Abstract
Myotonic dystrophy type 1 (DM1) is caused by the expansion of noncoding CTG repeats in the dystrophia myotonica-protein kinase gene. Mutant transcripts form CUG hairpins that sequester RNA-binding factors into nuclear foci, including Muscleblind-like-1 protein (MBNL1), which regulate alternative splicing and gene expression. To identify molecules that target toxic CUG transcripts in vivo, we performed a positional scanning combinatorial peptide library screen using a Drosophila model of DM1. The screen identified a D-amino acid hexapeptide (ABP1) that reduced CUG foci formation and suppressed CUG-induced lethality and muscle degeneration when administered orally. Transgenic expression of natural, L-amino acid ABP1 analogues reduced CUG-induced toxicity in fly eyes and muscles. Furthermore, ABP1 reversed muscle histopathology and splicing misregulation of MBNL1 targets in DM1 model mice. In vitro, ABP1 bound to CUG hairpins and induced a switch to a single-stranded conformation. Our findings demonstrate that ABP1 shows antimyotonic dystrophy activity by targeting the core of CUG toxicity.
Keywords: disease model, drug discovery, non-coding RNA disease, medicinal chemistry, RNA secondary structure
Myotonic dystrophy type 1 (DM1, OMIM #160900) is an autosomal dominant disease caused by the expansion of a CTG trinucleotide repeat in the 3′ untranslated region (UTR) of the dystrophia myotonica-protein kinase (DMPK) gene. Characteristic symptoms include myotonia, progressive muscle wasting, and cardiac conduction defects, among other systemic manifestations. The molecular mechanisms underlying DM1 pathogenesis are complex and affect a large number of cellular processes (1). However, most data suggest that the main triggering event is a toxic gain-of-function of the expanded CUG RNA. CUG-repeat expansions form double-stranded hairpins that are retained as inclusions within the nucleus. These hairpins recruit a number of transcription and splicing factors, including Muscleblind-like 1 (MBNL1) (2–6). Sequestration of MBNL1 originates a loss of function, which plays a key role in the development of DM1 symptoms. Mbnl1 knockout mice reproduced typical features of DM1, and overexpression of Mbnl1 in a mouse model that expressed CTG repeats reversed these phenotypes (7, 8). CTG-repeat expression in mice caused misregulation of at least 156 alternative splicing events. Of these, 128 also occurred in Mbnl1 knockout animals (9–12). The splicing factor CUG-binding protein 1 (CUGBP1) is another key component in the development of DM1 phenotypes. CUGBP1 antagonizes MBNL1 activity in the regulated use of alternative exons in a number of transcripts and is abnormally upregulated in patients with DM1, further contributing to splicing misregulation (13–15).
Mahadevan et al. provided the first in vivo proof-of-principle for a therapeutic strategy based on ablating toxic RNA molecules in DM1 (16). They demonstrated that expanded CTG-induced effects could be reverted if CTG-repeat transgene expression was interrupted in a DM1 mouse model. Several other groups developed synthetic molecules and (CAG)n oligonucleotides that disrupted MBNL1 interaction with expanded CUG repeats (17–22). However, those approaches may not address all the pathological consequences of expanded CUG RNAs. Moreover, unspecific MBNL1-RNA binding inhibition could affect other normal, MBNL1-dependent splicing events. Therefore, it would be desirable to identify molecules that target toxic repeats without interfering with cellular, nonpathological MBNL1-RNA interactions.
Results
In Vivo Screening of a Combinatorial Peptide Library in Drosophila Identified a Hexapeptide that Suppressed CUG Toxicity.
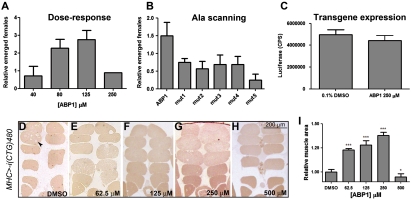
The targeted expression of 480 interrupted CTG repeats, UAS-i(CTG)480, to the Drosophila brain structure the mushroom bodies (MB) with the X-linked 103Y-Gal4 driver (103Y-Gal4/+; UAS-i(CTG)480/+) originates a female-specific, semilethal phenotype at pupal stage (23). To screen for molecules that suppressed that phenotype, we used a positional scanning synthetic combinatorial library (PS-SCL) of D-amino acid hexapeptides (24, 25). The library comprised 120 peptide mixtures, each of them containing approximately 2.5 million individual hexapeptides (195; D-Cys was omitted from the mixture positions). Each mixture addressed a separate, single position of the hexamer sequence. The 120 peptide mixtures were tested individually in vivo (80 μM) in nutritive media with 0.1% dimethyl sulfoxide (DMSO) (23). The number of female flies that emerged was compared to DMSO-treated control individuals, and 28 peptide mixtures that significantly suppressed CUG-induced lethality were identified. Based on the screen results, 16 individual peptides were generated and evaluated (Table S1). The hexapeptide Ac-ppyawe-NH2 (ABP1; lower case one-letter code for D-amino acids) significantly suppressed lethality in a dose-dependent manner (Fig. 1A). The individual contribution of each amino acid in ABP1 was determined using an alanine scanning, single substitution approach (26). Each mutant peptide (mut1 to mut5; see Table S2) was evaluated for its effect on the viability of 103Y-Gal4/+; UAS-i(CTG)480/+ female flies. None of these peptides suppressed CUG-induced lethality, indicating that all amino acids in the original ABP1 sequence were necessary for its activity (Fig. 1B). To examine whether ABP1 could nonspecifically reduce UAS-i(CTG)480 transcription, we measured Luciferase reporter activity in flies of the genotype Myosin heavy chain (MHC)-Gal4/+; UAS-luciferase/+ after treatment with DMSO or ABP1. The results revealed no significant differences in the Luciferase signal (Fig. 1C). In addition, the levels of i(CUG)480 transcripts in MHC-Gal4/+; UAS-i(CTG)480/+ flies were measured by semiquantitative RT-PCR. We detected no reduction in the amount of toxic RNA in ABP1-treated individuals (Fig. S1 A and B).
