Abstract
There are currently two distinct models proposed to explain why both MDM2 and MDMX are required in p53 control, with a key difference centered on whether these two p53 inhibitors work together or independently. To test these two competing models, we generated knockin mice expressing a point mutation MDMX mutant (C462A) that is defective in MDM2 binding. This approach allowed a targeted disassociation of the MDM2/MDMX heterocomplex without affecting the ability of MDMX to bind to p53, and while leaving the MDM2 protein itself completely untouched. Significantly, MdmxC462A/C462A homozygous mice died at approximately day 9.5 of embryonic development, as the result of a combination of apoptosis and decreased cell proliferation, as shown by TUNEL and BrdU incorporation assays, respectively. Interestingly, even though the MDMX mutant protein abundance was found slightly elevated in the MdmxC462A/C462A homozygous embryos, both the abundance and activity of p53 were markedly increased. A p53-dependent death was demonstrated by the finding that concomitant deletion of p53 completely rescued the embryonic lethality in MdmxC462A/C462A homozygous mice. Our data demonstrate that MDM2 and MDMX function as an integral complex in p53 control, providing insights into the nonredundant nature of the function of MDM2 and MDMX.
Keywords: knockin mouse model, p53 regulation
Under normal physiological conditions, wild-type p53 protein levels must be kept low owing to its growth-inhibitory activities, and this control is mainly modulated via regulation of p53 protein stability. Although a number of different regulators have been reported to be involved in this protein regulation, MDM2 has been shown to be the principal player in control of p53 turnover (1). MDM2 primarily functions as an E3 ubiquitin ligase targeting p53 for ubiquitination and subsequent degradation. At the same time, p53 induces the expression of the Mdm2 gene, forming a negative feedback loop (1). The importance of MDM2 in p53 control is highlighted by the finding that Mdm2 knockout results in p53-dependent embryonic lethality in mice (2, 3).
MDMX (also known as MDM4), which was originally isolated as a novel p53-interacting protein, shares substantial structural homology with MDM2 (4, 5). The highest sequence similarity between MDM2 and MDMX lies at the N terminus and contains a p53-binding domain, and the two also share high sequence homology in a RING-finger domain, a region that mediates the association between MDMX and MDM2 (6, 7). Genetic studies have demonstrated that like MDM2, MDMX is another essential negative regulator of p53 (8–10). Although it remains unclear why both MDM2 and MDMX are required for p53 control, a model has been proposed that these two proteins function independently. On the basis of the fact that unlike MDM2, MDMX lacks an intrinsic ubiquitin E3 ligase activity, it has been proposed that MDMX inhibits p53 chiefly by binding to the p53 transactivation domain and antagonizing p53 transcription activity, whereas MDM2 inactivates p53 primarily by working as an E3 ligase to control p53 turnover (11).
However, abundant evidence indicates an intricate interplay between MDM2 and MDMX in p53 regulation (12–16). Consistent with the prediction made by structural studies that the formation of MDM2 and MDMX heterocomplex is structurally favored over the homocomplex (17), the MDM2 and MDMX proteins were found to exist in cells mainly as a heterocomplex (14). It was also shown that MDM2 alone is a relatively ineffective E3 ligase (12–14) but can more efficiently ubiquitinate p53 after heterodimerization with MDMX (15, 16). Thus, a second model was proposed in which MDM2 and MDMX work together in p53 regulation (18–21).
