Abstract
Identifying protein–protein interactions (PPIs) is essential for understanding various disease mechanisms and developing new therapeutic approaches. Current methods for assaying cellular intermolecular interactions are mainly used for cells in culture and have limited use for the noninvasive assessment of small animal disease models. Here, we describe red light-emitting reporter systems based on bioluminescence resonance energy transfer (BRET) that allow for assaying PPIs both in cell culture and deep tissues of small animals. These BRET systems consist of the recently developed Renilla reniformis luciferase (RLuc) variants RLuc8 and RLuc8.6, used as BRET donors, combined with two red fluorescent proteins, TagRFP and TurboFP635, as BRET acceptors. In addition to the native coelenterazine luciferase substrate, we used the synthetic derivative coelenterazine-v, which further red-shifts the emission maxima of Renilla luciferases by 35 nm. We show the use of these BRET systems for ratiometric imaging of both cells in culture and deep-tissue small animal tumor models and validate their applicability for studying PPIs in mice in the context of rapamycin-induced FK506 binding protein 12 (FKBP12)-FKBP12 rapamycin binding domain (FRB) association. These red light-emitting BRET systems have great potential for investigating PPIs in the context of drug screening and target validation applications.
Keywords: optical imaging, reporter gene
Protein–protein interactions (PPIs) are a prerequisite to most cellular processes, and their pharmacologic control offers promising avenues for therapeutic intervention in a variety of diseases (1). Developing techniques to identify and analyze these transient protein–protein associations is, therefore, of high importance. Several methods are available to investigate PPIs in vitro and in cell culture (2, 3). However, ex vivo analysis of interacting proteins neglects the intricate effects of physiologic and pathophysiologic stimuli that occur in the native microenvironment within living organisms. Similarly, in vitro drug validation ignores the pharmacokinetic properties of prospective pharmacophores. To investigate PPIs in their actual biological context, imaging technologies that enable direct translation of cell culture assays to deep tissues of living subjects are required. Current approaches for monitoring PPIs rely on the two-hybrid system (4), FRET (5), split reporter protein complementation and reconstitution (6–8), and bioluminescence resonance energy transfer (BRET) (9–11). Because most of these PPI detection schemes are optical approaches, signal attenuation by tissues constitutes the primary impediment for studying PPIs in animal models, and it results in low sensitivity for these assays, especially in the context of deep-tissue tumor models.
Bioluminescence-based methods hold particular promise for imaging of PPIs in small living subjects (12, 13). Although both BRET and FRET detection schemes rely on the Förster resonance energy transfer mechanism (14), BRET systems provide enhanced sensitivity in living subjects, because autofluorescence, photobleaching, and tissue attenuation associated with fluorophore excitation are absent. Split luciferase complementation systems are advanced designs for in vivo investigation of PPIs. The luciferase fragments are nonfunctional in the absence of protein association; when the proteins interact, the luciferase regains its reporter function and generates a bioluminescent turn-on signal. However, BRET probes could potentially offer several advantages over split luciferase for interrogating PPIs in living subjects. First, the higher light output of BRET translates into enhanced sensitivity for small animal imaging; generally, split luciferases after complementation exhibit only 20–50% of the activity of corresponding intact luciferase (15). Second, BRET enables real-time two-color ratiometric measurements. Third, false-negative signals associated with misfolding of the reconstituted protein and false-positive signals arising from nonspecific association of the split fragments are significantly reduced for BRET relative to split protein reporters.
Standard BRET systems consist of a luciferase, which in the presence of its bioluminogenic substrate, acts as a resonance energy donor, and a fluorescent protein, which is the resonance energy acceptor. To successfully translate a BRET assay from cell culture to living subjects, it is critical that the BRET system is characterized by efficient energy transfer, excellent spectral resolution, a BRET donor with high bioluminescence quantum yield, and a red light-emitting BRET acceptor. Current BRET systems emit light mostly in the green to yellow region of the visible spectrum (510–570 nm), rendering them suboptimal for imaging in living subjects. Examples of such systems include BRET1 (16) and BRET2 (17), which consist of the donor Renilla reniformis luciferase [RLuc; λem = 480 nm for coelenterazine (CLZ) and λem = 395 nm for DeepBlueC] paired with either YFP (λex/λem = 514/530 nm) or GFP (λex/λem = 400/510 nm), respectively. Firefly luciferase (λem = 565 nm) combined with the red fluorescent protein (RFP) DsRed (λex/λem = 558/583 nm) provides a red-shifted signal; however, this system lacks sensitivity and spectral resolution (18, 19).
Recently, our laboratory has developed a red-shifted BRET system (BRET3) with improved spectral properties (11). BRET3 uses an improved RLuc variant, RLuc8 (20) (λem = 480 nm for CLZ substrate), as the BRET donor and mOrange (λex/λem = 548/564 nm) as the BRET acceptor protein. Because of its excellent spectral resolution (Δλem = 85 nm) and red-shifted emission (λem = 564 nm), BRET3 is currently the most promising BRET system for deep-tissue imaging applications (11). However, the significant tissue attenuation of light emitted at wavelengths < 600 nm (21) limits the application of the BRET3 system to superficial tumors. Here, we describe the development and characterization of greatly improved red light-emitting BRET systems that enable ratiometric measurements in living mice and show unprecedented performance for deep-tissue imaging. Moreover, using these improved BRET systems, we were able to successfully image the drug-mediated interaction between FK506 binding protein 12 (FKBP12) and FKBP12 rapamycin binding domain (FRB) in deep tissues of living mice.