Fig. 1.
ABP1 suppressed i(CTG)480-induced phenotypes in a dose-dependent manner. (A) Number of 103Y-Gal4/+; UAS-i(CTG)480/+ female flies emerged after ABP1 treatment relative to DMSO-treated controls. ABP1 concentrations of 40 μM, 80 μM, and 125 μM caused relative increments in the number of emerged flies of 0.7 (n = 120), 2.3 (n = 180), and 2.8 (n = 120), respectively, compared to DMSO-treated controls. ABP1 at 250 μM caused a drop in female viability, suggesting a toxic effect (n = 120). (B) Five mutant derivates of ABP1 (mut1-5), with single alanine substitutions, did not suppress i(CTG)480 toxicity (n = 60). (C) ABP1 did not reduce the expression of luciferase reporter in MHC-Gal4/+; UAS-luciferase/+ flies. The average luciferase light emission is shown for 9–10 replicates (total n = 27–30 flies). (D–H) Representative bright field microscopy images of transversal sections of resin-embedded adult IFM of MHC-Gal4/+; UAS-i(CTG)480/+ flies that had been (D) treated with DMSO or (E–H) with ABP1. ABP1 (62.5 μM, 125 μM and 250 μM) increased the muscle area relative to DMSO-treated flies (quantified in I) and fiber loss was reduced (arrowhead). ABP1 at 500 μM enhanced the phenotype, indicating a toxic effect. All graphs show the mean and SEM except graph A, which shows the sum of all replicates. * indicates p-value < 0.05, *** indicates p-value < 0.0001.
To confirm that ABP1 could suppress CUG-induced toxicity in muscles, we analyzed ABP1 activity on a muscle degeneration phenotype caused by the expression of i(CUG)480 in the indirect flight muscles (IFM) under the control of the MHC-Gal4 driver (23, 27). ABP1 increased total muscle area in a dose-dependent manner up to 37.8% compared to DMSO-treated flies (Fig. 1 D–I).
Transgenic Expression of ABP1 in Drosophila Suppressed CUG-Induced Phenotypes.
In vivo ABP1 activity relied on its ability to diffuse through tissues and membranes, and to counteract CUG toxicity. To separate both properties, we performed tissue-specific expression of three transgenes that encoded ABP1-derivatives, UAS-ABP1f, UAS-ABP1r, and UAS-ABP1c. The UAS-ABP1f construct encoded a 10-mer peptide with a methionine and three spacer glycines followed by the forward sequence of a natural L-amino acid ABP1 variant; UAS-ABP1r encoded a similar peptide with the sequence of ABP1 reversed (thus mimicking the spatial disposition of the side chains of ABP1); and UAS-ABP1c encoded a 20-mer peptide that linearly linked UAS-ABP1f and UAS-ABP1r. When expressed alone, none of these transgenic lines had a phenotype. However, when crossed to flies that expressed i(CUG)480 in the eye under the control of the glass multiple reporter (GMR)-Gal4 driver, the UAS-ABP1r and UAS-ABP1c (but not UAS-ABP1f) transgenes suppressed eye phenotypes (Fig. S2 A–C). This confirmed ABP1 activity on CUG toxicity and indicated that the spatial conformation of the ABP1 side chains was important for in vivo activity.
UAS-ABP1r and UAS-ABP1c also suppressed muscle degeneration in the IFM, with a 3.5-fold increase in total muscle area (Fig. S2 D–F). This effect was approximately nine times stronger than that observed upon oral administration. Therefore, improving the bioavailability of the peptide enhanced its activity. In all cases, coexpression of UAS-i(CTG)480 and UAS-GFP was used as control to guarantee comparable amounts of Gal4 protein available to activate each transgene. To rule out an effect of the transgenic reverse forms of ABP1 on UAS-i(CTG)480 transcription, we measured Luciferase reporter activity in flies of the genotype MHC-Gal4/+;UAS-luciferase/UAS-ABP1r and compared it with MHC-Gal4/+; UAS-luciferase/UAS-ABP1f flies. No significant differences in the luciferase signal were detected between genotypes (Fig. S1C).
ABP1 Bound to CUG Repeats in Vitro Without Displacing Muscleblind.