Results and Discussion
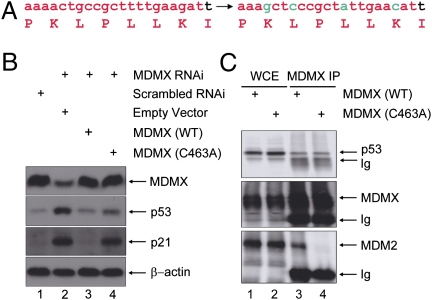
To directly test these two competing models, we sought to assess p53 activity under a condition whereby the interaction between MDM2 and MDMX was selectively disabled. We chose to use a MDMX RING mutant (C463A) that is defective in MDM2 binding (12–14). By using an MDMX mutant rather than an MDM2 mutant, the MDM2 RING domain, and thus the intrinsic E3 ligase activity of MDM2, was untouched. We initially tested this strategy in cell culture by replacing endogenous MDMX with MDMX(C463A). This was achieved by first generating cells that stably express MDMX(C463A), which was engineered for MDMXRNAi resistance (Fig. 1A), so that introduction of RNAi knocked down the expression of only endogenous MDMX. Cells stably expressing wild-type MDMX resistant to MDMXRNAi were included as a control. In agreement with our previous observation (16), MDMX depletion was associated with p53 activation (Fig. 1B, lane 2). Stably expressed wild-type MDMX efficiently blocked p53 activation (lane 3), as expected. Significantly, MDMX(C463A) was unable to rescue the loss of endogenous MDMX expression, as evident by the elevated p53 activity in these cells (lane 4). A co-immunoprecipitation (co-IP) experiment indicated that although the MDMX mutant failed to bind to MDM2 (Fig. 1C, Lower), its binding to p53 was similar to that of wild-type MDMX (Fig. 1C, Upper), indicating that the p53 activation in MDMX(C463A)-expressing cells was not caused by compromised p53-binding by the MDMX mutant. These in vitro results implicate that even with the binding of MDMX to p53, and the MDM2 protein unaltered, disassociation of the MDM heterocomplex is associated with p53 activation.
Fig. 1.
Substitution of wild-type MDMX with MDMX (C463A) results in p53 activation in vitro. (A) Oligonucleotide sequence targeting MDMX is shown in red. Base alterations were introduced that did not alter the coding amino acids (in green) to render the MDMX expressing plasmid resistant to the MDMXRNAi. (B) MCF-7 cells that were stably expressing an empty vector (lane 2), wild-type MDMX (lane 3), or MDMX (C463A) (lane 4) were infected with retroviral MDMXRNAi (lanes 2–4). The cells were harvested 60 h after infection and subjected to Western analysis using the indicated antibodies. Normal-growing MCF-7 cells were included as a control (lane 1). (C) MDMXRNAi MCF-7 cells reexpressing wild-type or MDMX(C463A) mutant MDMX as in B were subjected to IP with an anti-MDMX antibody. Immunoprecipitates were analyzed by Western blots with the indicated antibodies.
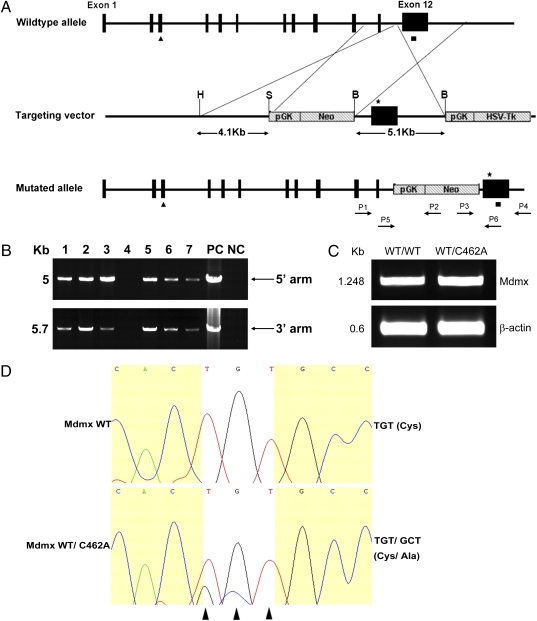
We next used a strategy as shown in Fig. 2A to generate a mouse Mdmx(C462A) mutant (equivalent to C463A in human MDMX). We targeted codon 462 of the Mdmx allele to replace TGT with GCT. ES cells were electroporated with the targeting vector, selected, and screened. Correct targeting was verified via PCR and sequencing. Three independent homologous recombinant ES clones were injected into C57BL/6 blastocysts to generate MdmxWT/C462A chimeras. From the germ line of MdmxWT/C462A chimera, progenies of heterozygous MdmxWT/C462A mice were produced and verified via PCR (Fig. 2B), which were viable and appeared phenotypically normal. RT-PCR analysis of the expression of target gene in the thymus showed a similar expression level of MDMX in both wild-type and heterozygous mice (Fig. 2C). Sequencing of RT-PCR product indicated that MdmxC462A allele was expressed, as expected (Fig. 2D). We went on to cross the heterozygous mice to obtain MdmxC462A/C462A homozygous mice. The offspring were genotyped by PCR of genomic DNA isolated from mouse tail biopsies. Among a cohort of 188 progenies generated from the intercrosses, 58 (30.9%) were wild-type for Mdmx, and 130 (69.1%) were heterozygous for MdmxWT/C462A. Although these data are consistent with the predicted 1:2 Mendelian ratios (Table 1) for wild-type over heterozygous embryos, no viable homozygous MdmxC462A/C462A mouse was obtained, suggesting that expression of MdmxC462A/C462A is associated with embryonic lethality in mice.