Results
Design of Red Light-Emitting BRET Systems.
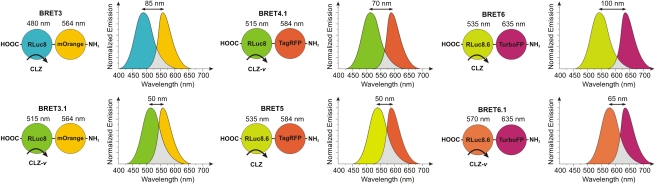
Great efforts have been made in the recent years to obtain brighter, more stable, and red-shifted mutants of the native luciferases (20, 22, 23). The RLuc mutants RLuc8 and red-shifted RLuc8.6 have been previously developed in our laboratory (20). Using these RLuc variants as donors, we generated a series of BRET systems with TagRFP (24) and TurboFP635 (25) as acceptors, two RFP variants that are derived from the WT RFP of the sea anemone Entacmaea quadricolor. These fluorescent proteins are excellent BRET acceptors because of their red light emission (λex/em = 555/584 and 588/635 nm, respectively) and have superior photophysical properties maintained in fusion constructs. In addition to the native CLZ substrate, we used the synthetic luciferases substrate CLZ-v (26–28), which further red shifts the emission maxima of RLuc8 and RLuc8.6 by 35 nm. This additional emission red shift allows for fine tuning of the spectral overlap between the donor emission and acceptor excitation for our BRET systems.
Characterization of the BRET Systems in Cultured Cells.
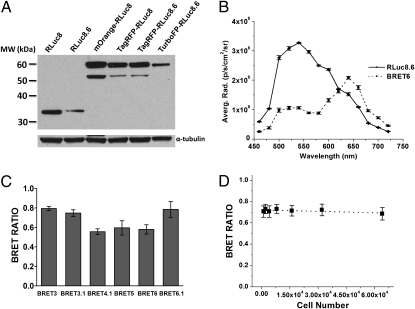
We constructed BRET fusion proteins to mimic the on-state of a BRET sensor system by fusing either RLuc8 or RLuc8.6 luciferases to the acceptors TagRFP or TurboFP635 through an 18-aa spacer (Fig. 1). We created HT1080 cells stably expressing these BRET fusion proteins and confirmed protein expression by Western blot analysis (Fig. 2A). Cells expressing the donor proteins RLuc8 or RLuc8.6 served for comparison and allowed for subtraction of donor-alone signal when calculating BRET ratios. Bioluminescence measurements of reporter cells were performed using appropriate filters, with 20-nm bandwidths for donor and acceptor emission maxima. These measurements provided quantitative assessment of energy transfer efficiency for all BRET systems (as fusion constructs) by calculating the BRET ratio. BRET ratio is defined as the ratio of acceptor and donor bioluminescence emission (A/D) from which A/D obtained with donor alone (RLuc8 or RLuc8.6) is subtracted to account for the spectral overlap of the donor and acceptor emissions (Eq. S1).
Fig. 1.
Schematic representation of the BRET fusion constructs used in this study. The bioluminescence spectra illustrate the emission spectra of the RLuc mutants and the red fluorescent acceptor proteins. All constructs contained an 18-aa linker inserted between the donor and acceptor proteins. Luciferase substrates used in each case were either CLZ or CLZ-v as indicated. Spectral resolution of each system is marked as a bidirectional arrow.
Fig. 2.
Characterization of the designed BRET systems. (A) Western blot analysis of protein expression for all BRET fusions and donor-only proteins contained in lysates of HT1080 cells stably expressing the reporter proteins. (B) Spectral imaging of HT1080 cells expressing either RLuc8.6 or BRET6 using 20-nm filers in the 460–720 nm range. EnduRen luciferase substrate was used for this experiment. Error bars = SD. (C) BRET ratios of the newly designed systems measured in cell culture. HT1080 cells (6.4 × 104), stably expressing each of the BRET fusion and donor-only proteins (RLuc8 or RLuc8.6), were plated in black 96-well plates and imaged on the addition of CLZ or CLZ-v. The graph shows the average BRET ratios calculated as the A/D of the BRET system minus the A/D of donor-only cells. (D) BRET ratios calculated for the BRET6 system using various number of HT1080 cells. Experiments were performed as described in C. The dotted line represents the linear fitting for the data points. Error bars = SEM.