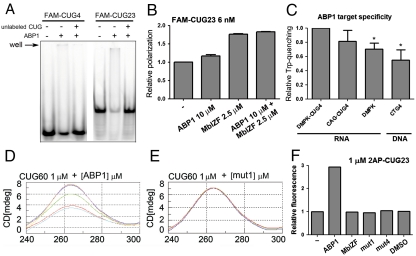
Binding of Muscleblind (Mbl) zinc finger domains to CUG repeats is strongly directed by the presence of aromatic residues (28). The sequence of the ABP1 hexapeptide is also enriched in aromatic amino acids. We therefore explored whether ABP1 activity relied on a direct binding to toxic CUG-repeat RNAs, and a potential competition with Mbl for binding. Using an electrophoretic mobility shift assay, we analyzed the binding of a synthetic, fluorescently labeled 23-CUG-repeat (FAM-CUG23) RNA and a 4-CUG-repeat (FAM-CUG4) RNA to ABP1. In these experiments, the peptide bound to both FAM-CUG23 and FAM-CUG4, and the interaction was reversed with excess nonlabeled RNA (Fig. 2A and Fig. S3B). However, the affinity was lower for the shorter repeats. The alanine scanning mutant peptides (mut1 to mut5) did not bind to FAM-CUG23 (Fig. S3A). This demonstrated that a single amino acid change in ABP1 disrupted its ability to bind to synthetic CUG repeats. Thus, in vitro RNA binding correlated with suppression of CUG-induced toxicity in vivo. A recombinant protein derived from the two zinc fingers of Drosophila Mbl (MblZF) also bound to CUG-repeat RNA (Fig. S3C). However, in our experimental conditions both ABP1 and MblZF formed complexes with the RNA that were unable to enter the gel, making qualitative and quantitative analyses of competition between ABP1 and MblZF not feasible. In order to study the effect of ABP1 on MblZF binding to FAM-CUG23, we used fluorescence polarization spectroscopy. In these experiments, both the peptide and the protein bound to the fluorescent RNA, increasing its polarization (Fig. 2B). The interaction was reversed with excess nonlabeled RNA, which reduced fluorescence polarization to that of free FAM-CUG23. However, when FAM-CUG23 was simultaneously incubated with ABP1 and MblZF, no significant reduction in FAM-CUG23 polarization was observed. In contrast, an additive effect was detected. These results suggested that ABP1 and MblZF had different binding sites in the CUG RNA.
Fig. 2.
ABP1 bound to CUG repeats and caused a switch in CUG secondary structure. (A) Electrophoretic mobility shift assay using a FAM-labeled 23 CUG-repeat RNA (FAM-CUG23; 60 nM) and a FAM-labeled 4 CUG-repeat RNA (FAM-CUG4; 60 nM). The gel shows a decrease in the free RNA band intensity. Binding of ABP1 (2.5 mM) to FAM-CUG23 was stronger than to CUG4. A nonlabeled CUG RNA (6 μM) competed both interactions. (B) Fluorescence polarization of FAM-CUG23. Both ABP1 (10 μM) and MblZF (2.5 μM) caused polarization of the RNA (6 nM). Coaddition of ABP1 and MblZF at nonsaturating concentrations did not reverse the RNA polarization but showed an additive effect. (C) Tryptophan fluorescence quenching (FQ) of ABP1. ABP1 (5 μM) was incubated with RNA (DMPK-CUG4, CAG•CUG4, DMPK) or DNA (CTG4) molecules at 2.5 μM, 5 μM, 7.5 μM, 10 μM, and 12.5 μM. The slopes of the curves obtained from plotting fluorescence emission values at 351 nm for each titration point were normalized to DMPK-CUG4 and are represented in bars as a measure of the FQ efficiency. (D) The circular dichroism (CD) spectrum of a 60 CUG-repeat RNA (CUG60; 1 μM) showed a major peak at 265 nm. This peak was reduced when the RNA was incubated with ABP1 at 0.1 μM (pink), 0.5 μM (dark blue), 1 μM (green), 10 μM (red), and 20 μM (light blue). (E) This change did not occur when CUG60 (red) was incubated with mutant derivatives of ABP1, including mut1 at 0.5 μM (dark blue) and 1 μM (green). (F) 2AP-fluorescence measurements of the 2AP-CUG23 RNA (1 μM). The addition of 100 μM peptide destabilized the hairpin, and caused a 2.9-fold increase in the fluorescence intensity. MblZF, DMSO, mut1, and mut4 peptides did not cause changes at the same concentration. All graphs show the mean and SEM values for at least four (B) or two measurements (C). * indicates p-value < 0.05.
ABP1 Bound with Higher Affinity to CUG Repeats than to Other Sequences.
ABP1 contains the fluorescent amino acid D-tryptophan. Changes in tryptophan fluorescence emission at 351 nm were detected upon ABP1 binding to RNA and DNA sequences. ABP1 tryptophan fluorescence was quenched when bound to the following: an RNA sequence that included 19 nucleotides (nt) from the 3′UTR of DMPK, followed by four CUG repeats (DMPK-CUG4); a single-stranded RNA sequence that included the 19-nt region of DMPK alone (DMPK); a perfect RNA hairpin with four CAG•CUG repeats (CAG•CUG4); a single-stranded DNA sequence that included the 19-nt region of DMPK (DMPK); and a double-stranded DNA with four CTG repeats (CTG4). This indicated that ABP1 could bind to different types of nucleic acids. However, DMPK-CUG4 RNA showed the highest quenching efficiency (Fig. 2C). Only the ABP1 interaction with CAG•CUG4 RNA showed a similar quenching rate, suggesting target selectivity for ABP1.