Fig. 2.
Creation of MdmxC462A knockin mice. (A) Schematic representation of the mouse Mdmx wild-type allele, targeting vector, and the predicted homologous recombination event. The targeting construct contains 4.1 kb of 5′ homology arm with exon 11, a loxP-flanked Neo for positive selection, 5.1 kb of 3′ homology arm with exon 12 harboring MdmxC462A mutation (star), and followed by an HSV-Tk negative selection marker. (B) Genotyping of MdmxWT/C462A heterozygous. The 5′ homology arm was identified by PCR using primers P1 and P2; the 3′ homologous arm was identified by PCR using primers P3 and P4. PC, ES gDNA as positive control; NC, wild-type mice gDNA as negative control. (C) Total RNAs were extracted from MdmxWT/WT and MdmxWT/C462A thymus, and RT-PCR analysis was performed using primers of Mdmx cDNA and β-actin. (D) PCR products from C were sequenced. White highlight shows that the codons that encode C462 of the wild-type mdmx are mutated in the MdmxWT/C462A heterozygous.
Table 1.
Analysis of mice from a MdmxWT/C462A × MdmxW/C462A cross
| Variable | Expected frequency, % (n) | Observed frequency, % (n) | P value |
| WT | 25 (47/188) | 30.9 (58/188) | 0.2061 |
| WT/C462A | 50 (94/188) | 69.1 (130/188) | 0.0002 |
| C462A/C462A | 25 (47/188) | 0/188 | <0.0001 |
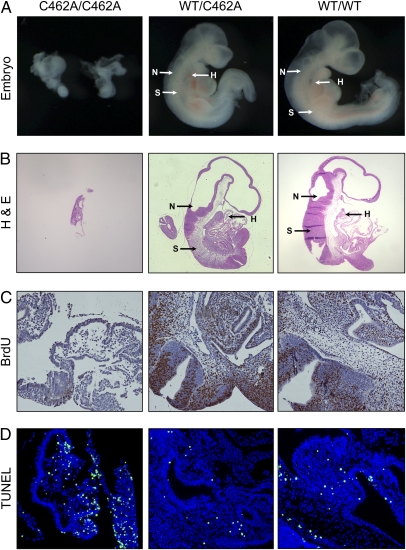
To determine the time of embryonic lethality in MdmxC462A/C462A mice, genomic DNA was isolated from embryos harvested at different stages of pregnant MdmxWT/C462A mice. Of 136 embryos isolated, we observed missing or abnormal embryos in 43 of 136 (31.6%) of the conceptuses (Table 2). Approximately 4.4% of the deciduae were empty. At day 9.5, normal embryos developed hearts, neural ectoderms, and somites, whereas the abnormal embryos developed none of these structures (Fig. 3 A and B). The abnormal embryos were smaller and detectable at day 10.5 and 11.5 but absorbed at approximately day 12.5 and 13.5. PCR-based genotyping of dissected embryos indicated that none of the Mdmx C462A/C462A embryos was normal. These results suggest that expression of the Mdmx(C462A) mutant results in embryonic lethality approximately at day 9.5.
