Abstract
Identification of the underlying molecular mechanisms for a derived phenotype by adaptive evolution is difficult. Here, we performed a systems-level inquiry into the metabolic changes occurring in the yeast Saccharomyces cerevisiae as a result of its adaptive evolution to increase its specific growth rate on galactose and related these changes to the acquired phenotypic properties. Three evolved mutants (62A, 62B, and 62C) with higher specific growth rates and faster specific galactose uptake were isolated. The evolved mutants were compared with a reference strain and two engineered strains, SO16 and PGM2, which also showed higher galactose uptake rate in previous studies. The profile of intermediates in galactose metabolism was similar in evolved and engineered mutants, whereas reserve carbohydrates metabolism was specifically elevated in the evolved mutants and one evolved strain showed changes in ergosterol biosynthesis. Mutations were identified in proteins involved in the global carbon sensing Ras/PKA pathway, which is known to regulate the reserve carbohydrates metabolism. We evaluated one of the identified mutations, RAS2Tyr112, and this mutation resulted in an increased specific growth rate on galactose. These results show that adaptive evolution results in the utilization of unpredicted routes to accommodate increased galactose flux in contrast to rationally engineered strains. Our study demonstrates that adaptive evolution represents a valuable alternative to rational design in bioengineering of improved strains and, that through systems biology, it is possible to identify mutations in evolved strain that can serve as unforeseen metabolic engineering targets for improving microbial strains for production of biofuels and chemicals.
In the field of industrial biotechnology, there is a need to develop efficient cell factories for the production of fuels and chemicals. Even though the concept of metabolic engineering (1) is frequently used in both academia and industry for the development of unique cell factories, evolutionary engineering methods are still widely performed (2). The power of adaptive evolution, sometimes in combination with metabolic engineering, is well illustrated in several recent examples (3, 4). Despite its advantages, conventional random mutagenesis and screening are hampered by the difficulties associated with finding the underlying molecular mechanisms for a derived phenotype and, hence, the combination of adaptive evolution with more rational approaches like metabolic engineering is attractive. Tools from systems biology and the ability to perform deep sequencing of several strains have offered new opportunities for establishing links between genotype and phenotype and, hereby, allow for combinations of random and rational approaches to strain improvement (5, 6).
Understanding the evolutionary strategies of a cell to metabolize nonfavored carbon sources is an integral part of strain development in cost efficient bioprocesses. Galactose is an abundant sugar in nonfood crops (7), and it is therefore interesting to generate strains that can efficiently use galactose as a carbon source. However, the yeast Saccharomyces cerevisiae, which is a frequently used cell factory in industrial biotechnology, grows at half the rate on galactose compared with glucose, despite the structural similarity between galactose and glucose (8). There is extensive knowledge on the regulation of the Leloir pathway in S. cerevisiae, the catabolic route for galactose metabolism, because this regulon has served as a paradigm for understanding eukaryotic transcription principles (9). Consequently, much information on the regulation and structure of the components involved in galactose metabolism has accumulated. There is also a vast amount of high throughput data available that elucidates galactose metabolism (10). Exploiting this abundant information, many elegant metabolic engineering approaches have been implemented to increase the galactose uptake rate by modification of transporters, regulators, metabolic genes, or a combination of them (11, 12). Based on analysis of these and other strains, it has been found that accumulation of metabolic intermediates in the galactose metabolism, such as galactose-1-phosphate and glucose-1-phosphate, may inhibit the flux through the Leloir pathway and, hence, lead to a lower galactose uptake rate (10). Therefore, successful strain development was performed by balanced expression of structural genes through modification of the regulatory system (12) or through overexpression of the final enzyme of galactose metabolism, PGM2, which converts glucose-1-phosphate to glucose-6-phospate (11). Both these engineered cells had lower concentrations of the intermediates and a higher galactose uptake rate (8). Despite these successes on improving galactose uptake in yeast through metabolic engineering, there has so far not been any description of using an evolutionary approach for improving galactose utilization.
Adaptive evolution of bacteria has enabled increasing the specific growth rate due to mutations that were not predicted by rational engineering (5). We therefore decided to apply the concept of adaptive evolution for the improvement of galactose utilization by yeast, with the objective to evaluate if unique strategies for improving galactose uptake could be identified. Furthermore, through detailed characterization of evolved mutants, we expected to expand our understanding of the galactose metabolism in yeast. We therefore characterized adaptively evolved mutants at the systems level for gaining better understanding of the molecular mechanisms that are responsible for acquired phenotypes. We evolved a laboratory strain of yeast for ≈400 generations in three different serial transfer lines and analyzed the changes in transcriptome, metabolome, and genome sequence that contribute to the phenotypic changes. Here, we present results of integrated analysis of the data from the three evolved mutants compared with wild-type yeast and two engineered strains that were developed by the rational approach in previous studies (11, 12). Based on our analysis of the evolved strains, unique strategies for improving galactose uptake were identified, and one of these strategies was proven to result in an improved galactose uptake. Furthermore, our analysis highlights that it is only by an integrated systems biology approach that it is possible to map out the mechanisms underlying evolved phenotypes.