To ensure that both donor and acceptor proteins maintain their characteristics as part of the BRET fusion constructs, we measured their spectral properties in the 460–720 nm range by bioluminescence imaging of cells using an IVIS-200 imaging system equipped with 20-nm bandwidth filters. Fusion of the donor and acceptor proteins induces no significant change to the spectral signature of these proteins (Fig. 2B and Fig. S1). As anticipated, the emission maximum of cells expressing BRET6 was red-shifted to 640 nm (emission of TurboFP) relative to the 540-nm RLuc8.6 signal (Fig. 2B). The BRET ratios for BRET4.1 (0.55 ± 0.02), BRET5 (0.59 ± 0.04), and BRET 6 (0.58 ± 0.02) (Fig. 2C), indicated similar energy transfer efficiencies. BRET6.1 (0.78 ± 0.04) showed slightly higher BRET efficiency than BRET6 but comparable with that of previously reported BRET3 (0.79 ± 0.01). In contrast, the use of CLZ-v (BRET3.1) provided a slightly lower BRET ratio (0.74 ± 0.02) than BRET3 in accordance with lower spectral resolution. Additionally, the calculated BRET ratios were independent of cell number in the given experimental setting, showing the ratiometric characteristic of the measurements (Fig. 2D). These results revealed that BRET6 and BRET6.1 systems hold the greatest potential for in vivo application because of their high BRET efficiencies, unprecedented red wavelength emission (640 nm), and high spectral resolution (100 and 65 nm, respectively).
Deep-Tissue Imaging in Living Mice Using Red Light-Emitting BRET Systems.
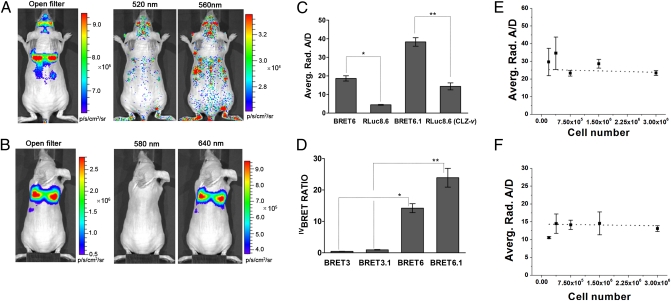
We performed bioluminescence imaging experiments in living mice to investigate the potential of the developed BRET systems to report PPIs in deep tissues. HT1080 cells expressing the BRET fusion proteins were injected through the tail vein in living mice to accumulate predominantly in the lungs and subsequently, were imaged using open and 20-nm bandwidth donor and acceptor filters (Fig. 3 A and B). Acceptor–donor emission ratios obtained from mice experiments using both BRET6 and BRET6.1 were 4.2 ± 0.4- and 2.7 ± 0.4-fold higher (P < 2 × 10−8), respectively, than the A/D values obtained from measurements with the corresponding donor protein alone (Fig. 3C), revealing that BRET signal can be monitored in living mice with high sensitivity. In vivo BRET (ivBRET) ratios were calculated by subtracting the corresponding donor contributions (Fig. 3D). The ivBRET ratios for BRET6 (14 ± 2) and BRET6.1 (24 ± 3) were significantly higher (P < 1 × 10−7) than those ratios of the BRET3 (0.45 ± 0.04) and BRET3.1 (0.95 ± 0.09) systems. The increase in ivBRET ratio was only minimal for BRET5 relative to BRET3.1; BRET4.1 provided an ivBRET ratio similar to BRET3 (Figs. S2 and S3). These findings show that our red light-emitting BRET systems (BRET6 and BRET6.1) have enhanced sensitivity for imaging in deep tissues, giving a brighter signal on all filters, including open filter, than BRET3 or BRET3.1, although the protein expression level of the mOrange-RLuc8 fusion protein was actually higher than the level of the TurboFP635-RLuc8.6 (Fig. 2A). This finding underlines the importance of red light-emitting systems for monitoring PPIs in living subjects.
Fig. 3.
Performance of the BRET systems for deep-tissue imaging in mice. (A and B) Representative bioluminescence images of HT1080 cells stably expressing BRET fusion proteins accumulated in the lungs of nude mice. Cells (3 × 106 in 150 μL PBS) were injected through the tail vein, resulting in significant trapping in the lungs. Mice were injected through the tail vein with luciferase substrate at 1.5 h after cell injection and imaged using sequentially open/donor/acceptor filters. Mice from (A) BRET3.1 (n = 10) and (B) BRET6.1 (n = 10) groups are shown. Average radiance from the thorax region was measured and used for calculating the BRET ratios. Substrate-only control mice (n = 5) were used for background subtraction. CLZ-v substrate was used. (C) Average A/Ds for BRET6, BRET6.1, RLuc8.6 (CLZ), and RLuc8.6 (CLZ-v) calculated from mice imaging experiments. *P = 3.1 × 10−9; **P = 3.5 × 10−10. (D) Average BRET ratios obtained from imaging of mice injected i.v. with cells expressing various BRET fusion proteins. A/D from mice injected with donor-only cells (RLuc8 or RLuc8.6) was subtracted. *P = 1.1 × 10−8; **P = 7.7 × 10−9. (E and F) Average A/Ds calculated from bioluminescence imaging of mice injected with increasing number of cells expressing either RLuc8.6 (n = 4 for each cell number) or BRET6.1 cells (n = 4 for each cell number). CLZ-v was used as substrate. The dotted line represents the linear fit for the data points. Error bars = SEM.