Table 2.
Analysis of progeny from a MdmxWT/C462A × MdmxWT/C462A cross
| Stage | No. of litters | Total no. of embryos | Phenotypes | Genotypes | |||
| Normal | Abnormal | MdmxWT/WT | MdmxWT/C462A | MdmxC462A/C462A | |||
| E8.5 | 2 | 15 | 9 | 6 | 4 | 6 | 3 |
| E9.5 | 4 | 41 | 26 | 15 | 14 | 12 | 11 |
| E10.5 | 2 | 20 | 10 | 10 | 8 | 4 | 6 |
| E11.5 | 3 | 22 | 15 | 7 | 11 | 5 | 4 |
| E12.5 | 2 | 18 | 16 | 2 | 7 | 9 | 1 |
| E13.5 | 2 | 20 | 17 | 3 | 3 | 14 | 0 |
| Total | 15 | 136 | 93 | 43 | 47 | 50 | 25 |
Many of the abnormal embryos could not be genotyped because of their small size; the ratios are thus skewed.
Fig. 3.
Embryonic lethal phenotype of MdmxC462A/C462A embryos. (A) Dissected Mdmx wild-type and mutant (heterozygous and homozygous) embryos at E9.5. (B) Histology of embryos from Mdmx wild-type and mutant (heterozygous and homozygous) mice at E9.5 with hematoxylin and eosin stain. Arrows indicate where hearts (H), neural ectoderms (N), or somites (S) are located. Analysis of proliferation and apoptosis in embryos at E9.5 from MdmxWT/C462A × MdmxWT/C462A cross via BrdU incorporation (C) or TUNEL assays (D), respectively.
To determine the mechanism underlying the embryonic death, we measured cell proliferation and apoptosis in the embryos at day 9.5 using BrdU incorporation and TUNEL assay, respectively. The embryos from both wild-type and MdmxWT/C462A mice displayed strong BrdU staining. In contrast, few cells from MdmxC462A/C462A embryos were BrdU positive (Fig. 3C), indicating a cease in cell proliferation. In addition, we observed substantial TUNEL-positive cells in the MdmxC462A/C462A embryos, whereas only a very low level of TUNEL staining was seen in wild-type and MdmxWT/C462A embryos (Fig. 3D). These results imply that a combination of decreased cell proliferation and induction of apoptosis contributed to the embryonic death of MdmxC462A/C462A mice.
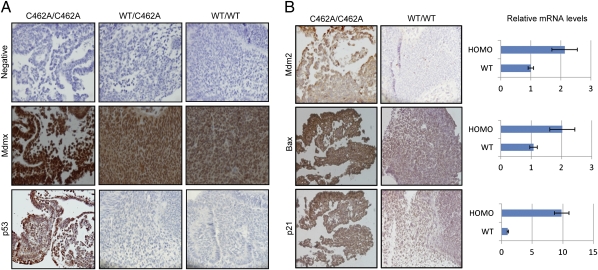
The embryonic lethal phenotype of MdmxC462A/C462A mice prompted us to examine whether the MDMX(C462A) mutant protein was properly expressed. We performed immunohistochemical staining of day-9.5 embryos for expression of MDMX and p53 (Fig. 4A). Consistent with previous reports that MDMX is expressed during embryonic development (8), our staining showed MDMX expression in both wild-type and MdmxWT/C462A embryos. Interestingly, the abundance of the MDMX mutant protein in MdmxC462A/C462A embryos was found to be higher than that in both wild-type and MdmxWT/C462A embryos. Conversely, we did not detect any increase of Mdmx mRNA level in MdmxC462A/C462A embryos. These results suggest that the steady-state level of MDMX is tightly regulated by an MDM2-dependent mechanism, even during the embryonic developmental stage. However, despite the elevated level of the MDMX mutant protein, there was a marked increase of p53 staining in MdmxC462A/C462A embryos, whereas p53 was barely detectable in wild-type and MdmxWT/C462A embryos. To determine the activity of p53, we examined the expression of its target genes. As shown in Fig. 4B, the MDM2, p21, and Bax protein abundance were significantly higher in MdmxC462A/C462A embryos than in wild-type embryos. Quantitative RT-PCR analysis revealed increased expression of these p53 genes at the transcription level (Fig. 4B, Right), indicating p53 activation. The increased levels of p21 and Bax indicate growth inhibition and apoptosis, consistent with decreased BrdU incorporation and increased TUNEL staining observed in the MdmxC462A/C462A embryos. The data collectively indicate that the expression of MDMX(C462A) mutant protein in mice, even to a level slightly higher than that of wild-type MDMX, was associated with increased p53 level as well as activity.
