Abstract
Trichomes are universal biological structures originating from the aerial epidermis, which serve as an excellent model to study plant differentiation at the cell level. Although the pathway regulating trichome formation in the Rosids has been well characterized, only very recently a few genes were identified for trichome initiation in the Asterids. In this study, we cloned Woolly (Wo), essential for trichome formation in tomato. Transgenic experiments revealed that the woolly phenotype is caused by the mutation in Wo which encodes a homeodomain protein containing a bZIP motif and a START domain. We identified three alleles of Wo and found that each allele contains a missense mutation, which respectively results in an amino acid substitution at the C terminus. Microarray and expression analysis showed that the expression of a B-type cyclin gene, SlCycB2, is possibly regulated by Wo, which also participates in trichome formation. Suppression of Wo or SlCycB2 expression by RNAi decreased the number of type I trichomes, and direct protein–protein interaction was detected between them, implying that both proteins may work together in the regulation of this type of trichome formation. Cytological observation and Wo transcript analysis in the developing seeds showed that embryo development was also correlated with Wo.
Keywords: cell cycle, multicellular trichome
The development of plants, like all multicellular eukaryotes, lies on the appropriate differentiation of distinct cell types (1). Cell differentiation requires that undifferentiated cells must be chosen before becoming committed to a specified cell (2). In recent years, many scientists have shifted the focus on plant developmental biology from the organization of the whole body or individual organs toward the cell fate determination of the smallest unit of organism, the single cell (3). Plant trichomes, which are found on the aerial epidermal surfaces of nearly all terrestrial plants, provide an excellent model system to study plant development at the single-cell level. Trichomes in the model plant Arabidopsis thaliana are single cells that distribute on the epidermis in a regular pattern (4). With numerous Arabidopsis mutants exhibiting trichome-related phenotypes, molecular studies have enabled the identification of several key regulators participating in trichome initiation. For example, the key gene GLABROUS1 (GL1) controlling trichome formation in Arabidopsis was among the first cloned (5). This gene encodes a protein belonging to the R2R3 MYB family, with two MYB repeats as the DNA binding domain (6). Another important regulator, TRANSPARENT TESTA GLABRA1 (TTG1), also required for trichome formation in Arabidopsis (7), encodes a protein containing four conserved WD repeats (8). Combination of these two regulators with another two basic helix–loop–helix proteins GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) forms a regulatory complex (9). This complex activates trichome formation by enhancing the expression of two targeted genes, GLABRA2 (GL2) and ENHANCER OF GLABRA2 (EGL2) (10, 11). Interestingly, the formation of cotton (Gossypium hirsutum) fibers resembling trichomes in Arabidopsis is controlled by GaMYB2, which shows high sequence homology to GL1 and can restore trichome formation in a gl1 mutant (12), indicating that these two species share a common pathway controlling trichome formation.
Both Arabidopsis and cotton belong to the clades of Rosids (13). Whether there is a similar pathway controlling trichome formation in the clades of Asterids is yet unknown. It was shown that overexpression of MIXTA, an MYB-related regulator controlling conical cell formation in snapdragon (Antirrhinum majus), could trigger trichome formation in A. majus (14) and tobacco (Nicotiana tabacum) (15). Another two MIXTA-like genes also function as activators, inducing trichome differentiation in woody nightshade (Solanum dulcamara) (16) and petunia (Petunia hybrida) (17). However, overexpression of GL1 in tobacco or ectopic expression of MIXTA in Arabidopsis does not induce trichome formation (15). In addition, ectopic expression of MIXTA in gl1-1 Arabidopsis mutant failed to rescue the trichome phenotype (15). These results suggested that trichome formation in the Asterid species such as snapdragon and petunia may engage a different regulatory pathway.
Homeodomain-leucine zipper (HD-Zip) proteins are unique to the plant kingdom, and contain two indispensable conserved domains, HD domain and bZIP motif (18). The former is responsible for DNA binding and the latter for protein–protein interaction (19). Based on gene structures and additional conserved domains, Ariel et al. classified HD-ZIP proteins into four subgroups (HD-Zip I–IV) (20). Each subgroup is involved in different biological processes. Results from Arabidopsis studies demonstrated that members of HD-Zip IV (characterized by a START domain) mainly regulated trichome formation and epidermal cell differentiation, such as GL2 (21) and PROTODERMAL FACTOR2 (PDF2) (22). However, it remains unknown whether trichome formation in tomato is also regulated by the proteins of this subgroup.
Woolly (Wo) is a spontaneous mutation in tomato (Solanum lycopersicum), which is responsible for trichome formation (23) and embryo lethality (24). Here we show that Wo encodes an HD-Zip protein containing a START domain. We further found that a B-type cyclin gene, SlCycB2, also participates in type I trichome formation in tomato, expression of which may be regulated by Wo. In addition, Wo physically interacts with SlCycB2. Our findings not only identify a mechanism for trichome formation in plants, but also shed light on the intrinsic relationship between trichome formation and embryo lethality in tomato.
Results
Map-Based Cloning of Wo.