Results
Physiological Changes in Evolved Mutants.
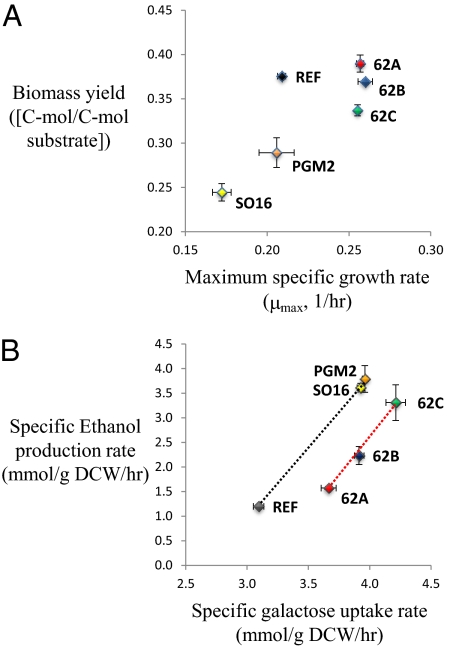
Three adaptively evolved strains that had 24% faster specific growth rates on galactose were obtained through 62-d serial transfers of cultures to fresh medium with galactose as the sole carbon source. The strains were isolated from the last culture and were designated 62A, 62B, and 62C. The gross phenotype of the three evolved strains was compared with those of the reference strain (CEN.PK113-7D) and two engineered strains characterized in earlier studies: SO16 with deletion of MIG1, GAL80, and GAL6 (12) and PGM2 with overexpression of PGM2 (11). All mutants showed an improved specific galactose uptake rate compared with the reference strain (Fig. 1 and Table S1). The difference between the evolved mutants and the engineered strains was found in the biomass yield, the specific ethanol production rate, and the specific growth rate (Fig. 1). The engineered strains exhibited the highest ethanol yield at the expense of the biomass yield, whereas the evolved mutants showed a similar biomass yield with the reference strain. The evolved mutants from the three lineages commonly exhibited a 24% increase in the maximum specific growth rate compared with the reference strain, whereas they differed in their specific galactose uptake rates and their specific ethanol production rates. The specific galactose uptake rates varied from an 18% increase in 62A strain to a 36% increase in 62C. The specific ethanol production rate in the evolved mutants was increased from 31 to 170%. In an earlier study, we found that several engineered strains were lying on a linear regression curve when their specific ethanol production rate was plotted against the specific galactose uptake rate (12). This experience led us to plot data from all of the strains in a similar kind of plot, but we found there to be a grouping pattern that clearly separated all of the strains into two groups (Fig. 1B). The reference and the two engineered strains were on the same regression curve (R2 = 0.99), whereas all of the evolved mutants were on a different regression curve (R2 = 0.98), indicating a common phenomenon underlying the increased galactose uptake rates. If data from all of the strains are included in the same linear regression, a rather poor correlation coefficient is obtained (R2 = 0.64).
Fig. 1.
Phenotypic changes of evolved mutant strains 62A, 62B, and 62C compare with the reference strain CEN.PK113-7D and the two engineered strains SO16 and PGM2. (A) Correlation between a maximum specific a growth rate and biomass yield. (B) Correlation between a specific galactose uptake rate and a specific ethanol production rate. The regression curves of the two lines (from right to left) have a slope of 2.953 and 3.187 and intercept of −7.9472 (R2 = 0.991) and −10.157 (R2 = 0.98), respectively.
Changes in the Transcriptome and the Metabolome.
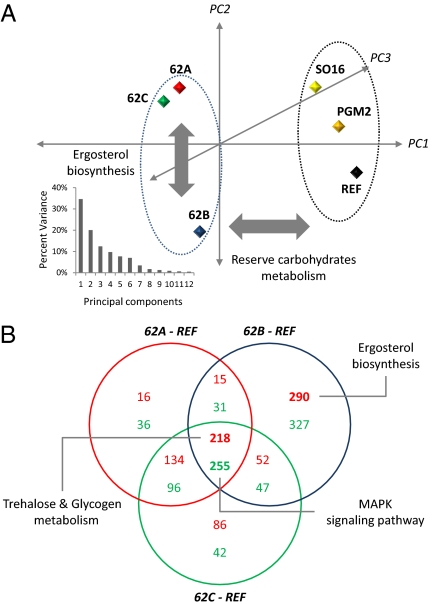
The evolved mutants showed clear separation from the reference and the two engineered strains in their transcriptome profile, using principal component analysis (PCA) (Fig. 2A). The first principal component (PC) (35%) separated the evolved mutants from the other strains. Among the differentially expressed genes between these two groups, the KEGG pathways involving trehalose and glycogen metabolism were overrepresented (P < 1e−4).
Fig. 2.