ABP1 Caused a Switch from Double-Stranded to Single-Stranded Conformation in the CUG RNA.
To study changes in RNA conformation induced by ABP1 we measured the circular dichroism (CD) of a 60 CUG-repeat RNA (CUG60) after incubation with increasing concentrations of the peptide. The CD spectrum of CUG60 alone showed a major peak at 265 nm. Addition of ABP1, but not the ABP1 alanine-substituted peptides mut1 (Fig. 2 D and E) and mut4 (Fig. S4C), caused a concentration-dependent decrease in the CD signal. This indicated that ABP1 specifically induced a reduction in RNA base stacking. The addition of MblZF did not reverse this effect (Fig. S4 A and B). The observed CD signal reduction was not caused by partial denaturation or degradation of the RNA during the time course of the experiment (Fig. S4D). These results suggested that ABP1 promoted a relaxation in the secondary structure of CUG60.
To confirm that ABP1 induced CUG-repeat RNA to change from a double-stranded to single-stranded conformation, we substituted a guanosine residue in a 23 CUG-repeat RNA with the adenosine analogue 2-amino purine (2AP-CUG23). The 2-amino purine fluorescence is strongly quenched in a base-paired structure, but it increases 2–3 fold in a single-stranded conformation (29). When different concentrations of MblZF and ABP1 were incubated with the RNA at molar ratios lower than 1∶5, neither the MblZF protein nor the ABP1 peptide disrupted the double-stranded RNA conformation (Fig. S4E). However, at a molar ratio of 1∶100 ABP1 caused a 2.9-fold increase in 2-AP fluorescence (Fig. 2F). This indicated a switch from predominantly double-stranded to predominantly single-stranded conformation. In contrast, addition of DMSO alone, mut1, or mut4 did not modify 2AP-CUG23 fluorescence, demonstrating the specificity of the effect. Because double-stranded CUG and single-stranded CUG RNAs coexist in vitro, and Muscleblind proteins are known to bind CUG hairpins (3), these results explained the additive behavior of ABP1 and MblZF observed in our polarization experiments.
ABP1 Reduced CUG–RNA Foci Formation and Muscleblind Aggregation in Drosophila.
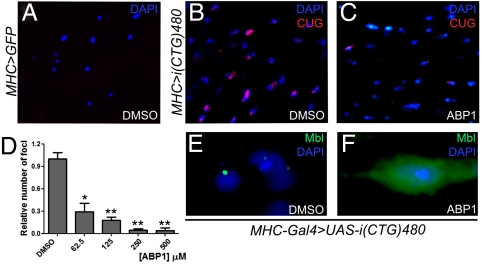
In vitro results suggested that ABP1 activity relied on its ability to reduce the formation of CUG–RNA hairpins. To study whether the peptide effectively decreased the number of toxic double-stranded CUG molecules in Drosophila, which accumulate forming nuclear foci, we studied CUG–RNA distribution in the IFM on longitudinal thorax sections of MHC-Gal4/+; UAS-i(CTG)480/+ flies that had been fed different concentrations of ABP1. Oral administration of the peptide significantly suppressed CUG foci formation at all concentrations assayed (Fig. 3 A–D), indicating that ABP1 also inhibits double-stranded RNA formation in vivo. This reduction was accompanied by a redistribution of Mbl protein within the cells. In the absence of peptide, CUG expression caused Mbl to accumulate into nuclear aggregates (Fig. 3E). However, ABP1 treatment induced a Mbl redistribution to a preferentially diffuse pattern, which also included the cytoplasm (Fig. 3F).
Fig. 3.
ABP1 reduced the number of CUG–RNA foci in Drosophila and released Mbl from nuclear aggregates. (A–C) Representative fluorescent in situ detection of i(CUG)480 transcripts in the IFM on thorax sections of MHC-Gal4/UAS-GFP (control; A) or MHC-Gal4/+; UAS-i(CTG)480/+ (B and C) flies orally treated with 0.12% DMSO (A and B) or different concentrations of ABP1 (250 μM is shown as an example in C). ABP1 treatment significantly reduced the number of CUG–RNA foci per section at all concentrations tested (D). (E and F) Immunofluorescent detection of Mbl (green) in the IFM of DMSO-treated (0.12%; control) and ABP1-treated (250 μM) MHC-Gal4/+; UAS-i(CTG)480/+ flies. Oral administration of ABP1 caused a switch in Mbl cellular distribution from aggregated (control; E) to disperse (treated; F). Nuclei are counterstained with DAPI (blue) in E and F. The graph in D shows the mean and SEM from ABP1-responsive animals. * indicates p-value < 0.05, ** indicates p-value < 0.0005.
ABP1 Reversed Missplicing and Muscle Histopathology in a DM1 Mouse Model.
To confirm the effect of ABP1 in mammals, we used transgenic DM1 model mice that carry 250 (HSALR) or 5 (HSASR) CTG repeats in the 3′UTR of the human skeletal actin open reading frame (30).