We have previously mapped Wo to an approximately 200-kb fragment by using an F2 population from a cross between an introgression line 2–3 and woolly mutant LA3186, in which 19 putative ORFs (ORFs 1–19) were predicted (Fig. S1). The predicted protein corresponding to ORF4 showed 73% amino acid sequence identity to a HD-like gene, PDF2, which encodes a transcription factor and regulates the shoot epidermal cell differentiation in Arabidopsis (22). As trichomes originate from the aerial epidermal cells, we considered this ORF (ORF4) as a good candidate for Wo. The allele of Wo in LA3186 is embryo-lethal when homozygous; therefore, this locus remains heterozygous. We amplified and sequenced the genomic and cDNA sequences of this candidate from LA3186, and identified two alleles. One allele is identical to the sequence from the cultivar Ailsa Craig (AC), and the other contains a missense mutation (Fig. S2). This finding further supported the inference that the candidate sequence was the gene responsible for the woolly phenotype.
Because Wo allele in LA3186 is a dominant mutation, we transformed nonwoolly plants with the mutant sequence and expected the transformants to be woolly. We prepared overexpression construct for this candidate gene and introduced this construct into nonwoolly segregants by stable transformation. Finally, we obtained seven independent T0 plants (hereafter referred to as ORF4-NW). Compared with the transgene-negative plants, all these T0 plants exhibited increased trichome density to various degrees. Two of these plants showed typical woolly phenotype, even with much higher trichome density on leaves than LA3186 (Fig. 1A). However, these T0 plants showed varying degrees of deformity, such as plant dwarfism, leaf curvature, flower bud abnormality (Fig. S3), and no fruit setting. In particular, the two T0 plants with typical woolly phenotype never grow up and have no lateral root formation. In addition, we also generated RNAi transgenic plants and found that they showed “no trichome” on the epidermis of all aerial parts (Fig. 1B), strongly suggesting ORF4 to be the candidate for Wo.
Fig. 1.
Transgenic analysis of the candidate ORF4. (A) Trichome phenotype of transgenic plants overexpressing ORF4 (Right, Middle) and negative plants (Left). (B) Trichome phenotype of transgenic plants by RNAi silencing of ORF4 (right two) and negative plants (left two). (Scale bars: 0.15 cm.)
As the ORF4-NW transgenic plants produce no fruit, cosegregation analysis cannot be carried out for them. Therefore, we examined just the trichome phenotype of the resulting T1 family from one RNAi T0 plants, which contain a single copy transgene. A total of 23 T1 plants were obtained, 17 of which were positive and the remaining six of which were negative. All the positive segregants showed no trichome, and negative segregants showed normal phenotype. Such perfect cosegregation between transgene and trichome phenotype confirmed that the ORF4 was the right candidate for Wo.
Trichome Type(s) Controlled by Wo.
Previous studies showed that there were seven trichome types (I–VII) on epidermis of tomato plants (25). To investigate which trichome type(s) causes the phenotypic changes, the trichomes on LA3186, nonwoolly segregants, and RNAi-knockdown plants were observed with SEM. Type I trichome density of LA3186 was much higher than that of the nonwoolly segregants, whereas the density of other trichome types was slightly reduced (Fig. S4). Type I trichomes were characterized by a multicellular base, a long multicellular stalk, and a small glandular tip (25). Trichome clusters of this type of trichomes were also observed at some trichome initiation sites on LA3186 (Fig. S4). However, this type of trichomes were nearly absent from the RNAi-knockdown plants (Fig. S4). Thus, we concluded that type I trichomes accounted for the woolly phenotype and that their formation was regulated by Wo. Different trichome types on the species that produce more than one trichome types are regulated by distinct pathways, such as tobacco and petunia (15, 16, 26). Our conclusion is consistent with this inference.
Intra- and Interspecific Allelic Polymorphism at Wo.
The predicted coding sequence of Wo was 2,193 bp based on the annotated genomic sequence of the cultivar Heinz 1706. The protein encoded by this sequence consisted of 730 aa. Through genomic and cDNA sequence comparison of Wo, we identified 10 exons and 9 introns. As mentioned earlier, there was one nucleotide substitution in the predicted coding sequence of Wo− (designated Wo− in LA3186), which caused one amino acid substitution from Pro-635 to Arg at its C terminus (Fig. S5). Besides LA3186, there are several other woolly mutants maintained in our laboratory, such as LA0053 (Wo−), LA1531 (Wov), MF802 (unknown), LA0258 (Wom), and LA1908 (Womz). According to the Tomato Genetics Resource Center (http://tgrc.ucdavis.edu), these mutants have different trichome phenotypes. To determine whether there were mutations in the allelic sequences of Wo from these mutants, we sequenced and aligned these alleles. By using the cDNA sequence of wo as the reference, all of these alleles contain one nucleotide substitution in their 3′ portion of the coding region, respectively (Fig. S6). Alleles of Wo in LA0053 and MF802 are identical with Wo−, including a nucleotide substitution at 1,904 bp downstream of the predicted translation initiation site (TIS). Alleles of Wo in LA1531 and LA0258 are identical and homozygous at this locus, including a nucleotide substitution at 1,700 bp downstream of the predicted TIS, which led to an amino acid substitution from Arg-567 to Leu (Fig. S5). Womz in LA1908 contains a point mutation at 2,075 bp downstream of the predicted TIS, resulting in an amino acid change from Ile-692 to Thr (Fig. S5). These data indicate that different mutant sites in these alleles result in different trichome phenotypes.