Results from transcriptome analysis. (A) Principle component analysis of all of the strains is used to highlight the differences among strains. The box shows how much variance is observed by the different principle components. The results are projected by the first three PCs, which covered 85% of the variance. (B) Differentially expressed genes (P < 0.01) are categorized in a Venn diagram. The functions of genes in each part are analyzed by hypergeometric model using KEGG, Reactome, and GO term database (P < 0.001). Red color letter means up-regulation; green one means down-regulation.
Transcriptional differences (P < 0.01) between the evolved mutants and the reference strain were categorized into those that commonly changed in the three lineages and those that were mutant-specific (Fig. 2B). Genes involved in trehalose and glycogen metabolism were commonly up-regulated in all of the evolved mutants, whereas genes encoding proteins involved in the MAPK signaling pathway (also based on KEGG pathway) were down-regulated, relative to the reference strain (cumulative hypergeometric probability, P < 1e−4). Interestingly, few genes appeared to be differentially expressed between 62A and 62C, even though they showed very different physiology. However, several genes (≈600 genes) were differentially expressed in 62B, and the genes involved in ergosterol biosynthesis were overrepresented among these genes.
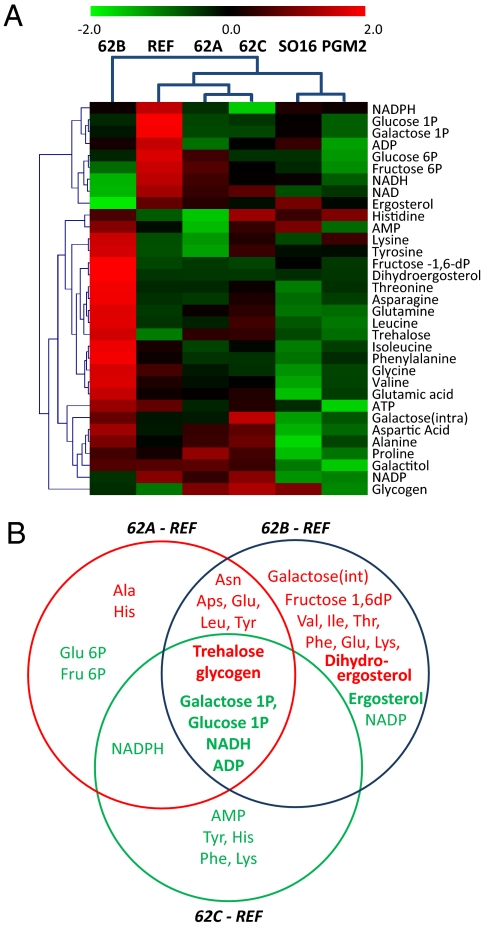
We also measured the concentration of intracellular metabolites of the Leloir pathway, redox cofactors, and amino acids. Prompted by the transcriptional changes in genes involved in glycogen, trehalose, and ergosterol metabolism (Table S2), we also measured the levels of these compounds in the cell. Hierarchical clustering of all of the metabolites separated 62B from the other two evolved mutants, similar to the transcriptome data (Fig. 3A). The evolved and engineered strains commonly showed lower intracellular concentrations of sugar phosphates than the reference strain. In general, the level of free amino acids was higher only in 62B, relative to the other strains. The two engineered ones exhibited the lowest levels of amino acids.
Fig. 3.
Analysis of the metabolite data from the evolved strains compared with the reference and the two recombinant strains. (A) Hierarchical clustering of all strains is computed after standardizing of metabolites concentration to z score. (B) Differentially produced metabolites are categorized in a Venn diagram by calculation of significance (P < 0.05) based on a Student t test. Red color letter means higher levels; green one means lower levels.
Significantly changed metabolites were classified as those whose intracellular concentration changed commonly in all of the evolved mutants or in one specific strain compared with the reference strain (P < 0.05) (Fig. 3B). Trehalose and glycogen were at a higher concentration only in all of the evolved mutants, whereas the concentrations of galactose-1-phosphate and glucose-1-phosphate, NADH, and ADP were lower in both the engineered and evolved strains. Interestingly, despite having increased transcriptional activity of the ergosterol pathway, the intracellular concentration of ergosterol was much lower in 62B. However, 62B was found to produce a higher amount of dihydroergosterol compared with any of the other strains, indicating a redirection of carbon flux in the ergosterol biosynthesis pathway to this metabolite.
Changes in Genotype.