Fig. 4.
Expression of Mdmx, Mdm2, or p53 in day-9.5 embryos. (A) Sections from indicated genotype embryos were stained with antibodies against MDMX and p53, respectively. (B) Sections from MdmxC462A/C462A homozygous or wild-type embryos were stained with anti-MDM2, Bax, or p21 antibodies. The homozygous (HOMO) or wild-type (WT) embryos were also examined for mRNA of MDM2, Bax, and p21 using quantitative RT-PCR (Right).
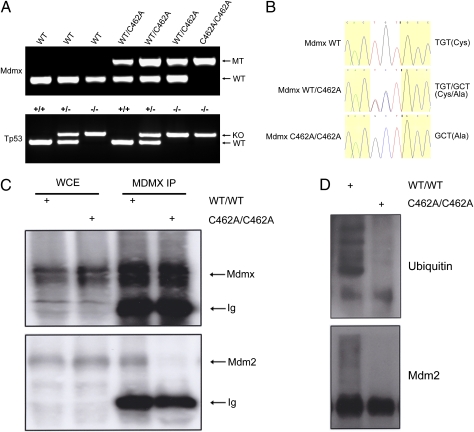
It has been well demonstrated that the lethality of Mdmx knockout mice can be completely rescued by concomitant p53 depletion. Having observed the increase of p53 activity in MdmxC462A/C462A embryos, we asked whether the lethality of MdmxC462A/C462A mice was caused by uncontrolled p53 activity. For this, we crossed the MdmxWT/C462A mice into a p53 null background (22, 23) to create MdmxWT/C462A/p53+/− mice, from which MdmxC462A/C462A/p53−/− mice were generated by further intercrossing (Fig. 5A). Significantly, MdmxC462A/C462A/p53−/− mice were born at the expected Mendelian ratio (Table 3) with the phenotypes almost identical to other littermates. The offspring were genotyped by PCR of genomic DNA (Fig. 5A) and confirmed by sequencing (Fig. 5B). This complete rescue clearly indicates a p53-mediated embryonic death. To ensure that the MDMX(C462A) mutation disrupted the interaction between MDMX and MDM2 in vivo, we carried out co-IP experiments with mouse embryonic fibroblasts (MEFs) isolated from MdmxC462A/C462A/p53−/− or MdmxWT/WT/p53+/+ mice. Indeed, the mutation resulted in disassociation of the binding between MDM2 and MDMX (Fig. 5C). Interestingly, the association with MDMX also affected the ubiquitin ligase activity of MDM2, as reflected by a much-reduced ubiquitin signal when comparing the MDM2 protein expressed in MdmxC462A/C462A/p53−/− mice with that in MdmxWT/WT/p53+/+ mice (Fig. 5D). Although MDM2 ubiquitination is not necessarily equivalent to p53 ubiquitination, the data are in agreement with published findings that MDMX enhances the E3 ligase activity of MDM2 (14, 15). Together, these results indicate that the association of MDM2 and MDMX is of great importance in p53 control during embryonic development, supporting the model in which MDM2 and MDMX work as an integral complex in p53 regulation.
Fig. 5.