The predicted Wo protein is a member of the HD gene family, which contains a bZIP motif and a START domain (Fig. S5). We further aligned the amino acid sequences encoded by these alleles with 11 homologues from other species, which were all annotated as HD proteins at the National Center for Biotechnology Information. Whereas the homologous gene in Vitis vinifera (XP_002266688) had a high amino acid sequence identity with Wo (75%), the one in Gossypium hirsutum (AAQ16126) had only a 46% sequence identity with Wo. The remaining nine homologues showed approximately 46% to 75% sequence identity to Wo. Notably, the sites of the mutation within their C terminus described earlier are highly conserved among these homologues from other species (Fig. S5), suggesting that these mutant sites may be functionally important.
Subcellular Localization and Expression Pattern.
To investigate the subcellular localization of Wo, we fused the coding sequence of Wo with that of EGFP driven by the CaMV 35S promoter (35S::Wo:EGFP) and bombarded the construct into onion epidermal cells. Transient expression of this construct showed that the fusion protein Wo-EGFP localized to the nucleus and membrane (Fig. S7C).
To examine the spatial expression pattern of Wo, we conducted RT-PCR analysis with total RNA extracted from several tissues of LA3186 and its nonwoolly segregants, including young stems, functional leaves, shoot apexes, flowers, green fruits, and roots. Wo is expressed in all tested tissues, being highest in the shoot apexes (Fig. S7A). We further carried out RNA in situ hybridization and Wo promoter-driven GUS-YFP transformation to analyze sites of Wo expression. The RNA in situ hybridization showed that the transcripts of Wo were mainly localized in the shoot apical meristem, leaf primordia, and epidermis of the young stem (Fig. S7B). There was no obvious difference in its expression pattern between LA3186 and its nonwoolly segregants, indicating that the woolly phenotype may be caused by amino acid substitution in Wo rather than its transcriptional changes. No phenotypic change in transgenic plants overexpressing wo cDNA under control of the CaMV 35S promoter also support this inference. GUS was strongly expressed in shoot apexes, axillary meristems, young stems, and adaxial young leaves (Fig. S8A). In addition, we also observed high GUS activity in the developing embryos (Fig. 2B) and the gland cells of type VII trichomes (Fig. 3B), and high YFP activity in the lateral root primordia (Fig. S8B). The expression pattern of Wo is consistent with its pleiotropic effects, which not only regulate trichome formation, but also participate in some other development processes, such as embryo development, lateral root differentiation, and even possibly synthesis of secondary metabolites in the type VII trichomes.
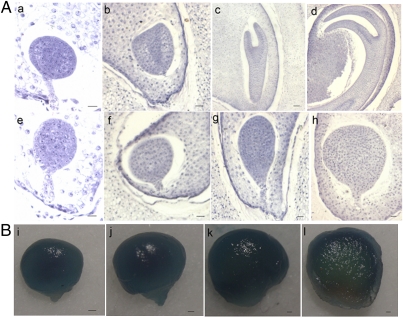
Fig. 2.
Observation of the embryo development in LA3186 fruits (A) and GUS activity in the developing seeds driven by Wo promoter (B). Upper: Normal embryos. Lower: Dying embryos: 7 DPA, a, e and i; 9 DPA, b, f, and j; 15 DPA, c, g, and k; and 23 DPA, d, h, and i. (Scale bars: 0.14 mm.)
Fig. 3.
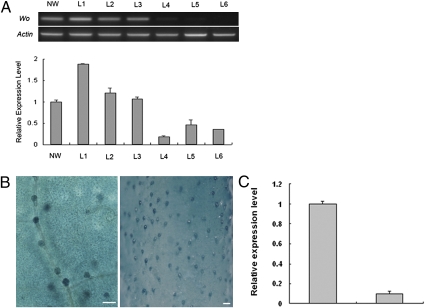
Expression of Wo and SlCycB2. (A) Upper: Expression of Wo in several transgenic lines by using semiquantitative RT-PCR; Lower: Expression of SlCycB2 in these transgenic lines by using quantitative RT-PCR. NW, transgenic negative plants with LA3186 nonwoolly segregants as the recipients; L1, L2, L3, transgenic lines overexpressing Wo; L4, L5, L6, transgenic lines suppressing Wo. (B) Wo promoter-GUS expression in trichomes on the leaf epidermis (Left) and young stem (Right). (Scale bars: 0.45 mm.) (C) Expression of SlCycB2 in the SlCycB2-RNAi transgenic plants (Right) and negative plants (Left) with LA3186 as the recipient by using quantitative RT-PCR. Data are displayed as the ratio of expression to tomato actin, given as mean ± SE (n = 3).
Identification of SlCycB2 as a Potential Target of Wo.
Because Wo is a transcription factor and can activate the formation of type I trichomes, each with a multicellular stalk, this suggests that it may promote the formation of this type of trichomes by controlling the expression of cell cycle-related genes. To test this hypothesis, we compared the transcriptomes of LA3186 and its nonwoolly segregants by using tomato TOM2 microarrays. To minimize the possible exaggerated difference caused by subsequent development, we extracted total RNA from the plantlets with only one true leaf for microarray hybridization. From this analysis, we detected approximate 150 genes with elevated transcript levels in LA3186 (Table S1). Many of these up-regulated genes participate in secondary metabolites synthesis, such as flavonol synthase (27), cytochrome P450 (28), and GST (29). As we expected, a unigene SGN-U226950 similar to AT5G06270.1, a hypothetical B-type cyclin (CYCB) in Arabidopsis, was found. CYCBs regulate the transition from gap 2 (G2) to mitosis (M) (30). Ectopic expression of CYCLIN B1;2 in trichomes can induce mitotic divisions and result in the formation of trichome clusters and multicellular trichomes (31). As leaves of LA3186 have obviously increased numbers of trichome clusters and multicellular trichomes (type I), we speculated that SGN-U226950 (referred to as SlCycB2) may function downstream of Wo and thereby regulate trichome formation in the tomato.
