Protein homeostasis within the endoplasmic reticulum (ER) is essential for cell viability, and its misregulation is implicated in a growing number of diseases, including Alzheimer's disease, diabetes, and cancer (1–3). Consequently, the pharmacological manipulation of this pathway is of broad scientific and clinical interest. A variety of drugs have been used to model specific types of stresses that impact protein folding in the ER. One of the most widely used is tunicamycin, a mixture of homologous compounds originally isolated from bacteria and found to have antibiotic and antiviral properties (4). Tunicamycin prevents the first committed step of N-linked glycosylation of proteins by DPAGT1 in the ER (5–7), which causes extensive protein misfolding and activation of the unfolded protein response (UPR). Despite its widespread use in the laboratory, the mechanism by which tunicamycin enters cells has remained a mystery. In PNAS, Reiling et al. (8) investigate the requirements for tunicamycin sensitivity in mammalian cells and identify MFSD2A as a putative plasma membrane transporter.
To identify factors required for tunicamycin-induced death, Reiling et al. (8) use an insertional mutagenesis strategy in the near-haploid human cell line KBM7. This remarkable technique, first reported by Carette et al. (9, 10), can yield complete gene disruptions at targeted loci, whose site of insertion can then be identified by sequencing. Reiling et al. (8) grow a pool of mutagenized KBM7 cells in the presence of tunicamycin to identify factors whose disruption conferred resistance to apoptosis induced by the drug.
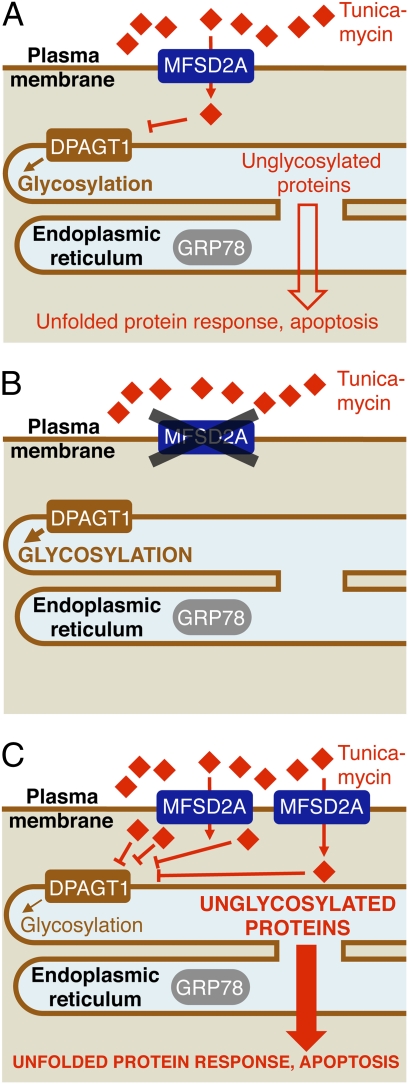
Surprisingly, Reiling et al. (8) identify only a single candidate—MFSD2A—as required for the toxic activity of tunicamycin. MFSD2A is a member of the major facilitator superfamily of proteins, which is the well-known transporter of diverse substrates from bacteria to human (reviewed in ref. 11). Reiling et al. (8) show that MFSD2A is localized to the plasma membrane and that this localization is required for its function, consistent with a role in transport (Fig. 1A). Depletion of MFSD2A by gene disruption or RNAi confers profound resistance to tunicamycin treatment (Fig. 1B), whereas overexpression of WT, but not mutant MFSD2A, leads to tunicamycin sensitivity and increased accumulation of tunicamycin in the cell as assessed by MS (Fig. 1C).
Fig. 1.
Model for tunicamycin uptake through MFSD2A. (A) Results by Reiling et al. (8) suggest that the human MFSD2A gene encodes a plasma membrane transporter that mediates uptake of the antibiotic tunicamycin (TM; red diamonds) into cells. After it is in the cytoplasm, TM inhibits DPAGT1, a protein required for N-linked protein glycosylation in the ER. Inhibition of N-linked protein glycosylation triggers the unfolded protein response and eventually, leads to apoptosis. (B) Reiling et al. (8) identify MFSD2A in a TM resistance screen using insertional mutagenesis in a haploid human cell line. MFSD2A-depleted cells are TM-resistant. Genetic interaction studies reveal that MFSD2A acts upstream of DPAGT1 and the ER chaperone GRP78. Depletion of DPAGT1 or GRP78 sensitizes WT cells to TM; this effect is blocked in MFSD2A-depleted cells. (C) Overexpression of MFSD2A enhances TM uptake and sensitivity, increasing the amount of unglycosylated, misfolded proteins.
How pleiotropic are the effects of MFSD2A deletion? One possible explanation for the ability of MFSD2A deletion to restore growth in the presence of tunicamycin is that the UPR is deficient in these cells. However, UPR induction by other drugs (brefeldin A, thapsigargin, and DTT) remains normal. Furthermore, Reiling et al. (8) analyze genetic interactions to show that MFSD2A function is required for increased sensitivity to tunicamycin induced by shRNA knockdown of the ER protein folding factors DPAGT1 or GRP78. Thus, MFSD2A acts upstream of ER folding in the tunicamycin pathway and more specifically, upstream of tunicamycin's direct drug target DPAGT1.