The BRET ratios for BRET6 and BRET6.1 measured in mice experiments largely differ from the BRET ratios obtained in cultured cells. This discrepancy is a result of the considerably higher tissue attenuation of shorter-wavelength light compared with longer-wavelength light (especially >600 nm), which is mainly associated with hemoglobin absorption (21). The different attenuation coefficients lead to an apparent increase of the BRET ratios in mice studies (Discussion). To confirm that the BRET ratio remains constant between cultured cells and mice, we analyzed the imaging results using the double ratio (DR) method, which partially corrects for signal attenuation (29, 30). The resulting DR is a dimensionless parameter independent of the total attenuation coefficient (Eq. S2). DRs of similar values were obtained for experiments performed in mice (0.95 ± 0.09 for BRET3.1 and 2.7 ± 0.4 for BRET6.1) and cell culture (0.74 ± 0.02 for BRET3.1 and 2.08 ± 0.07 for BRET6.1). All BRET ratios and DRs are summarized in Table 1. This agreement between the cell and mice DRs reveals that the mice imaging experiments are ratiometric in nature and that BRET imaging experiments can be confidently performed in deep tissues.
Table 1.
Comparison of BRET ratios and DRs obtained from cell culture and mice imaging experiments
| BRET ratio |
DR |
|||
| BRET system | Cell culture | Mice | Cell culture | Mice |
| BRET3 | 0.79 ± 0.01 | 0.45 ± 0.04 | 2.76 ± 0.04 | 2.5 ± 0.2 |
| BRET3.1 | 0.74 ± 0.02 | 0.95 ± 0.09 | 1.86 ± 0.03 | 2.9 ± 0.2 |
| BRET6 | 0.58 ± 0.02 | 14 ± 2 | 2.43 ± 0.07 | 4.2 ± 0.4 |
| BRET6.1 | 0.78 ± 0.04 | 24 ± 3 | 2.08 ± 0.07 | 2.7 ± 0.4 |
Values are given as mean ± SEM.
Moreover, the A/D measured in mice is independent of the reporter cell number (total bioluminescence signal) for both BRET6.1 (Fig. 3E) and RLuc8.6 (Fig. 3F) as long as the level of donor signal exceeds the background signal associated with spontaneous chemiluminescence of CLZ by at least twofold. The BRET6 system seems to reach the detection limit for <7.5 × 105 injected cells as indicated by the elevated error bars (Fig. 3E). These findings support that quantitative ratiometric measurements can be performed in living mice using the BRET systems described here.
Monitoring Rapamycin-Induced FRB-FKBP12 Association in Living Cells Using BRET6.
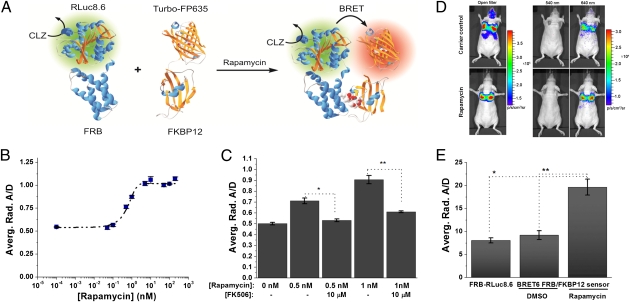
The imaging results obtained with the BRET6 fusion system in mice suggest that this system could provide the required sensitivity for reporting drug-mediated PPIs both in cells and living subjects. To validate the potential of BRET6 for imaging PPIs, we constructed a genetically encoded bimolecular FRB/FKBP12 sensor consisting of FRB-RLuc8.6 and FKBP12-TurboFP635 fusion fragments, whose association is under the control of the macrolide rapamycin. The rapamycin-mediated molecular interaction between the 11-kDa FRB domain of the mammalian target of rapamycin (mTOR) and the 12-kDa FKBP12 has been extensively characterized (31) and is a standard proof of principle system for PPIs assays (8, 32, 33). Rapamycin-induced interaction of FRB and FKBP12 juxtaposes the donor RLuc8.6 and acceptor TurboFP635, eliciting efficient energy transfer measurable as BRET signal (Fig. 4A). Our sensor constructs contained both donor and acceptor proteins at the C terminus of the binding unit (FRB-RLuc8.6 and FKBP12-TurboFP635). A schematic representation and mechanism of detection are shown in Fig. 4A. To characterize this BRET6 FRB/FKBP12 sensor in intact cells, we created HT1080 cells constitutively expressing both sensor subunits. The fusion proteins expression of the sensor components was confirmed by Western blot analysis (Fig. S4). Incubation of HT1080 cells expressing the BRET6 sensor with increasing concentrations of rapamycin generated a strong BRET signal, having higher acceptor to donor ratios than cells treated with carrier control. This result shows that the two components of BRET6 sensor, FRB-RLuc8.6 and FKBP12-TurboFP635, are brought together by the assembly of FRB and FKBP12 in the presence of rapamycin, eliciting BRET. The dependence of the BRET signal on rapamycin concentration followed a dose–response curve with an EC50 value of 0.7 ± 0.2 nM and a maximum BRET induction of 1.9-fold for rapamycin concentrations above 5 nM (Fig. 4B). Moreover, we verified the specificity of the BRET6 sensor by inhibiting the effect of rapamycin on the BRET6 sensor by the addition of FK506, an FKBP binding compound known to compete with rapamycin for FKBP12/FRB interactions (34). A/D calculations revealed that FK506 majorly perturbs the rapamycin-induced FRB/FKBP12 associations in BRET6 sensor cells treated with 1 nM rapamycin by 68% and cells treated with 0.5 nM rapamycin by 75% (Fig. 4C). These findings show that the observed BRET signal is caused by a specific molecular interaction between FRB and FKBP12 and indicate that our bimolecular FRB/FKBP12 BRET6 sensor performs well in cultured cells.