Embryonic lethal phenotype of MdmxC462A/C462A embryos was completely rescued by crossing with p53-null mice. (A) PCR genotyping of offspring from MdmxWT/C462A/p53+/− × MdmxWT/C462A/p53+/− crosses using primers P3, P5, and P6 specific for mutated and wild-type Mdmx alleles. (B) Sequences of PCR products from C. White highlight shows that TGT, the codon of the wild-type mdmx C462, is replaced with GCT in the MdmxWT/C462A heterozygous and MdmxC462A/C462A homozygous. (C) MEFs isolated from either MdmxC462A/C462A/p53−/− or MdmxWT/WT/p53+/+ mice were subjected to anti-MDMX IP. Immunoprecipitates were analyzed by Western blot with anti-MDMX and MDM2. (D) Anti-MDM2 IP was performed with the MEFs as in C and analyzed by Western blot with either anti-MDM2 or ubiquitin.
Table 3.
Genotypes of mice from a MdmxWT/C462A/p53+/− × MdmxWT/C462A/p53+/− cross
| Variable | MdmxWT/WT | MdmxWT/C462A | MdmxC462A/C462A | Total |
| p53+/+ | 3 | 6 | 0 | 9 |
| p53+/− | 5 | 13 | 0 | 18 |
| p53−/− | 2 | 9 | 5 | 16 |
| Total | 10 | 28 | 5 | 43 |
The current mainstream model of p53 regulation proposes that MDM2 inactivates p53 primarily as a ubiquitin E3 ligase and that MDMX inhibits p53 mainly by its physical binding to and blocking of the transactivation domain of p53. Even with these two activities preserved, the targeted disassociation of the MDM heterocomplex resulted in lethal p53 activity in vivo during embryonic development. Our data imply that a physical binding of MDMX to p53 is not sufficient for p53 inhibition. Consistent with this notion is the result from a recent study showing that targeted inactivation of MDM2 E3 ligase activity was associated with lethal p53 activity during development (24). However, the MDM2(C464A) mutant is, in addition to being inactive as an E3 ligase, also defective in MDMX binding. The available information implies that the intrinsic E3 ligase activity of MDM2 is essential but that its full potential in p53 control depends on its interaction with MDMX. This view is consistent with the findings that MDM2 alone is a relatively ineffective E3 ligase and that MDM2 is a more efficient E3 ligase after association with MDMX (Fig. 5D and refs. 12–16). The RING domain-mediated heterocomplex formation seems to be a common mechanism of regulation in RING domain-containing ubiquitin E3 ligase. A prototype of this mode of regulation is the activity of the RING E3 ligase Brca1, which depends on its association with another RING domain protein, BARD, to form a heterocomplex (25). Our genetic data imply that the MDM heterocomplex is the physiological E3 ligase for p53, at least at the developmental stage. Conditional mouse models will be necessary to investigate the importance of the MDM heterocomplex in p53 control at the adult stage and during various stress conditions. Our study also provides a strong rationale to target the MDM heterocomplex for p53 activation, which may have important therapeutic implications.
Materials and Methods
Targeting Construct.
A 9.2-kb fragment containing intron 11 through 3′ UTR of the Mdmx gene was isolated from 129 BAC clone bMQ-462H8. The targeting vector for generating the Mdmx mutation allele was designed to insert a pGK-neo gene upstream of exon 12. The copy of exon 12 in the 3′ arm of homology was mutated by changing the codon of Cys-462 to Alanine. The final targeting vector contains the Mario Cappeci-1 promoter (MC) herpes simplex virus thymidine kinase (MC-hsvTK) expression cassette for negative selection. The point mutation was confirmed by sequencing.
Generating and Genotyping the MdmxC462A Knockin Mice.
The targeting vector was linearized with NotI and electroporated into 129/Sv ES cells before being selected for neomycin resistance. Genomic DNA was isolated from individual ES colonies. Homologous recombination was confirmed by PCR screening with primers P1 and P2 for 5′ homologous recombination arm (5 kb), P3 and P4 for 3′ homologous recombination arm (5.7 kb). Three targeted clones were injected into C57BL/6J blastocysts, which were then implanted into pseudopregnant females. Germline transmission, through breeding chimeras with C57BL/6 mice, was confirmed by PCR using the same primers as ES colonies identification. The offspring were PCR genotyped using primers P5, P3, and P6 to distinguish between the wild-type and mutant Mdmx alleles. P53 knockout mice were purchased from Jackson Laboratories. Trp53 primers p53x7 and p53x6.5 recognize wild-type Trp53. Primers p53x7 and p53-Neo18.5 amplify the Trp53 mutant allele (Table 4).