We analyzed the expression of SlCycB2 in young leaves of LA0258, LA1908, and the Wo transgenic plants by using quantitative RT-PCR. SlCycB2 expression was also up-regulated in these two allelic lines (Fig. S9), and more importantly, it was significantly higher in Wo-overexpressing transgenic plants but lower in Wo-RNAi plants (Fig. 3A). Moreover, we investigated the expression pattern of SlCycB2 in different tissues of LA3186 by using RT-PCR. The analysis revealed no significant differences in these tissues between SlCycB2 and Wo (Fig. S9). These results further indicated that SlCycB2 may be regulated by Wo. Next, we down-regulated SlCycB2 by RNAi in LA3186 and found that the RNAi lines showed obvious decrease in trichome density (Fig. 4). SEM observation showed that the type I trichomes nearly disappeared, and the type V-like trichomes increased, most of which have many branches (Fig. 4). We therefore speculated that Wo promotes type I trichome formation by up-regulating the expression of SlCycB2, which may induce a shift from endoreduplication to mitosis.
Fig. 4.
Trichome phenotype of SlCycB2-RNAi transgenic positive (Right) and negative plants (Left). Trichome of total plants (A), young stems (B), old stems (C), magnified view of leaflet boxed in a (D), and SEM observation of leaf trichomes (E). (Scale bars: 0.5 cm.)
Interaction Between Wo and SlCycB2.
It has been shown that HD functions in DNA binding and bZIP in protein–protein interaction (19). Therefore, first, we detected the cis-element of the SlCycB2 promoter sequence by using the PLACE program (http://www.dna.affrc.go.jp) and found an 11-bp DNA motif (CTAATTGTTTA) encompassed in the promoter region, which has been demonstrated as a cis-element responsible for direct binding with HD-containing proteins (32). To examine whether SlCycB2 may be a direct target of Wo, we tested the binding activity of Wo with this motif by using a yeast one-hybrid assay. The cotransformants could not grow on the SD/-Leu/AbA 50 ng/mL medium, whereas the transformed positive control exhibited normal growth. This result indicated that Wo had no binding activity with this motif, suggesting that SlCycB2 may not be regulated by promoter binding with Wo.
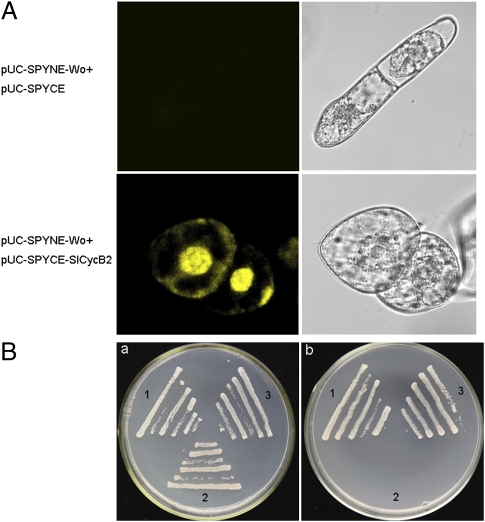
Subsequently, we used bimolecular fluorescence complementation (BiFC) to determine whether there was protein–protein interaction between Wo and SlCycB2. We transformed the tobacco BY-2 (N. tabacum cv. Bright Yellow 2) cells with sets of pUC-SPYNE-Wo/pUC-SPYCE-SlCycB2 and pUC-SPYNE-Wo/pUC-SPYCE. A strong YFP fluorescence was observed when pUC-SPYNE-Wo was coexpressed with pUC-SPYCE-SlCycB2, whereas no signal was detected in the control cells (Fig. 5A). This result suggested that Wo interacts with SlCycB2. To further confirm the interaction between Wo and SlCycB2, we performed yeast two-hybrid experiment. The transformants harboring pGBKT7-Wo and pGADT7-SlCycB2 were able to grow on SD/-Ade/-His/-Leu/-Trp medium, whereas the positive and negative controls showed the expected results (Fig. 5B). These data, together with the BiFC results, demonstrated that Wo can physically interact with SlCycB2.
Fig. 5.
Wo interacts with SlCycB2 in vivo. (A) Interaction between Wo and SlCycB2 in BiFC assays. YFP fluorescence was detected only when pUCSPYNE-Wo was coexpressed with pUCSPYCE-SlCycB2. (B) Interaction of Wo and SlCycB2 in transformed AH109 cells grown on SD/-Leu/-Trp (a) and SD/-Ade/-His/-Leu/-Trp (b). 1, pGBKT7-53+pGADT7-RecT (positive control); 2, pGBKT7-Lam+pGADT7-RecT (negative control); 3, pGBKT7-Wo+pGADT7-SlCycB2.
Embryo Lethality in Wo Homozygous Seeds.