Fig. 4.
Characterization of the genetically encoded FRB-FKBP12 BRET6 sensor. (A) Schematic illustration of the BRET6 sensor for monitoring the rapamycin-induced FRB-FKBP12 association. (B) Rapamycin dose–response curve for BRET6 FRB-FKBP12 sensor in HT1080 cells. HT1080 cells (1 × 105) expressing both FRB-RLuc8.6 and FKBP12-TurboFP635 sensor components were plated in black 24-well plates and incubated with increasing concentrations of rapamycin. After 6 h, the plates were imaged using donor- and acceptor-specific filters on an IVIS-200 imager. A/Ds were calculated and plotted against rapamycin concentration. The data were fitted to a sigmoidal curve fitting (EC50 = 0.7 ± 0.2 nM); error bars = SD. (C) Inhibition of rapamycin-induced FRB-FKBP12 association by FK506. HT1080 cells (1 × 105) were incubated for 6 h with rapamycin (0, 0.5, and 1 nM) and with and without FK506 (10 μM), and they were imaged as described above. A/Ds were calculated for each condition; error bars = SD. *P = 2.2 × 10−3; **P = 4.1 × 10−3. (D) Representative bioluminescence images of HT1080 cells stably expressing FRB-FKBP12 BRET6 sensor accumulated in the lungs of nude mice. Cells (3 × 106 in 150 μL PBS) were injected through the tail vein, resulting in significant trapping in the lungs. One group of mice (n = 8) was injected 2 h before cell injection with 40 μg rapamycin dissolved in 20 μL DMSO and further diluted in 130 μL PBS administered through the tail vein. A second group of mice (n = 8) was injected with DMSO (20 μL in 130 μL PBS). Two hours after cells injection, the mice were injected i.v. with CLZ luciferase substrate and sequentially imaged using open/donor/acceptor filters. Substrate-only control mice (n = 4) were used for background subtraction. (E) Average A/D values for BRET6 FRB-FKBP12 sensor (rapamycin and DMSO-treated groups) and donor-only FRB-RLuc8.6 calculated from mice lung-trapping model imaging experiments; error bars = SEM. *P = 1.7 × 10−4; **P = 2.7 × 10−4.
Imaging Rapamycin-Induced FRB-FKBP12 Association in Living Mice.
Next, we investigated if our red light-emitting BRET6 sensor enables monitoring the rapamycin-induced FRB/FKBP12 association in deep tissues of living mice. We created HT1080 cells expressing both sensor subunits (FRB-RLuc8.6 and FKBP12-TurboFP635) and injected them through the tail vein in nude mice, which led to cell trapping in the lungs. We imaged two groups of mice with 20-nm bandwidth donor (540 nm) and acceptor-specific filters (640 nm) (Fig. 4D). One group of mice received rapamycin through the tail vein (n = 8), and a control group was treated with DMSO (n = 8). An additional group of mice (n = 8) was injected with cells expressing only the FRB-RLuc8.6 donor fragment of the sensor. Representative images of mice treated with rapamycin and DMSO control are shown in Fig. 4D. We calculated A/Ds for all three groups of mice. The results show a clear BRET signal induction based on the high differences in A/Ds obtained between the rapamycin-treated group and the two control groups (Fig. 4E). On average, the rapamycin-treated mice gave 2.4-fold higher A/Ds than the donor-only (FRB-RLuc8.6) control group (P < 3 × 10−4). Moreover, no BRET activation was detected for the DMSO-treated mice control group (Fig. 4E); the A/D was 2.1-fold higher for the rapamycin-treated group than for the DMSO-treated group (P < 2 × 10−4). These results show that specific rapamycin-induced FRB/FKBP12 interactions can be monitored with high sensitivity in deep tissues of mice. The DR obtained from the mice imaging experiments against the DMSO control (2.1 ± 0.3) is within error range of the DR obtained from the rapamycin-induction experiments in cell culture (1.9 ± 0.04). This finding suggests that our measurements are ratiometric and that BRET signal achieves maximum fold induction in both intact cells and mice experiments. Overall, these results underline the advantage of the red light-emitting BRET systems for imaging of PPIs in deep tissues of living subjects.
Discussion
BRET technology offers a straightforward genetically encoded approach to ratiometric imaging of PPIs in cells and living subjects. Being a bioluminescence-based technique, which is excitation- and autofluorescence-free, BRET imaging rapidly extended to studies in living subjects. The ratiometric character of BRET imaging eliminates the need of introducing a second reporter for internal referencing, which is normally needed for split luciferases systems. Previous BRET systems were mainly limited to cell culture assays and s.c. tumors because of their green to yellow light emission, which is strongly absorbed by blood in the highly vascularized tissues of living subjects. Owing to their red light emission, the BRET systems presented here expand the scope of BRET techniques to imaging with high sensitivity in deep tissues of small animals. We have paired red-shifted RLuc variants as BRET donors with suitable RFPs (TagRFP or TurboFP635) based on spectral overlapping. The obtained BRET constructs showed high BRET efficiencies in cell culture and provided ratiometric BRET measurements in living mice.