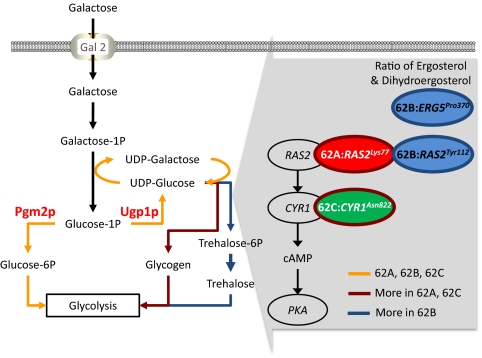
The genomes of the evolved mutants were sequenced and compared with that of the parent strain, CEN.PK113-7D (13) (Table S3). Only a small fraction of the genome of the reference strain (0.7–1.4%) was not covered by the reads for the three mutant strains, allowing for high quality mapping of the genome sequence of the three evolved mutants. Based on the raw sequence data, we identified in the order of 6,000 putative mutations in all of the three evolved strains, but after filtering the number of mutations, was reduced to 44, 334, and 40 in the 62A, 62B, and 62C, respectively (Table S4, Fig. S1, and Dataset S1). We aimed for 20× coverage in the sequencing, but we received a substantially higher coverage for strain 62B (≈55×) (Table S3). We applied a filtering process, and this resulted in maintaining a larger number of mutations in the 62B strain, which is likely due to the high sequencing coverage for this strain. Of the identified mutations, 21, 104, and 29 where single nucleotide polymorphism (SNPs), respectively, whereas the remainder were insertions and deletions (INDELs). Only about one-third of the SNPs were in coding regions, and only very few of the INDELs were in coding regions (none in 62A, 11 of 230 in 62B, and 3 of 11 in 62C). The number of mutations is much larger than observed in an earlier study on evolution of E. coli (14), but we believe that this difference could be explained by the rather poor growth of the reference strain on galactose and possible also by the larger genome size of S. cerevisiae. Surprisingly, no mutations were detected in galactose regulatory and structural genes, even in PGM2, which was considered the most beneficial target for increasing the galactose uptake rate in previous studies based on metabolic engineering (Dataset S2) (11). Furthermore, no mutations in the trehalose and glycogen pathway genes were found, which showed significant changes in the transcriptome and metabolome level in all of the evolved mutants. Only genes encoding proteins of the Ras/PKA signaling pathway were found to carry mutations in all three evolved mutants (Table 1). 62A and 62B had a mutation in RAS2 at different positions, whereas 62C had one mutation in CYR1, encoding adenylate cyclase. Both genes are related to the cAMP-dependent stress response signaling pathway. A mutation in ERG5 in 62B was identified, which is a gene encoding one of the enzymes of the ergosterol pathway. To evaluate whether these mutations are causing the increased specific growth rate on galactose, we reconstructed the mutation in position 112 of Ras2p resulting in an amino acid substitution of aspartate with tyrosine. The resulting strain was evaluated for its growth on galactose in shake flasks, and it was found to have a 10% higher specific growth rate than the reference strain (P = 0.05) (Table S5).
Table 1.
Genetic changes
| Strains | Mutations | Functions |
| Commonly mutated pathway | ||
| 62A | RAS2 [Gln77→Lys] | Ras/PKA signaling pathway |
| 62B | RAS2 [Asp112→Tyr] | |
| 62C | CYR1 [Asp822→Asn] | |
| Uniquely mutated genes | ||
| 62B | ERG5 [Arg370→Pro] | Ergosterol metabolism |
Discussion
In this study, we characterized adaptive evolution of yeast to acquire faster growth and galactose utilization. First, we analyzed the phenotypic differences by gross kinetic parameters such as maximum specific growth rate, specific galactose uptake rate, and yield coefficients. These values indicated that the three evolved mutants had obtained improved growth on galactose by obtaining different mutations and also by different means than the strategy applied in metabolic engineering studies for improving the galactose uptake. To understand the underlying metabolic changes that conferred the phenotypic differences of the evolved mutants, we measured the variation of the transcriptome and the metabolome of the evolved strains and compared these with the reference strain and the earlier constructed engineered strains. Changed transcription of genes involved in trehalose and glycogen metabolism were detected in all three evolved strains, and genes involved the ergosterol pathway were noticed to have uniquely changed expression in the 62B strain. From comparative genome analysis of the evolved mutants, we could find no mutations in galactose and reserve carbohydrates metabolisms, whereas proteins of the Ras/PKA signaling pathway was discovered to contain mutations in all of the evolved mutants. Furthermore, 62B had a unique missense mutation in ERG5.
Adaptive Evolution Achieves Improved Galactose Availability with Different Physiology.
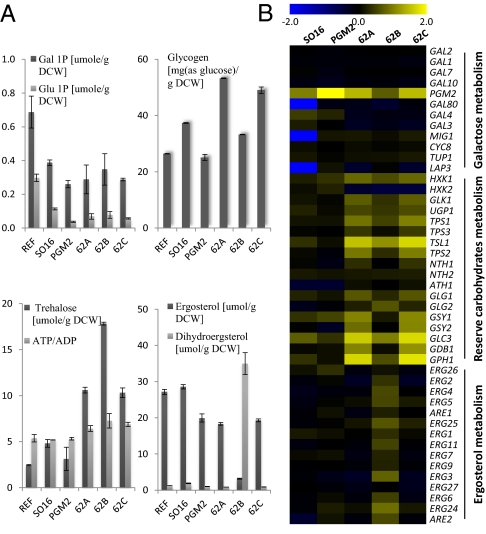
All mutants, including the evolved mutants and the two engineered strains, showed lower CO2 yield than the reference strain, even though they had a higher ethanol yield (Table S1). The lower CO2 yield along with the higher ethanol yield indicates a reduced TCA cycle activity. The significant decrease in the NADH concentration partially supports this phenomenon because NADH is the main cofactor of the TCA cycle (Fig. 3B). All mutants showed lower levels of sugar phosphates, such as galactose-1-phosphate and glucose-1-phosphate, than the reference strain (Fig. 4A). Decrease in their concentration is likely due to an increased flux downstream of these metabolites. In the case of the two engineered strains, this reduction is realized solely by amplification of PGM2, which is a final enzyme in galactose metabolism converting glucose-1-phosphate to glucose-6-phosphate (11). For the evolved mutants there is, however, besides increased expression of PGM2, likely to be an increased flux through trehalose and glycogen.