Table 4.
Sequence of primers
| Primer | Sequence |
| P1 | 5′ GTATGTCCCAGCATTCACACTCTG 3′ |
| P4 | 5′ TTCCTTAGTGTTCAAACAAACCAAC 3′ |
| P3 | 5′ GCCTTCTTGACGAGTTCTTCTG 3′ |
| P2 | 5′ AGTCATAGCCGAATAGCCTCTC 3′ |
| P5 | 5′ CTTGTAAAGGTTTTTTGCTTTGT 3′ |
| P6 | 5′ GCCTAACAACAGGAGCTGAAA 3′ |
| P53-Neo18.5 | 5′ TCCTCGTGCTTTACGGTATC 3′ |
| P53x7 | 5′ TATACTCAGAGCCGGCCT 3′ |
| P53x6.5 | 5′ ACAGCGTGGTGGTACCTTAT 3′ |
| cDNA-up | 5′ TAACAAGAAGACGGTGGAGG 3′ |
| cDNA-down | 5′ ATGTACACCTGTGTTACCTGA 3′ |
| β-actin-F | 5′ AACGAGCGGTTCCGATGCCCTGAG 3′ |
| β-actin-R | 5′ TGTCGCCTTCACCGTTCCAGTT 3′ |
Semiquantitative RT-PCR.
Total RNA of thymus from MdmxWT and MdmxWT/C462A mice were extracted using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. cDNA was generated by reverse transcription using AMV Reverse Transcriptase (TaKaRa). RT-PCR was performed with Mdmx cDNA and β-actin primers indicated in Table 4. PCR products were analyzed by 1% agarose gels. The amplification fragments were verified by DNA sequencing.
Quantitative RT-PCR.
Total RNA was extracted from embryos at embryonic day 9.5 (E9.5) with genotype of MdmxWT, MdmxWT/C462A, and MdmxC462A/C462A. Quantitative PCR was carried out with the SYBR Green PCR kit according to the manufacturer's instructions (TaKaRa). Amplifications were performed in ABI PRISM 7500 Sequence Detection System (Applied Biosystems). Relative transcript quantities were calculated using the ΔΔCt method with β-actin as the endogenous reference gene. The value of each genotype was identified by three samples, and each sample was repeated three times independently.
Histological and Immunohistochemical Analysis.
Before embedding in paraffin, embryo specimens were fixed in 4% paraformaldehyde in PBS and dehydrated. For histological analysis, 6-μm sections were cut and stained with hematoxylin and eosin according to the standard procedure. For immunohistochemical analysis, paraffin-embedded sections were deparaffinized with xylene, treated with gradually decreasing concentrations of ethanol, and then processed in 10 mM citrate buffer (pH 6.0) and heated to 92–96 °C for 30 min for antigen retrieval. Tissue sections were treated with 3% hydrogen peroxidase in PBS for 10 min to block endogenous peroxidase activity. Sections were incubated with blocking serum for 1 h and then incubated with primary antibody overnight at 4 °C. The staining procedure followed the manufacturer's instructions for the ABC staining system (Santa Cruz Biotechnology). Rat anti-BrdU, rabbit anti-p53, and anti-MDM2 were from Santa Cruz Biotechnology. Mouse anti-MDMX was purchased from Sigma. Rabbit anti-p21 and anti-bax were purchased from AbCam.
Immunoprecipitation and Western analysis were performed as previously described (14, 16).
BrdU Staining.
Cell proliferation in embryos was measured by the incorporation of BrdU, which was administered i.p. to mice at a dosage of 100 μg/g body weight 2 h before killing. BrdU incorporation was detected on sections by immunohistochemistry.
TUNEL.