Light attenuation by tissue constitutes the major difficulty for ratiometric analysis of PPIs by a BRET system. Light attenuation varies with the wavelength of the emitted photons and the depth of the tissue, making the A/D (and ivBRET) a complex function of the emission spectra of the donor and acceptor and spatial localization of the reporter protein. Indeed, the A/Ds and consequently, the ivBRET ratios obtained with BRET6 and BRET6.1 were significantly higher in the mouse deep-tissue model than in cell culture, which is in agreement with lesser tissue attenuation of the light emitted at 640 nm (acceptor) than at 540 nm (donor). However, we have observed excellent consistency of the ivBRET ratio among different mice, showing that our lung cell-trapping model offers sufficient spatial control to retain the ratiometric characteristic of a BRET sensor. Furthermore, when calculating the turn-on ratios for the BRET systems (DRs), the signal attenuation factors cancel out (using the approximation that the attenuation coefficients are constant throughout the entire thorax area and are the same among mice), and these DR values should remain constant, independent of tissue depth. This hypothesis is confirmed by the similar DRs measured for the BRET systems in mice and cells. We observed that DR values obtained in cell culture and mice vary to some extent (although they are close in value), mostly because the DR correction is unable to account for all attenuation and scattering factors. For example, to calculate a dimensionless DR, we assumed that the attenuation coefficient is constant for all mice and identical over the entire thorax area; this approximation introduces a certain degree of inaccuracy in the calculated DR values. Nevertheless, the A/Ds used to calculated the DRs remained in a near margin of the error for different mice, and we show that they are independent of the number of reporter cells used (total light output), indicating that ratiometric measurements can be performed in deep tissues of mice. The DR method provides a depth-independent measure of the BRET signal; however, both donor and acceptor signals used to calculate the DRs decrease with tissue depth. To alleviate some of these tissue attenuation differences, one can further improve the BRET6 system by using both donor and acceptor proteins emitting at wavelengths >600 nm. This parameter could be realized, for example, by pairing the click beetle red luciferase (CBR; λem = 615 nm, d-luciferin) with the newly developed near-IR fluorescent protein eqFP670 (λex/em = 605/670 nm) (35).
To show the potential of these BRET systems for monitoring PPIs in deep tissues of small animals, we generated a bimolecular FRB/FKBP12 system of BRET6, which acts under the control of rapamycin. Our sensor represents a model of a drug-mediated association of two proteins, and we successfully imaged this PPI in deep tissues of mice. The BRET6 FRB/FKBP12 sensor noninvasively reports the rapamycin-induced interaction, which is shown by a significant increase of the A/D in the presence of rapamycin relative to control experiments. This effect is equally detectable in cultured cells and mice with similar DRs. Blocking the rapamycin effect in cells using the well-known inhibitor FK506, we showed, under cell culture conditions, that the observed ratiometric difference reflects a specific interaction between FRB and FKBP12.
Owing to its ability to detect specific interactions in living mice, the BRET6 system provides a valuable platform to monitor PPIs in their native environment. The red-shifted BRET6 sensor can potentially be translated to study PPIs in other cancer models in small animals, especially in mice. Our BRET6 sensor could also be used for BRET imaging in highly vascularized tissues such as the liver; however, further optimization of the reporter's brightness in the desired cells expressing the BRET sensor may be needed to balance for the higher tissue attenuation in the liver. If necessary, wider (50-nm) donor/acceptor bandpass emission filters can be used to increase the signal output.
The ratiometric characteristic of BRET approaches allows for drug screening and mechanism/target validation directly in living subjects. The BRET6 system also compares favorably with other systems for imaging PPIs. For example, a split luciferases system (split CBR), with emission maxima at 615 nm, has been recently reported (36). This system could also enable PPIs imaging in small animal models; however, the reassembled CBR luciferase exhibits a relatively low light output, which may be insufficient for applications requiring sensitive detection of PPIs in deep tissues. A direct comparison between the red light-emitting BRET system and the red light-emitting split luciferase system remains to be assessed. Nonetheless, our red light-emitting BRET systems are optimized for deep-tissue imaging and represent an unprecedented development of RET technologies to monitor PPIs directly in disease models in living subjects.
Materials and Methods
Plasmid construction, cell culturing, transfection, clonal isolation, and Western blot analysis protocols are described in SI Materials and Methods.
Bioluminescence Cell Spectral Imaging.
HT1080 cells expressing various BRET fusion proteins and RLuc8 or RLuc8.6 were plated in 96-well plates in increasing variable numbers (200–60,000). After 24 h, the luciferase substrate (2 μg/well), CLZ (Nanolight Technology), or CLZ-v (Promega) was added. Cells were imaged immediately after the addition of substrate using an IVIS-200 or IVIS-Spectrum imaging system equipped with a cooled charge-coupled device camera (Caliper). Sequential imaging was performed using open/donor/acceptor/open filters. Spectral imaging was performed from 460 to 720 nm in 20-nm increments (each spectral filter used spans for 20-nm wavelength and is denoted by the midpoint); when EnduRen (Promega) was used as substrate, it was added 4 h before imaging. Regions of interest were drawn over each well for all filter sets, and the average radiance was determined. The equations used for calculating BRET ratios and DRs are provided in SI Materials and Methods.
Rapamycin-Induced FRB-FKBP12 Association in Cell Culture.