Fig. 4.
Changes in the galactose, reserve carbohydrates, and ergosterol metabolism in the evolved mutants are illustrated by changes in the concentration of metabolites and fold changes of transcriptome compared with the other strains. (A) The concentrations of sugar phosphates, storage carbohydrates, and sterols and the ratio of ATP to ADP. (B) Fold changes of all genes involved in galactose, reserve carbohydrates, and ergosterol metabolism are compared with the reference strain.
Another common feature of the evolved mutants and the engineered strains is that in a plot of the specific ethanol production rate versus the specific galactose uptake rate, the three evolved mutants lie on one regression line with a slope of ≈3 and the engineered strains lie on another regression line together with the reference strain also with a slope of ≈3 (Fig. 1B). The results for the engineered strains are consistent with our earlier findings that engineering of the GAL-regulon results in a slope >2 in this kind of plot (12). If an increase in galactose uptake resulted solely in ethanol production, this slope should be 2, but because both sets of strains are ≈3, it shows that an increase in the galactose uptake results in a redirection of flux from respiratory metabolism, i.e., TCA cycle, to fermentative metabolism. Thus, when the galactose uptake is increasing, then carbon catabolite repression of the respiratory systems sets in, but the transcriptome analysis did not provide any indication of increased carbon catabolite repression and/or decreased respiratory metabolism in the metabolically engineered strains compared with the reference strain and in 62C and 62B compared with 62A. Despite the similarity in slope for the two sets of strains, it is interesting that the evolved mutants seems to find a different metabolic operation that allows a higher galactose uptake without increasing the ethanol production, probably due to the redirection of flux through the storage carbohydrates glycogen and trehalose.
Up-Regulation of PGM2 and Activation of Reserve Carbohydrates Metabolism Are Detected as Common Changes in All Evolved Mutants.
The significant changes in the levels of metabolic intermediates of the Leloir pathway did not arise because of transcriptional differences between the strains, except for PGM2 (Fig. 4B). The concentration of galactose-1-phosphate and glucose-1-phosphate were much lower in all of the evolved mutants compared with the reference strain (Fig. 4A). Both the changes in the levels of sugar phosphates and expression of PGM2 were common with the two engineered strains. These results reveal that adaptive evolution used partially the same strategy as designed using rational engineering. However, for the evolved strains, trehalose and glycogen metabolism showed significant change in the transcriptome and metabolome data in all three evolved mutants compared with the other strains (Figs. 2 and 3). The concentration of glycogen and trehalose were clearly increased (Fig. 4A), and the transcription level of structural genes in the metabolism of these storage carbohydrates was remarkably increased (Fig. 4B). The induction of this pathway for increasing galactose utilization could be explained simply by the fact that glucose-1-phosphate is used as a precursor for the production of these reserve carbohydrates. Ugp1p converts glucose-1-phosphate to UDP-glucose, which is a branch point metabolite in the trehalose and glycogen metabolism.
Trehalose and glycogen are reserve carbohydrates for maintaining the energy charge of yeast cells (15). Their metabolism is very closely linked to the concentration of glucose, which is the most favored carbon source to yeast (16). Upon sensing glucose limitation, yeast mobilizes energy reserve to balance the rate of glycolytic activity and retain the energy charge. Trehalose production is induced especially under stress condition. However, galactose seems not to trigger activation of glycogen and trehalose metabolism. Wild-type yeast showed no change of the concentration of trehalose and the glycogen during growth on galactose (17). The evolved mutants probably have higher energy charge and, therefore, are capable of having an increased specific growth rate. This interpretation is supported by higher ratio of ATP/ADP in these cells (Fig. 4A; ref. 18).
Adaptive Evolution of Yeast on Galactose Generates No Mutations in Galactose and Reserve Carbohydrates Metabolism.