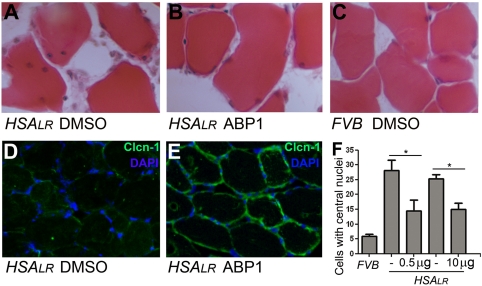
A histological hallmark of both DM1 and HSALR muscle fibers is the presence of central nuclei (30). Based on initial ABP1 toxicity analyses in wild-type animals (FVB; see Table S4), we injected 0.5 μg (in 0.2% DMSO) or 10 μg (in 2% DMSO) of ABP1 into the right hind limb tibialis anterioris (TA) muscle. In both cases, ABP1 significantly reduced the percentage of fibers with central nuclei in three out of five HSALR mice (60% of the animals; Fig. 4 A–C and F and Table S5; referred to as “ABP1-responsive animals”) one month after injection (a/i) compared to DMSO controls. Another characteristic of DM1 muscles is a reduction of functional Chloride Chanel-1 (ClC-1) protein. A loss of ClC-1 activity in the muscle membranes is the cause of myotonia (30). Immunohistochemical studies of Clcn-1 levels on muscle sections of ABP1-treated (10 μg) mice showed a qualitatively evident increase in the amount of Clcn1 protein detected in 80% of the animals one month a/i (Fig. 4 D–E and Table S5).
Fig. 4.
ABP1 suppressed histological defects in DM1 mouse muscles. Intramuscular injection was carried out in the hind limb TA muscles of mice that expressed human skeletal actin flanked by 250 CTG repeats (HSALR) (30) or wild-type controls (FVB). (A–C) Representative histological TA sections from animals injected with 0.2 or 2% DMSO (control left limb in HSALR and right limb in FVB; A and C), or 0.5 or 10 μg of ABP1 (right limb in HSALR; B), respectively. The images show a significant decrease in the percentage of cells with central nuclei one month after ABP1 injection (F). (D and E) Immunofluorescent detection of Clcn1 protein (green) in HSALR mouse TA sections one month a/i. In 2% DMSO-treated limbs (D) the Clcn1 signal was almost absent, whereas the fluorescent signal was significantly stronger in the contralateral ABP1-treated limb (10 μg) (E). Nuclei are counterstained with DAPI (blue) in D and E. The graph in F shows the mean and SEM from ABP1-responsive animals. * indicates p-value < 0.05.
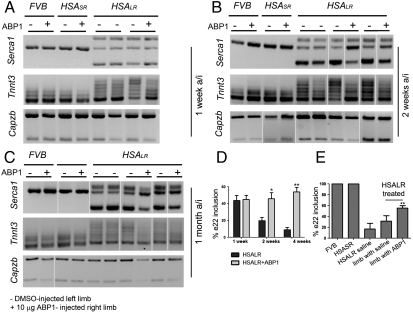
Alternative splicing is severely misregulated in DM1 patients and in HSALR mice (11, 12). We analyzed alternative splicing patterns in transcripts of two direct Mbnl1 targets: the muscle sarcoplasmic/endoplasmic reticulum Ca2+ ATPase (Serca1) and fast skeletal muscle troponin T (Tnnt3). Exon 22 of Serca1 is included in adult muscle but excluded in neonatal, adult DM1, and mouse HSALR muscles (31). Semiquantitative RT-PCR on muscle tissues from FVB and DM1 mice showed that ABP1 injection (10 μg) in the TA of HSALR animals caused a significant increase in Serca1 exon 22 inclusion in 60% of the treated animals (mice 2, 3, and 4; Table S5). The degree of reversion to normal splicing was quantified in ABP1-responsive animals at two and four weeks a/i yielding a 25.9% and 38.3% increase in exon 22 inclusion, respectively. At the later time point, we also detected a 14.7% increase in exon 22 inclusion in the DMSO-injected left limb of ABP1-responsive animals injected with ABP1 in the right limb compared to animals that received DMSO in both limbs (Fig. 5). These results suggested that ABP1 had a systemic effect. A qualitative assessment of Tnnt3 missplicing showed that ABP1 also reversed the splicing defects in HSALR animals one week a/i (40% of the animals), two weeks a/i (60% of the animals), and four weeks a/i (80% of the animals). Importantly, ABP1 did not change the splicing pattern of Serca1 or Tnnt3 transcripts in control FVB or HSASR mice. This demonstrated that ABP1 did not compromise Mbnl1 function in vivo. Finally, the peptide did not interfere with the activity of other alternative splicing factors, because it did not affect the Mbnl1-independent alternative splicing of F-actin-capping protein subunit beta (Capzb) exon 8, the regulation of which is unaffected in HSALR mice. Taken together, these results validate the potential of ABP1 as a suppressor of CUG-repeat RNA-induced toxicity in mammals.
Fig. 5.