HT1080 cells stably expressing BRET6 FRB/FKBP12 sensor were plated in black 24-well plates (5 × 104 cells/well) and incubated overnight under standard culture conditions. Next day, rapamycin in increasing concentrations was added (0–200 nM) together with EnduRen. Cells were imaged after 6 h as described above. The same protocol was used for the FK506 inhibition experiment; in addition, some cells were treated with 10 μM FK506 (Cell Signaling Technology).
Bioluminescence Animal Imaging.
All animal handling was performed in accordance with Stanford University's Animal Research Committee guidelines. HT1080 cells constitutively overexpressing a BRET fusion protein, RLuc8 or RLuc8.6, were trypsinized, pelleted, and resuspended in PBS at 3 × 106 cells in 150 μL. The PBS cell suspension was injected through the tail vein in a set of anesthetized (2% isoflurane oxygen) female nude mice (Nu/Nu; Charles River). After cell injection, mice were removed from the anesthesia. CLZ or CLZ-v (35 μg/mouse) diluted in PBS (150 μL total volume) was injected through the tail vein 1.5 h after cell suspension injection. Mice were imaged immediately using an IVIS-200 or IVIS-Spectrum equipped with a charge-coupled device camera. Imaging was performed in sequence luminescence scan mode using either open or 20-nm bandwidth spectral filters appropriate for each donor/acceptor system, with 1 min acquisition time at each filter. Regions of interest were drawn over the mice lungs area for each image. The signal from control mice injected only with the luciferase substrate was subtracted from the total signal. All data were analyzed using Living Image 3.1 software. For experiments using various cell numbers of reporter cells, imaging was preformed as described above. Although the reporter cell number was varied, the total number of cells injected in mice was kept constant (3 × 106) by adding regular HT1080 cells. Detailed procedures for imaging the rapamycin-induced FRB/FKBP12 interaction in mice are given in SI Materials and Methods.
Statistical Analysis.
All cell culture and mice group comparisons were performed using the Student t test (two-sided and paired). Values of P < 0.01 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank T. Doyle for technical assistance with bioluminescence imaging experiments and R. Paulmurugan and J. Ronald for technical advice and assistance with cloning experiments. We thank J. Unch (Promega) for providing the coelenterazine-v substrate. T.F.M. received partial salary support from the National Institutes of Health Research Cambridge Biomedical Research Centre in the United Kingdom. We acknowledge funding support from National Cancer Institute Grants ICMIC P50 CA114747 and CA151459 CCNE-T U54 (to S.S.G.), National Institutes of Health Grant R01 CA082214 (to S.S.G.), and the Canary Foundation (to S.S.G.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100923108/-/DCSupplemental.
References
- 1.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: Progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 2.Guan H, Kiss-Toth E. Advanced technologies for studies on protein interactomes. Adv Biochem Eng Biotechnol. 2008;110:1–24. doi: 10.1007/10_2007_092. [DOI] [PubMed] [Google Scholar]
- 3.Phizicky EM, Fields S. Protein-protein interactions: Methods for detection and analysis. Microbiol Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luker GD, Sharma V, Piwnica-Worms D. Visualizing protein-protein interactions in living animals. Methods. 2003;29:110–122. doi: 10.1016/s1046-2023(02)00285-2. [DOI] [PubMed] [Google Scholar]
- 5.Truong K, Ikura M. The use of FRET imaging microscopy to detect protein-protein interactions and protein conformational changes in vivo. Curr Opin Struct Biol. 2001;11:573–578. doi: 10.1016/s0959-440x(00)00249-9. [DOI] [PubMed] [Google Scholar]
- 6.Massoud TF, Paulmurugan R, De A, Ray P, Gambhir SS. Reporter gene imaging of protein-protein interactions in living subjects. Curr Opin Biotechnol. 2007;18:31–37. doi: 10.1016/j.copbio.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villalobos V, Naik S, Piwnica-Worms D. Current state of imaging protein-protein interactions in vivo with genetically encoded reporters. Annu Rev Biomed Eng. 2007;9:321–349. doi: 10.1146/annurev.bioeng.9.060906.152044. [DOI] [PubMed] [Google Scholar]
- 8.Massoud TF, Paulmurugan R, Gambhir SS. A molecularly engineered split reporter for imaging protein-protein interactions with positron emission tomography. Nat Med. 2010;16:921–926. doi: 10.1038/nm.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De A, Gambhir SS. Noninvasive imaging of protein-protein interactions from live cells and living subjects using bioluminescence resonance energy transfer. FASEB J. 2005;19:2017–2019. doi: 10.1096/fj.05-4628fje. [DOI] [PubMed] [Google Scholar]
- 10.De A, Loening AM, Gambhir SS. An improved bioluminescence resonance energy transfer strategy for imaging intracellular events in single cells and living subjects. Cancer Res. 2007;67:7175–7183. doi: 10.1158/0008-5472.CAN-06-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De A, Ray P, Loening AM, Gambhir SS. BRET3: A red-shifted bioluminescence resonance energy transfer (BRET)-based integrated platform for imaging protein-protein interactions from single live cells and living animals. FASEB J. 2009;23:2702–2709. doi: 10.1096/fj.08-118919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bacart J, Corbel C, Jockers R, Bach S, Couturier C. The BRET technology and its application to screening assays. Biotechnol J. 2008;3:311–324. doi: 10.1002/biot.200700222. [DOI] [PubMed] [Google Scholar]