Embryo lethality occurs when Wo becomes homozygous in LA3186. To detect at which stage the defect is noticeable, we harvested and fixed seeds from the developing fruits of LA3186 from 5 d postanthesis (DPA) to 23 DPA, sequentially stained them with hematoxylin, and sectioned them. No obvious abnormality in embryo was observed until 7 DPA. Embryos at 7 DPA were arrested at the globular stage and stopped further development, and subsequently the cell number increased; on the contrary, normal embryos continued to differentiate into mature embryos (Fig. 2A). Our observation was partly consistent with an earlier report that WoWo embryos ceased to grow before any organogenesis (24). To detect whether there is a direct cause–consequence relationship between Wo and embryo lethality, we analyzed the temporal expression of Wo in the developing embryo. GUS activity maintained a high level in the 7-DPA embryo and gradually weakened (Fig. 2B), suggesting Wo transcription is associated with embryo lethality.
Discussion
Genetic analysis in Arabidopsis has established a regulatory pathway that controls trichome initiation. Although trichome structures of different species are convergent, they are regulated by independent networks (13). Exploration of the mechanisms involved in trichome formation in tomato can provide new insights into understanding the diversity of cell differentiation. In this study, we showed several lines of evidence to suggest that type I trichome formation is controlled by Wo in tomato. GL2, which was placed in the pathway inducing trichome initiation in Arabidopsis (9, 21), showed much lower identities with Wo than a shoot epidermal cell differentiation-related gene PDF2 with it. The closest homologue of Wo in Arabidopsis (PDF2) has not been shown to be involved in trichome formation (22), suggesting functional diversification among Wo (PDF2) genes in different plant species. This also suggested that trichome formation in tomato may be regulated by a mechanism distinct from that characterized in Arabidopsis. Moreover, it would be interesting to see whether the Wo homologues in other species of the Asterid division such as petunia, tobacco, and snapdragon are also involved in trichome formation.
Wo contains three conserved domains: a HD domain, a bZIP motif, and a START domain. The HD domain is involved in DNA binding and the bZIP motif is involved in protein homo- or heterodimerization (19). Interestingly, all the amino acid substitutions identified among Wo alleles occurred at their C terminus outside these three domains. Thus, it can be deduced that functional differentiation during these alleles is not caused by any defect in likely protein–DNA and protein–protein interactions. In addition, because Wo functions as a transcription factor, it could be inferred that these point mutations might play various roles in the regulation of its downstream genes, showing the functional diversity. It is worth noting that the polymorphic sites in Wo—amino acids 567, 635, and 692—are highly conserved in Wo homologues from a wide range of organisms. Arg-567 (hydrophilic) is replaced by Leu (hydrophobic) in Wom, Pro-635 (hydrophobic) is replaced by Arg (hydrophilic) in Wo−, and Ile-692 (hydrophobic) is replaced by Thr (hydrophilic) in Womz. These mutations may change the stability and activity of the proteins. However, how these residues are functionally implicated in the trichome formation and embryo development remains to be elucidated.
HD-Zip transcription factors are supposed to act as activators of many downstream target genes controlling specific developmental processes. However, the majority of these potential target genes are still not known. We showed that SlCycB2, a B-type cyclin gene, also participates in type I trichome formation. The SlCycB2 promoter contains an 11-bp motif that has been identified as a cis-element recognized by homeodomain proteins (32). However, there was no direct interaction between Wo and this motif, indicating that Wo may not regulate SlCycB2 expression by binding to this element. Suppression of either of them by using RNAi resulted in an obvious decrease of the number of type I trichomes, and protein–protein interaction was detected by BiFC and yeast two-hybrid assays, demonstrating that they may work together in the regulation of this type of trichome formation. In addition, SlCycB2 was up-regulated in woolly mutants and Wo-overexpressing transgenic plants but down-regulated in Wo-RNAi plants, suggesting that SlCycB2 may be regulated by Wo. However, whether and how SlCycB2 expression was regulated by Wo through direct interaction between their encoded proteins remain unknown. This possible regulatory model distinct from that characterized in Arabidopsis, may be a unique mechanism underlying trichome formation. It is also conceivable that Wo controls other developmental processes through regulation of different targets.
B-type cyclins function at the G2/M transition during mitosis. Enhancement of their expression could induce the shift from endoreduplication to mitosis. Therefore, it is easy to understand that the Wo gene results in a cell number increase in dying embryos by activating the expression of SlCycB2. However, it is still not known how SlCycB2 induces the formation of type I trichome. Schnittger el al. found that CYCLIN B1;2 expression under control of GL2 promoter can induce mitotic divisions, resulting in trichome cluster and multicellular trichome formation (31). There was no significant similarity between SlCycB2 and CYCLIN B1;2, but there was a 53% sequence identity between SlCycB2 and AT5G06270.1, a hypothetical B-type cyclin. It demonstrated that SlCycB2 has just a similar function to CYCLIN B1;2, but is absolutely a distinct gene. Trichome branch number is correlated with DNA content in Arabidopsis (33). Formation of multibranch trichomes in SlCycB2-RNAi transgenic plants indicated that suppression of SlCycB2 expression can activate a shift from mitosis to endoreduplication. Consequently, we inferred that a dramatic increase in type I trichome number may be attributed to the enhancement of mitosis in the epidermal cells destined to become trichomes.