ABP1 suppressed splicing defects in DM1 mouse muscles. ABP1 (10 μg) was injected intramuscularly in the right hind limb TA of HSALR mice, animals that expressed short (5) CTG repeats (HSASR) (30) or wild-type animals (FVB). Semiquantitative RT-PCR performed one week a/i (A), two weeks a/i (B), and four weeks a/i (C) showed that ABP1 partially reversed splicing defects in muscle transcripts of Serca1 (quantified in D and E) and Tnnt3 genes in HSALR mice. ABP1 did not change the splicing patterns of Serca1 and Tnnt3 in FVB or HSASR strains. A muscular transcript that undergoes Mbnl1-independent alternative splicing and is unaffected in this model, Capzb, was not altered by ABP1 injection in any strain. Each pair of lanes in the gels represent the left, 2% DMSO-injected leg (-) and the right, 10 μg ABP1-injected leg (+) of the same animal. All graphs show the mean and SEM from ABP1-responsive animals. * indicates p-value < 0.05.
Discussion
ABP1 efficiently suppressed CUG-induced phenotypes in three different Drosophila tissues: brain, muscle, and eye. Moreover, the peptide significantly reduced the number of CUG–RNA foci and caused a Mbl subcellular redistribution in the fly muscle. Two findings confirmed the specificity of ABP1 in vivo. First, ABP1 effects were dose-dependent in fly muscle, brain (oral administration), and eyes (transgenic expression). Second, alanine-substituted peptide derivatives of ABP1 had no effect on CUG-induced lethality in flies. Thus, CUG toxicity was specifically suppressed by ABP1, and each amino acid in the peptide sequence was necessary. Moreover, the suppression of CUG-induced phenotypes by transgenic expression of a reverse ABP1 sequence (ABP1r and ABP1c) indicated that the in vivo activity of ABP1 resided in the spatial disposition of its side chains. Interestingly, ABP1 has two aromatic amino acids in its sequence (tryptophan and tyrosine). The side chains of aromatic amino acids in a number of proteins have been found to be critical for enabling RNA binding, including the CCCH-type zinc finger protein Tis11, and MBNL1(28, 32).
Both the fly and mouse models used in this work expressed CUG-repeat transcripts independently of DMPK. Therefore, in vivo, ABP1 could target either the toxic CUG RNA or a pathogenic target downstream of the expanded repeats. However, the downstream effects of CUG-repeat RNA expression are likely to vary among different tissues. The fact that ABP1 suppressed CUG-repeat associated toxicity in at least three different fly tissues indicated that its activity was independent of environmental factors and only depended on the toxicity of CUG-repeat RNA. This notion is supported by our in vitro experiments, which demonstrate that ABP1 directly binds to CUG-repeat RNAs and suggest that ABP1 might be effective to treat other diseases caused by CUG expansions.
Based in our in vitro and in vivo results, in this study we propose that the molecular mechanism underlying ABP1 activity relies on the induction of a conformational shift in CUG secondary structure (see Fig. S5). Causing a switch from predominantly double-stranded CUG RNA to predominantly single-stranded CUG molecules in the nuclei should, in principle, restore homeostasis to CUG-expressing cells. Our in vitro results suggested that ABP1 did not prevent MblZF from binding to CUG hairpins. In contrast, their binding to the CUG repeats was additive, most likely because they bind and stabilize different coexisting conformations of the RNA. Importantly, the Mbnl1 splicing function on its direct targets Serca1 and Tnnt3 was partially restored in ABP1-treated HSALR mice. This demonstrates that the suppression of CUG-dependent phenotypes and recovery of Mbnl1 activity can be achieved without directly competing with Muscleblind binding to CUG repeats but by reducing the fraction of double-stranded CUG RNA from the cellular pool of toxic RNA, hence preventing sequestration of the protein, which requires CUG RNA hairpins. This is a unique mechanism that contrasts with that of other recently characterized molecules that target the binding of MBNL1 to toxic RNAs (17–22, 33). Disrupting Muscleblind binding to RNA could affect its function as a splicing factor in non-CUG-expressing cells. Our results suggest that ABP1 does not interfere with natural Mbnl1 splicing activity, because Tnnt3 and Serca1 transcripts were not misspliced in FVB and HSASR mice. Moreover, ABP1 does not alter the function of other splicing factors, because the Mbnl1-independent transcript Capzb was not affected by ABP1 treatment.
An important advantage of ABP1 is that it is composed of D-amino acids, which are not recognized by proteases. This, coupled with its small molecular weight (< 900 Da), confers ABP1 with a potentially higher systemic bioavailability compared to larger L-amino acid peptides. The protease resistance of ABP1 also confers stability in the organism, and therefore it may exhibit long-lasting effects. This could account for our observation that the reversal of splicing defects and muscle histopathology in mice lasted for at least four weeks after one single injection. Based on our results, we propose that ABP1 represents a promising approach in the generation of new effective treatments for DM1.
Materials and Methods
Drosophila Stocks.
MHC-Gal4 flies were a gift of Eric Olson, University of Texas, Southwestern Medical Center, Texas. UAS-ABP1f, UAS-ABP1r, and UAS-ABP1c flies were generated with synthetic DNA oligonucleotides (Metabion; Table S3) cloned into the Drosophila pUAST vector. Transgenes were microinjected into y1w1118 embryos (BestGene). Other fly stocks were described in ref. 23.
Eye and Muscle Histology.