- 13.Pfleger KD, Eidne KA. Illuminating insights into protein-protein interactions using bioluminescence resonance energy transfer (BRET) Nat Methods. 2006;3:165–174. doi: 10.1038/nmeth841. [DOI] [PubMed] [Google Scholar]
- 14.Förster R. Intermolecular energy transfer and fluorescence. Ann Phys. 1948;437:55–75. [Google Scholar]
- 15.Paulmurugan R, Umezawa Y, Gambhir SS. Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc Natl Acad Sci USA. 2002;99:15608–15613. doi: 10.1073/pnas.242594299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y, Piston DW, Johnson CH. A bioluminescence resonance energy transfer (BRET) system: Application to interacting circadian clock proteins. Proc Natl Acad Sci USA. 1999;96:151–156. doi: 10.1073/pnas.96.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertrand L, et al. The BRET2/arrestin assay in stable recombinant cells: A platform to screen for compounds that interact with G protein-coupled receptors (GPCRS) J Recept Signal Transduct Res. 2002;22:533–541. doi: 10.1081/rrs-120014619. [DOI] [PubMed] [Google Scholar]
- 18.Arai R, Nakagawa H, Kitayama A, Ueda H, Nagamune T. Detection of protein-protein interaction by bioluminescence resonance energy transfer from firefly luciferase to red fluorescent protein. J Biosci Bioeng. 2002;94:362–364. doi: 10.1263/jbb.94.362. [DOI] [PubMed] [Google Scholar]
- 19.Yamakawa Y, Ueda H, Kitayama A, Nagamune T. Rapid homogeneous immunoassay of peptides based on bioluminescence resonance energy transfer from firefly luciferase. J Biosci Bioeng. 2002;93:537–542. [PubMed] [Google Scholar]
- 20.Loening AM, Wu AM, Gambhir SS. Red-shifted Renilla reniformis luciferase variants for imaging in living subjects. Nat Methods. 2007;4:641–643. doi: 10.1038/nmeth1070. [DOI] [PubMed] [Google Scholar]
- 21.Zhao H, et al. Emission spectra of bioluminescent reporters and interaction with mammalian tissue determine the sensitivity of detection in vivo. J Biomed Opt. 2005;10:41210. doi: 10.1117/1.2032388. [DOI] [PubMed] [Google Scholar]
- 22.Loening AM, Dragulescu-Andrasi A, Gambhir SS. A red-shifted Renilla luciferase for transient reporter-gene expression. Nat Methods. 2010;7:5–6. doi: 10.1038/nmeth0110-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Branchini BR, et al. Thermostable red and green light-producing firefly luciferase mutants for bioluminescent reporter applications. Anal Biochem. 2007;361:253–262. doi: 10.1016/j.ab.2006.10.043. [DOI] [PubMed] [Google Scholar]
- 24.Merzlyak EM, et al. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 25.Shcherbo D, et al. Bright far-red fluorescent protein for whole-body imaging. Nat Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 26.Inouye S, Shimomura O. The use of Renilla luciferase, Oplophorus luciferase, and apoaequorin as bioluminescent reporter protein in the presence of coelenterazine analogues as substrate. Biochem Biophys Res Commun. 1997;233:349–353. doi: 10.1006/bbrc.1997.6452. [DOI] [PubMed] [Google Scholar]
- 27.Shimomura O, Musicki B, Kishi Y. Semi-synthetic aequorin. An improved tool for the measurement of calcium ion concentration. Biochem J. 1988;251:405–410. doi: 10.1042/bj2510405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stepanyuk GA, et al. Coelenterazine-v ligated to Ca2+-triggered coelenterazine-binding protein is a stable and efficient substrate of the red-shifted mutant of Renilla muelleri luciferase. Anal Bioanal Chem. 2010;398:1809–1817. doi: 10.1007/s00216-010-4106-9. [DOI] [PubMed] [Google Scholar]
- 29.Sinaasappel M, Sterenborg HJCM. Quantification of the hematoporphyrin derivative by fluorescence measurementusing dual-wavelength excitation and dual-wavelength detection. Appl Opt. 1993;32:541–548. doi: 10.1364/AO.32.000541. [DOI] [PubMed] [Google Scholar]
- 30.Bradley RS, Thorniley MS. A review of attenuation correction techniques for tissue fluorescence. J R Soc Interface. 2006;3:1–13. doi: 10.1098/rsif.2005.0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banaszynski LA, Liu CW, Wandless TJ. Characterization of the FKBP.rapamycin.FRB ternary complex. J Am Chem Soc. 2005;127:4715–4721. doi: 10.1021/ja043277y. [DOI] [PubMed] [Google Scholar]
- 32.Luker KE, et al. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc Natl Acad Sci USA. 2004;101:12288–12293. doi: 10.1073/pnas.0404041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Remy I, Michnick SW. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat Methods. 2006;3:977–979. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- 34.Chen J, Zheng XF, Brown EJ, Schreiber SL. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci USA. 1995;92:4947–4951. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shcherbo D, et al. Near-infrared fluorescent proteins. Nat Methods. 2010;7:827–829. doi: 10.1038/nmeth.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villalobos V, et al. Dual-color click beetle luciferase heteroprotein fragment complementation assays. Chem Biol. 2010;17:1018–1029. doi: 10.1016/j.chembiol.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.