Future studies will be directed to study whether and how Wo activates SlCycB2 expression through protein–protein interaction between their predicted proteins and then type I trichome formation, to determine whether the amino acid polymorphism in Wo mutant alleles correlates with functional differentiation, and to investigate how Wo may control different developmental processes. As Wo expression in homozygous status is associated with embryo lethality, it will be interesting to dissect the likely distinct roles of Wo and SlCycB2 in trichomes formation and embryo development.
Materials and Methods
Plant Materials.
Tomato genotypes LA3186, LA0053, LA0258, LA1531, and LA1908 are woolly mutants, and were provided by the Tomato Genetics Resource Center. AC and MF802 are commercial cultivars.
Sequence Alignment.
We amplified (with the primers in Table S2) and sequenced the genomic and cDNA sequences of ORF4 from LA3186, LA0053, LA0258, LA1531, LA1908, MF802, and AC. These sequences were aligned with ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2) and edited with GeneDoc.
Transgenic Analysis.
Full-length coding sequences of two alleles corresponding to ORF4 (amplified with the primers in Table S2) were inserted into the pMV2 vector, which we rebuilt in our laboratory based on pHellsgate 8 vector under control of the CaMV 35S promoter. Suppression expressions of Wo and SlCycB2 were performed by using RNAi vector pHellsgate 2, which were ligated with the fragment of Wo and SlCycB2 (amplified with the primers in Table S2) by using Clonase BP reaction (Invitrogen). Both constructs of Wo were transformed into nonwoolly plants segregated from LA3186 and the construct of SlCycB2 into LA3186, mediated by Agrobacterium tumefaciens strain C58. These empty vectors were also transformed as controls. Transgene copy number was detected by Southern blot hybridization. T1 populations obtained from the T0 plants harboring a single-copy transgene were amplified for the neomycin phosphotransferase II gene fragment for segregation analysis (primers in Table S2).
Transcript Test.
Total RNA was extracted with TRIzol reagent (Invitrogen) and converted into first-strand cDNA by using PrimeScript reverse transcriptase (TaKaRa). RT-PCR was carried out to amplify an approximately 400 bp fragment of Wo with 31 cycles using the first-strand cDNA as a template. In addition, actin was amplified with 24 cycles as an internal control. Quantitative RT-PCR was performed in a total volume of 20 μL containing 8 μL of the first-strand cDNA, 8 μL SYBR Green I Master Mix (Roche), and 1 μL gene-specific primers (10 μM/μL) with the Roche LightCycler 480 system by an initial incubation of 10 min at 50 °C, followed by 45 cycles of 10 s at 95 °C, 15 s at 58 °C, and 20 s at 72 °C. Relative gene expression was calculated with Microsoft Excel. All primers used in this analysis are listed in Table S2.
Subcellular Localization and Tissue Distribution.
To analyze the cellular localization of Wo (allele in LA3186), we created a CaMV 35S::Wo-EGFP fusion construct, and bombarded it into onion epidermal cells using Biolistic PDS-1000 (Bio-Rad). Samples were observed with a Leica TCSST2 confocal laser microscope. We cloned a 3.5-kb Wo endogenous promoter region upstream of the start codon from LA3186 genomic DNA (amplified with the primers in Table S2) to construct the promoter Wo:GUS-YFP fusion vector. Transformants carrying this construct were obtained as described earlier. These transformants were submerged in a solution containing 25 mM phosphate buffer (NaH2PO4 and Na2HPO4), 1.25 mM K3[Fe(CN)6], 1.25 mM K4[Fe(CN)6], 0.25% Triton X-100, 0.5 mM EDTA, and 1.25 mg/mL X-Gluc. After vacuum-infiltrating for 3 to 5 min, the transformants were incubated at 37 °C overnight. Chlorophyll was removed after staining by incubation in 75% ethanol, and YFP fluorescence was observed by confocal microscopy.
RNA in Situ Hybridization.
We amplified a 453-bp fragment from the cDNA of woolly plants with the primers in Table S2 for an RNA probe and cloned it into the pEASY-T3 cloning vector (TransGen). Sense and antisense RNA probes were synthesized from linearized pEASY-T3 plasmid by using T7 and SP6 RNA polymerase with a DIG RNA Labeling Kit (SP6/T7; Roche). Probe sensitivity was checked with a comparison of labeled probes and labeled control RNA with the dot blot method. Paraffin sections of shoot apexes and young stems of LA3186 and its nonwoolly segregants were prepared by using the methods described by Narváez-Vásquez and Ryan (34).
SEM Observation.
Young leaves and stems were fixed with 2% glutaraldehyde for approximately 24 h. After washing in a 0.1 M cacodylate buffer, these samples were dehydrated in a graded ethanol series, dried in a desiccator (HCP-2; Hitachi), and coated with a film of gold. Observations were carried out on a JSM-6390/LV scanning electron microscope.
Microarray Analysis.
To compare the transcriptomes between LA3186 and its nonwoolly segregants, triplicate RNA samples with five seedlings of each line were isolated from the young tomato shoots with only one true leaf and sent for microarray hybridization by tomato TOM2 oligo microarray (CapitalBio). Unigenes with a false discovery rate (35) less than 0.05 and a fold change greater than or equal to two were identified as differentially expressed genes. We convert these unigenes to corresponding probe ID and then carry out probe annotation at the Tomato Expression Database (http://ted.bti.cornell.edu/). Some unannotated unigenes were further analyzed at the National Center for Biotechnology Information.