SEM imaging was described in ref. 23. Ten-day-old Drosophila thoraces were embedded in Epon for transversal, semithin sectioning (34). Muscle area was quantified by binarizating images (equal dimensions) and quantifying pixels (%) within muscle tissue with Image-J software. Frozen sections (10 μm) of mouse TA were stained with H&E or immunostained using antibody against the ClC-1 C terminus (Alpha Diagnostic; 1∶50) as described in ref. 7. For comparison of treated and untreated muscle, sections were stained on the same slide, and images were obtained using the same settings with a confocal microscope (Leica TCS SP2 DM-R).
MblZF Expression and Purification.
DNA encoding 98 amino acids of Drosophila Mbl was amplified by PCR from OrR flies (Table S3) and cloned into the pET-15b expression vector. Transformed Escherichia coli BL21 (DE3) cells, grown in LB medium supplemented with 2% glucose and 50 μM ZnCl2, were induced with 1 mM of IPTG for 2 h at 16 °C. Cells were collected, resuspended (50 mM Tris-HCl pH 7.5, 250 mM NaCl, 50 μM ZnCl2, 10% glycerol, and 1 mM DTT), and sonicated. MblZF was purified from the soluble fraction in an FPLC ÄKTA™ with 1 ml HisTrapHP columns (Amersham Biosciences).
Fluorescence Polarization.
CUG RNA (23 repeats) labeled with carboxyfluorescein (FAM-CUG23; 6 nM) was incubated with ABP1, MblZF, or DMSO (free RNA controls) in binding buffer (25 mM Tris-HCl pH 7.5, 100 mM NaCl, 5 mM MgCl2, 50 μM ZnCl2, and 10% glycerol) on ice (20 min) in the dark. Polarization was measured in a Multilabel Counter (excitation filter FP480, emission filter FP535).
Fluorescence Electrophoretic Mobility Shift Assay.
FAM-CUG23 and FAM-CUG4 RNAs were heated at 70 °C (5 min), diluted (60 nM) in binding buffer, kept at 37 °C (5–10 min), incubated with ABP1, MblZF or DMSO (free RNA controls) at 37 °C (20 min), and loaded into prerun 6% polyacrylamide nondenaturing gels. Electrophoresis at 4 °C (approximately 1 h) at 150–200 V was followed by visualization with 520 BP/526 SP filters on a Typhoon 9400 scanner (Amersham).
Tryptophan Fluorescence Quenching.
Fluorescence of ABP1 (5 μM in binding buffer), incubated with increasing concentrations of DNA or RNA, was measured at room temperature with a Jasco FP6500 spectrofluorimeter (excitation 290 nm, emission 351 nm). Emission measurements were normalized to those of peptide alone. At least two measurements were taken per titration point.
2-aminopurine Fluorescence.
2AP-CUG23 RNA (1 μM in binding buffer) was heated at 90 °C (3 min), placed on ice, and fluorescence was measured (35). Increasing amounts of ABP1 and MblZF were added and fluorescence changes were measured three times per titration point. Emissions (375 nm) of ABP1 (or MblZF) alone were subtracted from RNA spectra. Measurements were normalized by dividing the emission values at 375 nm by those of 2AP-CUG23 RNA alone.
Transgenic Mice and Peptide Administration.
Mouse handling and experimental procedures conformed to the European law of laboratory animal care and experimentation (2003/65/CE). Transgenic HSALR (line 20 b) and HSASR (line 05) mice were described (30). FVB mice were used as wild-type animals. The right and left hind limb TAs of five five-week-old gender-matched mice were injected intramuscularly with 10 μl of peptide or DMSO (internal control), or DMSO was injected on both sides ( HSALR controls). One, two, or four weeks after injection, animals were sacrificed and TAs were dissected in two equal pieces for histological and splicing analyses.
Supplementary Material
Acknowledgments.
We thank Valentia Biopharma for the UAS-luciferase flies; E. Lavara and members of the E. Pérez-Payá laboratory for technical support; and C. Thornton for feedback on the manuscript and for providing HSALR model mice. A.G.-L. was awarded a predoctoral fellowship from the Ministerio de Educación y Ciencia. This work and B.L. were supported by a Muscular Dystrophy Association research grant to R.A. (4127). Additional funding was provided by the Acciones de Articulación IIARC02004-A-51 from the Generalitat Valenciana (GV) and the BFU2009-10940 grant from the Ministerio de Ciencia e Innovación (MICINN) to R.A, and by the BIO2007-60066 (MICINN) and ACOMP2009-011 grants (GV) to E.P.-P. Emissions relating to this work, estimated at some 6.9 CO2 tones, have been compensated through a reforestation project.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. H.M.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018213108/-/DCSupplemental.