In general, when microorganisms face a new carbon source, they evolve the structural or regulatory genes that are related to the metabolism of it. For example, all adaptively evolved mutants of E. coli on glycerol had mutations commonly in glpK, glycerol kinase (5). However, in our case, no mutations were identified in the galactose metabolism, including promoter regions of the GAL genes (there were also no considerable changes in transcription levels). Furthermore, no mutations were detected in genes involved in the reserve carbohydrates metabolism, but there were significant changes in both the transcripts and the levels of trehalose and glycogen. These findings indicate that the phenotypic changes are consequences of mutations in regulatory systems and, indeed, all evolved mutants had mutations in proteins involved in the Ras/PKA signaling pathway, which is related to PGM2 overexpression and activation of reserve carbohydrates metabolism (19). Reduced activity of Ras2 or Cyr1 lead to a decreased concentration of cAMP, and lower levels of this metabolite can release the blocking effect of PKA on the transcription factors Msn2/4. Release of the Msn2/4 transcription factors from PKA control results in up-regulation of genes having STRE elements in their promoter region, such as PGM2 and UGP1. 62A and 62B have mutations commonly in RAS2 (RAS2Lys77 and RAS2Tyr112, respectively), whereas 62C has a mutation in CYR1 (CYR1Asn822). RAS2Lys77 in 62A and CYR1Asn822 in 62C appear to exert similar control, as reflected by the close patterns in their transcriptome and metabolome profiles. In contrast, 62B showed a very different pattern in its omics data, even though it had mutations in RAS2 like 62A. The trehalose concentration in this strain was the highest among the evolved mutants, whereas the concentration of glycogen was the lowest (Fig. 4A). Also, the transcript level of this strain in reserve carbohydrates metabolism was much lower than for the other two evolved mutants, the 62A and 62C strains (Fig. 4B). These unique features of 62B indicate that the changes of the reserve carbohydrates metabolism caused a mechanism that is different from that in 62A and 62C. It is interesting to note that the ergosterol biosynthesis and the concentration of ergosterol showed marked changes in this strain (Fig. 4A). A unique mutation was identified in ERG5Pro370. It has been found that a knockout mutant of ERG5 (desaturase) produces dihydroergosterol instead of ergosterol (20). Because 62B had low levels of ergosterol and high levels of dihydroergosterol, it seems plausible that the mutation in ERG5 results in reduced activity of Erg5p, and we speculate that this declined activity may also be closely linked to the increased trehalose production. Both ergosterol and trehalose are regarded as protectants for stress response (21), and a changed sterol composition in the cell membrane can induce trehalose accumulation. The 62B strain will have changed cell membrane rigidity, because dihydroergosterol has more loosened structure than ergosterol because of the loss of one double bond (22), and this looseness may trigger the accumulation of trehalose and, consequently, affect the galactose metabolism by consuming glucose-1-phosphate as a precursor of trehalose. The effect of one of the identified mutations in the Ras/PKA signaling pathway was evaluated, and site-directed mutagenesis of amino acid 112 in Ras2p resulted in an increased specific growth rate on galactose.
The different strategies of adaptive evolution of yeast for improving galactose metabolism are summarized in Fig. 5. As mentioned above, increasing the flux through trehalose and glycogen results in a drain of glucose-1-phosphate, which may have a positive effect on the galactose uptake. Furthermore, the increased flux toward glycogen and trehalose may lead to increased levels of UDP-glucose, which is a cosubstrate in the conversion of galactose-1-phospate to glucose-1-phospate, and this reaction may lead to improved conversion of galactose-1-phospate, which also acts as a feed-forward inhibitor of Pgm2p (8). That increased concentration of UDP-glucose may have a positive effect on the flux through the Leloir pathway is partly supported by earlier findings that the Gal7/Gal10 enzyme system is not controlling the flux through the pathway (23).
Fig. 5.
Summary of evolution changes in the three evolved mutants; 62A, 62B, and 62C. Color circular boxes indicate genes having genetic mutations. Color lines indicate activated fluxes inferred from transcriptome and metabolome analysis.
In conclusion, our study showed that through adaptive evolution the cells may find different ways to ensure an increased flux through the Leloir pathway. This pathway is, despite its few steps, quite complex because it involves the conversion of galactose-1-phospate to glucose-1-phospate with the cocurrent conversion of UDP-glucose to UDP-galactose by galactose-1-phospate uridylyltransferase (encoded by GAL7), and further regeneration of UDP-glucose from UDP-galactose by UDP-glucose 4-epimerase (encoded by GAL10). Because there is further feed-forward inhibition of galactose-1-phospate on phosphoglucomutase (encoded by PGM2), it is clear that activity of the Gal7/Gal10 enzyme system is very important for proper function of the pathway. We, however, earlier found that overexpression of Gal7/Gal10, either alone or together with the other structural GAL genes, does not result in an improved flux through the pathway (on the contrary the flux was decreased), indicating a high level of metabolic regulation. Through adaptive evolution, the cells find a way to circumvent this regulation. It up-regulates expression of PGM2, which partly takes care of the problem of feed-forward inhibition by galactose-1-phospate, as in the engineered strains, but it further up-regulates the flux from glucose-1-phospate to glycogen and trehalose. This alteration is likely to result in increased levels of UDP-galactose that may allow an increased flux through the Gal7/Gal10 enzyme system and, hence, an increased flux through the pathway. Thus, as in other studies on adaptive evolution, it is clear that this strategy allows the generation of new strategies that cannot be found from a rational approach. The most crucial advantage of adaptive laboratory evolution is the finding of unpredictable and unexpected beneficial mutations. In E. coli, this phenomenon is well known (5). Thus, when this bacterium was evolved for improved growth on glycerol, it resulted in mutations in glk (glycerol kinase) that is a key enzyme in the glycerol pathway. However, more effective mutations were detected in unpredicted genes such as RNA polymerase b and b′ subunits (24). In the case of yeast, adaptive laboratory evolution on galactose generated no mutations in the galactose pathway but generated unforeseen ones in other pathways such as carbon regulatory pathway and ergosterol biosynthesis. The main hurdle of galactose metabolism in yeast may therefore not be in Leloir pathway, and increased expression of the GAL genes results in reduced galactose metabolism (12). The beneficial changes are detected in the consuming reaction of glucose-1-phosphate that is directly linked to glycolysis and reserve carbohydrates metabolism, and these changes comes around through mutations in the regulatory PKA pathway.