Yeast One-Hybrid Assay.
Yeast one-hybrid assay was carried out by using the Matchmaker Gold Yeast One-Hybrid Library Screening System (Clontech). Oligonucleotides (33 bp) with three tandem repeats of the 11-bp DNA motif CTAATTGTTTA from the SlCycB2 promoter was cloned into pAbAi. Plasmid was linearized and transformed into Y1HGold. Positive yeast cells were then transformed with pGADT7-AD, which contains Wo. The DNA–protein interaction was determined based on the growth ability of the cotransformants on SD/-Leu medium with 0 to 100 ng/mL aureobasidin A.
BiFC Assay.
The full-length coding sequence of Wo and SlCycB2 (without their stop codons) were cloned into pUC-SPYNE and pUC-SPYCE, respectively. Plasmids were bombarded into tobacco BY-2 cells, and YFP fluorescence was imaged after incubation at 24 °C for 18 h as described earlier. The tobacco BY-2 cells containing pUC-SPYNE-Wo and pUC-SPYCE were also analyzed as control.
Yeast Two-Hybrid Assay.
For yeast two-hybrid analysis, we created the constructs pGBKT7-Wo and pGADT7-SlCycB2 by inserting Wo and SlCycB2 into pGBKT7 and pGADT7 vectors separately. The two plasmids were cotransformed into Saccharomyces cerevisiae strain AH109. Simultaneously, we also transformed pGBKT7-53 and pGADT7-RecT as positive control, and pGBKT7-Lam and pGADT7-RecT as negative control, which were provided with the BD Matchmaker library construction and screening kits. The transformants were then tested on SD/-Leu/-Trp and SD/-Ade/-His/-Leu/-Trp mediums.
Paraffin Section.
All seeds in the developing fruits (7 to 23 DPA) of LA3186 were harvested and fixed in formalin/acetic acid/alcohol for 24 h, and then stained in hematoxylin, dehydrated with each grade of ethanol, infiltrated with paraffin with chloroform used as the solvent, and gradually embedded with paraffin. Then, the specimens were cut into 8- to 10-μm sections and mounted on microscope slides. For removal of paraffin, slides were immersed in xylene (twice for 20 min) and enveloped with neutral resin. The finished slides were cured in a 42 °C oven until dry. Photomicrographs were taken by using an Olympus microscope.
Supplementary Material
Acknowledgments
We thank Drs. H. Kuang, S. Xiao, and L. Peng for critical reading of this manuscript and suggestions. We thank Dr. C. Chen for technical assistance. We are grateful to the Tomato Genetic Resource Center for supplying the tomato seeds of the woolly mutants. This work was supported by 973 Project Grant 2011CB100600, National Natural Science Foundation Grant 30971997, and China Agricultural Research System Grant CARS-25-A-02.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100532108/-/DCSupplemental.
References
- 1.Schiefelbein J. Cell-fate specification in the epidermis: a common patterning mechanism in the root and shoot. Curr Opin Plant Biol. 2003;6:74–78. doi: 10.1016/s136952660200002x. [DOI] [PubMed] [Google Scholar]
- 2.Larkin JC, Marks MD, Nadeau J, Sack F. Epidermal cell fate and patterning in leaves. Plant Cell. 1997;9:1109–1120. doi: 10.1105/tpc.9.7.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hülskamp M. Plant trichomes: A model for cell differentiation. Nat Rev Mol Cell Biol. 2004;5:471–480. doi: 10.1038/nrm1404. [DOI] [PubMed] [Google Scholar]
- 4.Pesch M, Hülskamp M. Creating a two-dimensional pattern de novo during Arabidopsis trichome and root hair initiation. Curr Opin Genet Dev. 2004;14:422–427. doi: 10.1016/j.gde.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Marks MD, Feldmann KA. Trichome development in Arabidopsis thaliana. I. T-DNA tagging of the GLABROUS1 gene. Plant Cell. 1989;1:1043–1050. doi: 10.1105/tpc.1.11.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell. 1991;67:483–493. doi: 10.1016/0092-8674(91)90523-2. [DOI] [PubMed] [Google Scholar]
- 7.Koornneef M. The complex syndrome of ttg mutants. Arabidopsis Inf Serv. 1981;18:45–51. [Google Scholar]
- 8.Walker AR, et al. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell. 1999;11:1337–1350. doi: 10.1105/tpc.11.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao M, Morohashi K, Hatlestad G, Grotewold E, Lloyd A. The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development. 2008;135:1991–1999. doi: 10.1242/dev.016873. [DOI] [PubMed] [Google Scholar]
- 10.Larkin JC, Brown ML, Schiefelbein J. How do cells know what they want to be when they grow up? Lessons from epidermal patterning in Arabidopsis. Annu Rev Plant Biol. 2003;54:403–430. doi: 10.1146/annurev.arplant.54.031902.134823. [DOI] [PubMed] [Google Scholar]
