ATM-repleted cells (Fig. 3A), indicating that ATM contributes to the survival of cells in response to NK314. Moreover, cells lacking BRCA2 or XRCC3, two other proteins in the HR pathway that interact with Rad51 (Bishop et al., 1998; Davies et al., 2001), were sensitized to NK314 (Fig. 3, C and D). The consistency of these results indicating the participation of the HR pathway in response to NK314 is somewhat in contrast to those of an earlier study (Toyoda et al., 2008) that showed only a small sensitization in cells deficient in Rad54, another HR participant. Because both results were generated using clonogenic assays, it may be that there are redundant activities for the function of Rad54. Consistent with our studies in deficient cell lines, inhibitors of DNA-PK and ATM, NU7441 and KU55933, respectively, mimicked these findings by increasing the cytotoxicity of NK314. This suggests that the pharmacological inhibition of these DSB sensors may be a useful strategy for mechanism-based combinations with NK314. Therefore, both the NHEJ and HR pathways are likely to be involved in the repair of NK314-induced DNA damage and contribute to the survival or resistance of the cells.
Supplementary Material
 The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported by the National Institutes of Health National Cancer Institute, Department of Health and Human Services [Grants CA28596, CA100632, CA16672].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.109.057125.
- DSB
- DNA double-strand break
- ATM
- ataxia telangiectasia mutated
- ATR
- ataxia telangiectasia mutated and rad3-related
- DNA-PK
- DNA-dependent protein kinase
- DNA-PKcs
- DNA-dependent protein kinase catalytic subunit
- FBS
- fetal bovine serum
- HR
- homologous recombination
- NHEJ
- nonhomologous end-joining
- PFGE
- pulsed-field gel electrophoresis
- NU7441
- 8-(4-dibenzothienyl)-2-(4-morpholinyl)-4H-1-benzopyran-4-one
- KU55933
- 2-(4-morpholinyl)-6-(1-thianthrenyl)-4H-pyran-4-one
- MEM
- minimum essential medium
- PBS
- phosphate-buffered saline
- m-AMSA
- 4′-(9-acridinylamino)methanesulfon-m-anisidide.
Authorship Contributions
Participated in research design: Guo, Liu, and Plunkett.
Conducted experiments: Guo, Liu, and Jiang.
Performed data analysis: Guo, Liu, Jiang, and Plunkett.
Wrote or contributed to the writing of the manuscript: Guo, Liu, and Plunkett.
Other: Nishikawa contributed vital chemicals for this study and critically reviewed the manuscript.
References
- Ahnesorg P, Smith P, Jackson SP. (2006) XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell 124:301–313 [DOI] [PubMed] [Google Scholar]
- Anderson CW, Dunn JJ, Freimuth PI, Galloway AM, Allalunis-Turner MJ. (2001) Frameshift mutation in PRKDC, the gene for DNA-PKcs, in the DNA repair-defective, human, glioma-derived cell line M059J. Radiat Res 156:2–9 [DOI] [PubMed] [Google Scholar]
- Azarova AM, Lyu YL, Lin CP, Tsai YC, Lau JY, Wang JC, Liu LF. (2007) Roles of DNA topoisomerase II isozymes in chemotherapy and secondary malignancies. Proc Natl Acad Sci USA 104:11014–11019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer WR. (1978) Structure and reactions of closed duplex DNA. Annu Rev Biophys Bioeng 7:287–313 [DOI] [PubMed] [Google Scholar]
- Bishop DK, Ear U, Bhattacharyya A, Calderone C, Beckett M, Weichselbaum RR, Shinohara A. (1998) Xrcc3 is required for assembly of Rad51 complexes in vivo. J Biol Chem 273:21482–21488 [DOI] [PubMed] [Google Scholar]
- Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. (2008) GammaH2AX and cancer. Nat Rev Cancer 8:957–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter BZ, Milella M, Altieri DC, Andreeff M. (2001) Cytokine-regulated expression of survivin in myeloid leukemia. Blood 97:2784–2790 [DOI] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Sung P, Tomkinson AE. (2000) Interactions of the DNA ligase IV-XRCC4 complex with DNA ends and the DNA-dependent protein kinase. J Biol Chem 275:26196–26205 [DOI] [PubMed] [Google Scholar]
- Collis SJ, DeWeese TL, Jeggo PA, Parker AR. (2005) The life and death of DNA-PK. Oncogene 24:949–961 [DOI] [PubMed] [Google Scholar]
- Cortez D, Wang Y, Qin J, Elledge SJ. (1999) Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science 286:1162–1166 [DOI] [PubMed] [Google Scholar]
- Cowell IG, Durkacz BW, Tilby MJ. (2005) Sensitization of breast carcinoma cells to ionizing radiation by small molecule inhibitors of DNA-dependent protein kinase and ataxia telangiectsia mutated. Biochem Pharmacol 71:13–20 [DOI] [PubMed] [Google Scholar]
- Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC. (2001) Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell 7:273–282 [DOI] [PubMed] [Google Scholar]
- Ding J, Miao ZH, Meng LH, Geng MY. (2006) Emerging cancer therapeutic opportunities target DNA-repair systems. Trends Pharmacol Sci 27:338–344 [DOI] [PubMed] [Google Scholar]
- Durocher D, Jackson SP. (2001) DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr Opin Cell Biol 13:225–231 [DOI] [PubMed] [Google Scholar]
- Ewald B, Sampath D, Plunkett W. (2007) H2AX phosphorylation marks gemcitabine-induced stalled replication forks and their collapse upon S-phase checkpoint abrogation. Mol Cancer Ther 6:1239–1248 [DOI] [PubMed] [Google Scholar]