References
- 1.Llamusi B, Artero R. Molecular effects of the CTG repeats in mutant dystrophia myotonica protein kinase gene. Curr Genomics. 2008;9:509–516. doi: 10.2174/138920208786847944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim DH, et al. HnRNP H inhibits nuclear export of mRNA containing expanded CUG repeats and a distal branch point sequence. Nucleic Acids Res. 2005;33:3866–3874. doi: 10.1093/nar/gki698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller JW, et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fardaei M, et al. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- 5.Ebralidze A, Wang Y, Petkova V, Ebralidse K, Junghans RP. RNA leaching of transcription factors disrupts transcription in myotonic dystrophy. Science. 2004;303:383–387. doi: 10.1126/science.1088679. [DOI] [PubMed] [Google Scholar]
- 6.Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanadia RN, et al. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc Natl Acad Sci USA. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanadia RN, et al. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 9.Orengo JP, et al. Expanded CTG repeats within the DMPK 3′ UTR causes severe skeletal muscle wasting in an inducible mouse model for myotonic dystrophy. Proc Natl Acad Sci USA. 2008;105:2646–2651. doi: 10.1073/pnas.0708519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osborne RJ, Thornton CA. RNA-dominant diseases. Hum Mol Genet. 2006;15:R162–R169. doi: 10.1093/hmg/ddl181. [DOI] [PubMed] [Google Scholar]
- 11.Lin X, et al. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 12.Du H, et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalsotra A, et al. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci USA. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang GS, Kearney DL, De Biasi M, Taffet G, Cooper TA. Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J Clin Invest. 2007;117:2802–2811. doi: 10.1172/JCI32308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahadevan MS, et al. Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat Genet. 2006;38:1066–1070. doi: 10.1038/ng1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wheeler TM, et al. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulders SA, et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc Natl Acad Sci USA. 2009;106:13915–13920. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gareiss PC, et al. Dynamic combinatorial selection of molecules capable of inhibiting the (CUG) repeat RNA-MBNL1 interaction in vitro: Discovery of lead compounds targeting myotonic dystrophy (DM1) J Am Chem Soc. 2008;130:16254–16261. doi: 10.1021/ja804398y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pushechnikov A, et al. Rational design of ligands targeting triplet repeating transcripts that cause RNA dominant disease: Application to myotonic muscular dystrophy type 1 and spinocerebellar ataxia type 3. J Am Chem Soc. 2009;131:9767–9779. doi: 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arambula JF, Ramisetty SR, Baranger AM, Zimmerman SC. A simple ligand that selectively targets CUG trinucleotide repeats and inhibits MBNL protein binding. Proc Natl Acad Sci USA. 2009;106:16068–16073. doi: 10.1073/pnas.0901824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warf MB, Nakamori M, Matthys CM, Thornton CA, Berglund JA. Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proc Natl Acad Sci USA. 2009;106:18551–18556. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Lopez A, et al. Genetic and chemical modifiers of a CUG toxicity model in Drosophila. PLoS One. 2008;3:e1595. doi: 10.1371/journal.pone.0001595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinilla C, et al. A review of the utility of soluble peptide combinatorial libraries. Biopolymers. 1995;37:221–240. doi: 10.1002/bip.360370306. [DOI] [PubMed] [Google Scholar]
- 25.Blondelle SE, Houghten RA, Perez-Paya E. Identification of inhibitors of melittin using nonsupport-bound combinatorial libraries. J Biol Chem. 1996;271:4093–4099. doi: 10.1074/jbc.271.8.4093. [DOI] [PubMed] [Google Scholar]
- 26.Lefevre F, Remy MH, Masson JM. Alanine-stretch scanning mutagenesis: A simple and efficient method to probe protein structure and function. Nucleic Acids Res. 1997;25:447–448. doi: 10.1093/nar/25.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Haro M, et al. MBNL1 and CUGBP1 modify expanded CUG-induced toxicity in a Drosophila model of myotonic dystrophy type 1. Hum Mol Genet. 2006;15:2138–2145. doi: 10.1093/hmg/ddl137. [DOI] [PubMed] [Google Scholar]
- 28.Teplova M, Patel DJ. Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat Struct Mol Biol. 2008;15:1343–1351. doi: 10.1038/nsmb.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Datta K, Johnson NP, von Hippel PH. Mapping the conformation of the nucleic acid framework of the T7 RNA polymerase elongation complex in solution using low-energy CD and fluorescence spectroscopy. J Mol Biol. 2006;360:800–813. doi: 10.1016/j.jmb.2006.05.053. [DOI] [PubMed] [Google Scholar]
- 30.Mankodi A, et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 31.Kimura T, et al. Altered mRNA splicing of the skeletal muscle ryanodine receptor and sarcoplasmic/endoplasmic reticulum Ca2+-ATPase in myotonic dystrophy type 1. Hum Mol Genet. 2005;14:2189–2200. doi: 10.1093/hmg/ddi223. [DOI] [PubMed] [Google Scholar]
- 32.Brown RS. Zinc finger proteins: Getting a grip on RNA. Curr Opin Struct Biol. 2005;15:94–98. doi: 10.1016/j.sbi.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 33.Lee MM, et al. Controlling the specificity of modularly assembled small molecules for RNA via ligand module spacing: Targeting the RNAs that cause myotonic muscular dystrophy. J Am Chem Soc. 2009;131:17464, 17472. doi: 10.1021/ja906877y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomlinson A, Ready DF. Cell fate in the Drosophila ommatidium. Dev Biol. 1987;123:264–275. doi: 10.1016/0012-1606(87)90448-9. [DOI] [PubMed] [Google Scholar]
- 35.Warf MB, Diegel JV, von Hippel PH, Berglund JA. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc Natl Acad Sci USA. 2009;106:9203–9208. doi: 10.1073/pnas.0900342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.