To detect apoptotic nuclei, paraffin sections were analyzed by the DeadEnd Fluorometric TUNEL System (Promega) according to the manufacturer's instructions.
Statistical Analysis.
All analyses were conducted using SAS 9.12 (SAS Institute).
Acknowledgments
We thank Professor Jingsheng Feng for his expert help with embryo histology analysis; Ying Kuang and Long Wang for embryonic stem cell clone injection; and Xiaojin Wang for statistical analysis. This work was partially supported by National Natural Science Foundation of China Grants 30871363 and 81071666; New Century Excellent Talents in University and the Scientific Research Foundation for the Returned Overseas Chinese Scholars of State Education Ministry Grant NCET-08-0349; Ministry of Science and Technology of China Grant 2006BAI23B02; E-Institutes of Shanghai Municipal Education Commission Grant E03003; and National Institutes of Health Grants NCI R01 CA85679 and RO1 CA125144 (to Z.-M.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.H.V. is a guest editor invited by the Editorial Board.
References
- 1.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 3.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 4.Shvarts A, et al. MDMX: A novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996;15:5349–5357. [PMC free article] [PubMed] [Google Scholar]
- 5.Shvarts A, et al. Isolation and identification of the human homolog of a new p53-binding protein, Mdmx. Genomics. 1997;43:34–42. doi: 10.1006/geno.1997.4775. [DOI] [PubMed] [Google Scholar]
- 6.Sharp DA, Kratowicz SA, Sank MJ, George DL. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J Biol Chem. 1999;274:38189–38196. doi: 10.1074/jbc.274.53.38189. [DOI] [PubMed] [Google Scholar]
- 7.Tanimura S, et al. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 1999;447:5–9. doi: 10.1016/s0014-5793(99)00254-9. [DOI] [PubMed] [Google Scholar]
- 8.Parant J, et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet. 2001;29:92–95. doi: 10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- 9.Migliorini D, et al. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol. 2002;22:5527–5538. doi: 10.1128/MCB.22.15.5527-5538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finch RA, et al. mdmx is a negative regulator of p53 activity in vivo. Cancer Res. 2002;62:3221–3225. [PubMed] [Google Scholar]
- 11.Marine JC, Dyer MA, Jochemsen AG. MDMX: From bench to bedside. J Cell Sci. 2007;120:371–378. doi: 10.1242/jcs.03362. [DOI] [PubMed] [Google Scholar]
- 12.Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J. 2007;26:102–112. doi: 10.1038/sj.emboj.7601469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poyurovsky MV, et al. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 2007;26:90–101. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawai H, Lopez-Pajares V, Kim MM, Wiederschain D, Yuan ZM. RING domain-mediated interaction is a requirement for MDM2’s E3 ligase activity. Cancer Res. 2007;67:6026–6030. doi: 10.1158/0008-5472.CAN-07-1313. [DOI] [PubMed] [Google Scholar]
- 15.Linares LK, Hengstermann A, Ciechanover A, Müller S, Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci USA. 2003;100:12009–12014. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu J, et al. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J Biol Chem. 2002;277:19251–19254. doi: 10.1074/jbc.C200150200. [DOI] [PubMed] [Google Scholar]
- 17.Linke K, et al. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008;15:841–848. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- 18.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marine JC, et al. Keeping p53 in check: Essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ. 2006;13:927–934. doi: 10.1038/sj.cdd.4401912. [DOI] [PubMed] [Google Scholar]
- 20.Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 21.Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 2010;20:299–309. doi: 10.1016/j.tcb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan L, Liu JG, Hoja MR, Lightfoot DA, Höög C. The checkpoint monitoring chromosomal pairing in male meiotic cells is p53-independent. Cell Death Differ. 2001;8:316–317. doi: 10.1038/sj.cdd.4400828. [DOI] [PubMed] [Google Scholar]
- 23.Jacks T, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 24.Itahana K, et al. Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell. 2007;12:355–366. doi: 10.1016/j.ccr.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Hashizume R, et al. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem. 2001;276:14537–14540. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]