Besides our findings on unique strategies for improving the galactose uptake, our study clearly provides two key lessons for success in terms of identifying the underlying genotypes of mutants with improved phenotypes:
It is essential to combine detailed phenotypic analysis, e.g., involving transcriptome and metabolome analysis, with genome sequencing. Each of these techniques do not allow for drawing solid conclusions, but combined they provide a clear picture of the consequences of identified mutations.
It is important to analyze several evolved mutants with different control strains because this comparison allows for identification of conserved mutations that result in the same phenotype. Each of the three evolved mutants has several mutations that probably do not contribute to the evolved phenotype, but by identifying conserved mutations, a clear picture emerged.
From these two lessons, we are confident that there is opened up for wider use of systems biology and genome-sequencing for identifying the underlying genotypes for evolved phenotypes in eukaryotic cells.
Materials and Methods
Yeast Strains and Adaptive Evolution.
S. cerevisiae CEN.PK113-7D was used as a reference and starting strain for the adaptive evolution. Two engineered strains, SO16 (Δgal6 Δgal80 Δmig1) and PGM2 (overexpression of PGM2) were constructed in previous study (11, 12). Three adaptively evolved strains, 62A, 62B, and 62C, were generated from CEN.PK 113–7D after daily serial dilution for 62 d (≈400 generations) on galactose (20 g/L) minimal media. Three culture lines were independently carried out at 30 °C and × 0.76 g in cotton-covered 500-mL Erlenmeyer flasks with baffles with 100 mL of media. The cells were cultivated until they reached midexponential phase, before they were transferred to new fresh media. Single clone isolates were obtained from the last shake flasks. The construction of site-directed mutant (RAS2Tyr112) and growth rate measurements in a flask is explained in SI Materials and Methods.
Batch Fermentation and Measurement of Cell Mass and Extracellular Metabolites.
Biological duplication of all strains was performed from seed culture to sample preparation for omics data analyses. Additional details are provided in SI Materials and Methods. The dry cell weight and extracellular metabolites were determined as described (12).
Transcriptome Analysis.
Affymetrix Yeast Genome 2.0 Array was used for transcriptome analysis. Gene expression data were deposited to the Gene Expression Omnibus (GEO) database with accession number GSE27185. Detailed methods are included in SI Materials and Methods.
Metabolome Analysis.
Quenching and extraction of intracellular metabolites were done with biological duplicates of all strains as described (8). Venn diagram was used to represent common and specific features of all of the evolved mutants.
Illumina/Solexa Genome Sequencing.
Genome sequencing was performed by Fasteris SA who used Illumina/Solexa technology. Details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Aldo Jesorka for assistance with dihydroergosterol analysis and Rahul Kumar for valuable discussion. We thank CJ CheilJedang Doctoral Fellowship for financial support (to K.-K.H.). We acknowledge the Chalmers Foundation, The Knut and Alice Wallenberg Foundation, the European Union funded projects UNICELLSYS (Contract 201142) and SYSINBIO (Contract 212766), and European Research Council Grant 247013 for financial contributions to this project.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE27185).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103219108/-/DCSupplemental.