Why do Reiling et al. (8) find only a single gene whose deletion could confer resistance to tunicamycin? One possibility is that, although deleting other genes could have minimized the effects of tunicamycin (e.g., by increasing chaperone levels or preventing the proper localization of MFSD2A), these genes could be essential, and their complete loss would cause apoptosis. In this case, a complementary approach would be to conduct an RNAi screen to identify genes that, when reduced, would allow cells to survive in the presence of tunicamycin.
The physiological role of MFSD2A is under active investigation. Interestingly, MFSD2A was previously shown to confer fusogenic properties on cells, and it acts as a receptor for the human endogenous retrovirus protein Syncytin-2 on the cell surface (12). This activity is proposed to play a role during trophoblast cell fusion in the formation of the syncytiotrophoblast during placental development.
Are there other functions of the MFSD2A channel? One recent study suggests that MFSD2A may have tumor suppressor function and play a role in matrix attachment (13). The up-regulation of MFSD2A in brown adipose tissue on fasting and cold exposure (14) suggests additional roles, but the identities of native ligands or transport substrates remain to be elucidated.
Beyond its biological findings, the study by Reiling et al. (8) illustrates the usefulness of the insertional mutagenesis approach in a quasi-haploid human cell line pioneered by Carette et al. (9, 10). It is complementary to both RNAi-based approaches, which allow partial knockdown of essential genes, and genetic interaction studies that reveal how individual factors act together in pathways. While the awesome power of yeast genetics made yeast the model system of choice for cell biologists in the 20th century, the recent advances in insertional mutagenesis and RNAi have given 21st century scientists equally powerful genetic tools to investigate human cells directly.
Footnotes
The authors declare no conflict of interest.
See companion article on page 11756.
References
- 1.Ma Y, Hendershot LM. The role of the unfolded protein response in tumour development: Friend or foe? Nat Rev Cancer. 2004;4:966–977. doi: 10.1038/nrc1505. [DOI] [PubMed] [Google Scholar]
- 2.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 3.Zhang K, Kaufman RJ. The unfolded protein response: A stress signaling pathway critical for health and disease. Neurology. 2006;66(Suppl 1):S102–S109. doi: 10.1212/01.wnl.0000192306.98198.ec. [DOI] [PubMed] [Google Scholar]
- 4.Takatsuki A, Arima K, Tamura G. Tunicamycin, a new antibiotic. I. Isolation and characterization of tunicamycin. J Antibiot (Tokyo) 1971;24:215–223. doi: 10.7164/antibiotics.24.215. [DOI] [PubMed] [Google Scholar]
- 5.Heifetz A, Keenan RW, Elbein AD. Mechanism of action of tunicamycin on the UDP-GlcNAc:dolichyl-phosphate Glc-NAc-1-phosphate transferase. Biochemistry. 1979;18:2186–2192. doi: 10.1021/bi00578a008. [DOI] [PubMed] [Google Scholar]
- 6.Keller RK, Boon DY, Crum FC. N-acetylglucosamine-1-phosphate transferase from hen oviduct: Solubilization, characterization, and inhibition by tunicamycin. Biochemistry. 1979;18:3946–3952. doi: 10.1021/bi00585a016. [DOI] [PubMed] [Google Scholar]
- 7.Brandish PE, et al. Modes of action of tunicamycin, liposidomycin B, and mureidomycin A: Inhibition of phospho-N-acetylmuramyl-pentapeptide translocase from Escherichia coli. Antimicrob Agents Chemother. 1996;40:1640–1644. doi: 10.1128/aac.40.7.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reiling JH, et al. A haploid genetic screen identifies the major facilitator domain containing 2A (MFSD2A) transporter as a key mediator in the response to tunicamycin. Proc Natl Acad Sci USA. 2011;108:11756–11765. doi: 10.1073/pnas.1018098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carette JE, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 10.Carette JE, et al. Global gene disruption in human cells to assign genes to phenotypes by deep sequencing. Nat Biotechnol. 2011;29:542–546. doi: 10.1038/nbt.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Law CJ, Maloney PC, Wang DN. Ins and outs of major facilitator superfamily antiporters. Annu Rev Microbiol. 2008;62:289–305. doi: 10.1146/annurev.micro.61.080706.093329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esnault C, et al. A placenta-specific receptor for the fusogenic, endogenous retrovirus-derived, human syncytin-2. Proc Natl Acad Sci USA. 2008;105:17532–17537. doi: 10.1073/pnas.0807413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spinola M, et al. MFSD2A is a novel lung tumor suppressor gene modulating cell cycle and matrix attachment. Mol Cancer. 2010;9:62. doi: 10.1186/1476-4598-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Angers M, Uldry M, Kong D, Gimble JM, Jetten AM. Mfsd2a encodes a novel major facilitator superfamily domain-containing protein highly induced in brown adipose tissue during fasting and adaptive thermogenesis. Biochem J. 2008;416:347–355. doi: 10.1042/BJ20080165. [DOI] [PMC free article] [PubMed] [Google Scholar]