- 11.Serna L. A network of interacting factors triggering different cell fates. Plant Cell. 2004;16:2258–2263. doi: 10.1105/tpc.104.160931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S, et al. Control of plant trichome development by a cotton fiber MYB gene. Plant Cell. 2004;16:2323–2334. doi: 10.1105/tpc.104.024844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serna L, Martin C. Trichomes: different regulatory networks lead to convergent structures. Trends Plant Sci. 2006;11:274–280. doi: 10.1016/j.tplants.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 14.Glover BJ, Perez-Rodriguez M, Martin C. Development of several epidermal cell types can be specified by the same MYB-related plant transcription factor. Development. 1998;125:3497–3508. doi: 10.1242/dev.125.17.3497. [DOI] [PubMed] [Google Scholar]
- 15.Payne T, Clement J, Arnold D, Lloyd A. Heterologous myb genes distinct from GL1 enhance trichome production when overexpressed in Nicotiana tabacum. Development. 1999;126:671–682. doi: 10.1242/dev.126.4.671. [DOI] [PubMed] [Google Scholar]
- 16.Glover BJ, Bunnewell S, Martin C. Convergent evolution within the genus Solanum: the specialised anther cone develops through alternative pathways. Gene. 2004;331:1–7. doi: 10.1016/j.gene.2004.01.027. [DOI] [PubMed] [Google Scholar]
- 17.Avila J, Nieto C, Cañas L, Benito MJ, Paz-Ares J. Petunia hybrida genes related to the maize regulatory C1 gene and to animal myb proto-oncogenes. Plant J. 1993;3:553–562. doi: 10.1046/j.1365-313x.1993.03040553.x. [DOI] [PubMed] [Google Scholar]
- 18.Schena M, Davis RW. HD-Zip proteins: members of an Arabidopsis homeodomain protein superfamily. Proc Natl Acad Sci USA. 1992;89:3894–3898. doi: 10.1073/pnas.89.9.3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee YH, Chun JY. A new homeodomain-leucine zipper gene from Arabidopsis thaliana induced by water stress and abscisic acid treatment. Plant Mol Biol. 1998;37:377–384. doi: 10.1023/a:1006084305012. [DOI] [PubMed] [Google Scholar]
- 20.Ariel FD, Manavella PA, Dezar CA, Chan RL. The true story of the HD-Zip family. Trends Plant Sci. 2007;12:419–426. doi: 10.1016/j.tplants.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Rerie WG, Feldmann KA, Marks MD. The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 1994;8:1388–1399. doi: 10.1101/gad.8.12.1388. [DOI] [PubMed] [Google Scholar]
- 22.Abe M, Katsumata H, Komeda Y, Takahashi T. Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development. 2003;130:635–643. doi: 10.1242/dev.00292. [DOI] [PubMed] [Google Scholar]
- 23.Shilling PR. An investigation of the hereditary character, woolly in the tomato. Ohio J Sci. 1959;59:289–302. [Google Scholar]
- 24.Huang PC, Paddock EF. The time and site of the semidominant lethal action of ‘Wo’ in Lycopersicon esculentum. Am J Bot. 1962;49:388–393. [Google Scholar]
- 25.Luckwill LC. Aberdeen, UK: Aberdeen Univ; 1943. The genus Lycopersicon: A historical, biological and taxonomic survey of the wild and cultivated tomatoes. PhD thesis. [Google Scholar]
- 26.Goodspeed TH. The genus Nicotiana. In: Verdoorn F, editor. Chronica Botanica. Waltham, MA: Chronica Botanica; 1954. pp. 102–131. [Google Scholar]
- 27.Owens DK, et al. Functional analysis of a predicted flavonol synthase gene family in Arabidopsis. Plant Physiol. 2008;147:1046–1061. doi: 10.1104/pp.108.117457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang E, et al. Suppression of a P450 hydroxylase gene in plant trichome glands enhances natural-product-based aphid resistance. Nat Biotechnol. 2001;19:371–374. doi: 10.1038/86770. [DOI] [PubMed] [Google Scholar]
- 29.Vanhaelen N, Haubruge E, Lognay G, Francis F. Hoverfly glutathione S-transferases and effect of brassicaceae secondary metabolites. Pestic Biochem Physiol. 2001;71:170–177. [Google Scholar]
- 30.Ishida T, Kurata T, Okada K, Wada T. A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol. 2008;59:365–386. doi: 10.1146/annurev.arplant.59.032607.092949. [DOI] [PubMed] [Google Scholar]
- 31.Schnittger A, Schöbinger U, Stierhof YD, Hülskamp M. Ectopic B-type cyclin expression induces mitotic cycles in endoreduplicating Arabidopsis trichomes. Curr Biol. 2002;12:415–420. doi: 10.1016/s0960-9822(02)00693-0. [DOI] [PubMed] [Google Scholar]
- 32.Korfhage U, Trezzini GF, Meier I, Hahlbrock K, Somssich IE. Plant homeodomain protein involved in transcriptional regulation of a pathogen defense-related gene. Plant Cell. 1994;6:695–708. doi: 10.1105/tpc.6.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perazza D, et al. Trichome cell growth in Arabidopsis thaliana can be derepressed by mutations in at least five genes. Genetics. 1999;152:461–476. doi: 10.1093/genetics/152.1.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narváez-Vásquez J, Ryan CA. The cellular localization of prosystemin: a functional role for phloem parenchyma in systemic wound signaling. Planta. 2004;218:360–369. doi: 10.1007/s00425-003-1115-3. [DOI] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. JR Stat Soc. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.