References
- 1.Nielsen J. Metabolic engineering. Appl Microbiol Biotechnol. 2001;55:263–283. doi: 10.1007/s002530000511. [DOI] [PubMed] [Google Scholar]
- 2.Patnaik R. Engineering complex phenotypes in industrial strains. Biotechnol Prog. 2008;24:38–47. doi: 10.1021/bp0701214. [DOI] [PubMed] [Google Scholar]
- 3.Wisselink HW, Toirkens MJ, Wu Q, Pronk JT, van Maris AJ. Novel evolutionary engineering approach for accelerated utilization of glucose, xylose, and arabinose mixtures by engineered Saccharomyces cerevisiae strains. Appl Environ Microbiol. 2009;75:907–914. doi: 10.1128/AEM.02268-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.José Manuel Otero DC, Patil KR, Poulsen SG, Olsson L, Nielsen J. Industrial systems biology of Saccharomyces cerevisiae enables novel succinic acid cell factory. PLoS ONE. 2011 doi: 10.1371/journal.pone.0054144. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herring CD, et al. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet. 2006;38:1406–1412. doi: 10.1038/ng1906. [DOI] [PubMed] [Google Scholar]
- 6.Ikeda M, Ohnishi J, Hayashi M, Mitsuhashi S. A genome-based approach to create a minimally mutated Corynebacterium glutamicum strain for efficient L-lysine production. J Ind Microbiol Biotechnol. 2006;33:610–615. doi: 10.1007/s10295-006-0104-5. [DOI] [PubMed] [Google Scholar]
- 7.Albers E, Larsson C. A comparison of stress tolerance in YPD and industrial lignocellulose-based medium among industrial and laboratory yeast strains. J Ind Microbiol Biotechnol. 2009;36:1085–1091. doi: 10.1007/s10295-009-0592-1. [DOI] [PubMed] [Google Scholar]
- 8.de Jongh WA, et al. The roles of galactitol, galactose-1-phosphate, and phosphoglucomutase in galactose-induced toxicity in Saccharomyces cerevisiae. Biotechnol Bioeng. 2008;101:317–326. doi: 10.1002/bit.21890. [DOI] [PubMed] [Google Scholar]
- 9.Ramsey SA, et al. Dual feedback loops in the GAL regulon suppress cellular heterogeneity in yeast. Nat Genet. 2006;38:1082–1087. doi: 10.1038/ng1869. [DOI] [PubMed] [Google Scholar]
- 10.Ideker T, et al. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 11.Bro C, Knudsen S, Regenberg B, Olsson L, Nielsen J. Improvement of galactose uptake in Saccharomyces cerevisiae through overexpression of phosphoglucomutase: example of transcript analysis as a tool in inverse metabolic engineering. Appl Environ Microbiol. 2005;71:6465–6472. doi: 10.1128/AEM.71.11.6465-6472.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ostergaard S, Olsson L, Johnston M, Nielsen J. Increasing galactose consumption by Saccharomyces cerevisiae through metabolic engineering of the GAL gene regulatory network. Nat Biotechnol. 2000;18:1283–1286. doi: 10.1038/82400. [DOI] [PubMed] [Google Scholar]
- 13.Otero JM, et al. Whole genome sequencing of Saccharomyces cerevisiae: From genotype to phenotype for improved metabolic engineering applications. BMC Genomics. 2010;11:723–740. doi: 10.1186/1471-2164-11-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrick JE, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 15.François J, Parrou JL. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 2001;25:125–145. doi: 10.1111/j.1574-6976.2001.tb00574.x. [DOI] [PubMed] [Google Scholar]
- 16.Hazelwood LA, et al. Identity of the growth-limiting nutrient strongly affects storage carbohydrate accumulation in anaerobic chemostat cultures of Saccharomyces cerevisiae. Appl Environ Microbiol. 2009;75:6876–6885. doi: 10.1128/AEM.01464-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Brink J, et al. Energetic limits to metabolic flexibility: responses of Saccharomyces cerevisiae to glucose-galactose transitions. Microbiology. 2009;155:1340–1350. doi: 10.1099/mic.0.025775-0. [DOI] [PubMed] [Google Scholar]
- 18.Boer VM, Crutchfield CA, Bradley PH, Botstein D, Rabinowitz JD. Growth-limiting intracellular metabolites in yeast growing under diverse nutrient limitations. Mol Biol Cell. 2010;21:198–211. doi: 10.1091/mbc.E09-07-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Estruch F. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol Rev. 2000;24:469–486. doi: 10.1111/j.1574-6976.2000.tb00551.x. [DOI] [PubMed] [Google Scholar]
- 20.Pierce AM, Unrau AM, Oehlschlager AC, Woods RA. Azasterol inhibitors in yeast. Inhibition of the delta 24-sterol methyltransferase and the 24-methylene sterol delta 24(28)-reductase in sterol mutants of Saccharomyces cerevisiae. Can J Biochem. 1979;57:201–208. doi: 10.1139/o79-025. [DOI] [PubMed] [Google Scholar]
- 21.Swan TM, Watson K. Stress tolerance in a yeast lipid mutant: membrane lipids influence tolerance to heat and ethanol independently of heat shock proteins and trehalose. Can J Microbiol. 1999;45:472–479. doi: 10.1139/w99-033. [DOI] [PubMed] [Google Scholar]
- 22.Kelly SL, et al. Purification and reconstitution of activity of Saccharomyces cerevisiae P450 61, a sterol delta 22-desaturase. FEBS Lett. 1995;377:217–220. doi: 10.1016/0014-5793(95)01342-3. [DOI] [PubMed] [Google Scholar]
- 23.Ostergaard S, Olsson L, Nielsen J. In vivo dynamics of galactose metabolism in Saccharomyces cerevisiae: Metabolic fluxes and metabolite levels. Biotechnol Bioeng. 2001;73:412–425. doi: 10.1002/bit.1075. [DOI] [PubMed] [Google Scholar]
- 24.Applebee MK, Herrgård MJ, Palsson BO. Impact of individual mutations on increased fitness in adaptively evolved strains of Escherichia coli. J Bacteriol. 2008;190:5087–5094. doi: 10.1128/JB.01976-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.