- Furuta T, Takemura H, Liao ZY, Aune GJ, Redon C, Sedelnikova OA, Pilch DR, Rogakou EP, Celeste A, Chen HT, et al. (2003) Phosphorylation of histone H2AX and activation of Mre11, Rad50, and Nbs1 in response to replication-dependent DNA double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J Biol Chem 278:20303–20312 [DOI] [PubMed] [Google Scholar]
- Guo L, Liu X, Nishikawa K, Plunkett W. (2007) Inhibition of topoisomerase IIalpha and G2 cell cycle arrest by NK314, a novel benzo[c]phenanthridine currently in clinical trials. Mol Cancer Ther 6:1501–1508 [DOI] [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. (2004) Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res 64:9152–9159 [DOI] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. (2006) ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 8:37–45 [DOI] [PubMed] [Google Scholar]
- Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, Craig RW. (1991) Participation of p53 protein in the cellular response to DNA damage. Cancer Res 51:6304–6311 [PubMed] [Google Scholar]
- Khanna KK, Jackson SP. (2001) DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 27:247–254 [DOI] [PubMed] [Google Scholar]
- Leahy JJ, Golding BT, Griffin RJ, Hardcastle IR, Richardson C, Rigoreau L, Smith GC. (2004) Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg Med Chem Lett 14:6083–6087 [DOI] [PubMed] [Google Scholar]
- Liu LF. (1989) DNA topoisomerase poisons as antitumor drugs. Annu Rev Biochem 58:351–375 [DOI] [PubMed] [Google Scholar]
- Long BH, Musial ST, Brattain MG. (1985) Single- and double-strand DNA breakage and repair in human lung adenocarcinoma cells exposed to etoposide and teniposide. Cancer Res 45:3106–3112 [PubMed] [Google Scholar]
- Morrison C, Sonoda E, Takao N, Shinohara A, Yamamoto K, Takeda S. (2000) The controlling role of ATM in homologous recombinational repair of DNA damage. EMBO J 19:463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. (2003) A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet 33:497–501 [DOI] [PubMed] [Google Scholar]
- Okuhara K, Ohta K, Seo H, Shioda M, Yamada T, Tanaka Y, Dohmae N, Seyama Y, Shibata T, Murofushi H. (1999) A DNA unwinding factor involved in DNA replication in cell-free extracts of Xenopus eggs. Curr Biol 9:341–350 [DOI] [PubMed] [Google Scholar]
- Onda T, Toyoda E, Miyazaki O, Seno C, Kagaya S, Okamoto K, Nishikawa K. (2008) NK314, a novel topoisomerase II inhibitor, induces rapid DNA double-strand breaks and exhibits superior antitumor effects against tumors resistant to other topoisomerase II inhibitors. Cancer Lett 259:99–110 [DOI] [PubMed] [Google Scholar]
- Osheroff N. (1989) Effect of antineoplastic agents on the DNA cleavage/religation reaction of eukaryotic topoisomerase II: inhibition of DNA religation by etoposide. Biochemistry 28:6157–6160 [DOI] [PubMed] [Google Scholar]
- Pommier Y, Covey JM, Kerrigan D, Markovits J, Pham R. (1987) DNA unwinding and inhibition of mouse leukemia L1210 DNA topoisomerase I by intercalators. Nucleic Acids Res 15:6713–6731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273:5858–5868 [DOI] [PubMed] [Google Scholar]
- Ross GM, Eady JJ, Mithal NP, Bush C, Steel GG, Jeggo PA, McMillan TJ. (1995) DNA strand break rejoining defect in xrs-6 is complemented by transfection with the human Ku80 gene. Cancer Res 55:1235–1238 [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. (2008) Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77:229–257 [DOI] [PubMed] [Google Scholar]
- Schwartz DC, Cantor CR. (1984) Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell 37:67–75 [DOI] [PubMed] [Google Scholar]
- Shiloh Y. (2001) ATM and ATR: networking cellular responses to DNA damage. Curr Opin Genet Dev 11:71–77 [DOI] [PubMed] [Google Scholar]
- Singal PK, Iliskovic N. (1998) Doxorubicin-induced cardiomyopathy. N Engl J Med 339:900–905 [DOI] [PubMed] [Google Scholar]
- Story MD, Mendoza EA, Meyn RE, Tofilon PJ. (1994) Pulsed-field gel electrophoretic analysis of DNA double-strand breaks in mammalian cells using photostimulable storage phosphor imaging. Int J Radiat Biol 65:523–528 [DOI] [PubMed] [Google Scholar]
- Tewey KM, Rowe TC, Yang L, Halligan BD, Liu LF. (1984) Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science 226:466–468 [DOI] [PubMed] [Google Scholar]
- Toyoda E, Kagaya S, Cowell IG, Kurosawa A, Kamoshita K, Nishikawa K, Iiizumi S, Koyama H, Austin CA, Adachi N. (2008) NK314, a topoisomerase II inhibitor that specifically targets the alpha isoform. J Biol Chem 283:23711–23720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Chen J. (2001) Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem 276:47759–47762 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Thomas HD, Batey MA, Cowell IG, Richardson CJ, Griffin RJ, Calvert AH, Newell DR, Smith GC, Curtin NJ. (2006) Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res 66:5354–5362 [DOI] [PubMed] [Google Scholar]
- Ziv Y, Bar-Shira A, Pecker I, Russell P, Jorgensen TJ, Tsarfati I, Shiloh Y. (1997) Recombinant ATM protein complements the cellular A-T phenotype. Oncogene 15:159–167 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.