Abstract
Due to the lack of relevant animal models, development of effective treatments for human mitochondrial diseases has been limited. Here we establish a rapid, yeast-based assay to screen for drugs active against human inherited mitochondrial diseases affecting ATP synthase, in particular NARP (neuropathy, ataxia, and retinitis pigmentosa) syndrome. This method is based on the conservation of mitochondrial function from yeast to human, on the unique ability of yeast to survive without production of ATP by oxidative phosphorylation, and on the amenability of the yeast mitochondrial genome to site-directed mutagenesis. Our method identifies chlorhexidine by screening a chemical library and oleate through a candidate approach. We show that these molecules rescue a number of phenotypes resulting from mutations affecting ATP synthase in yeast. These compounds are also active on human cybrid cells derived from NARP patients. These results validate our method as an effective high-throughput screening approach to identify drugs active in the treatment of human ATP synthase disorders and suggest that this type of method could be applied to other mitochondrial diseases.
Keywords: budding yeast, drug screening, transcription profiling, NARP cybrid
Although our understanding of the molecular mechanisms underlying mitochondrial diseases has considerably improved, the development of effective treatments has still been extremely limited (1). The insufficiency of relevant disease models in conjunction with the absence of high-throughput drug screening assays may, at least in part, explain this failure.
Among these disorders, some are associated with primary deficiencies in the mitochondrial ATP synthase (2), an enzyme that catalyzes the final steps of mitochondrial ATP production. To date, seven point mutations in the mitochondrial ATP6 gene encoding subunit a of ATP synthase have been associated with a group of maternally inherited neurodegenerative syndromes with onset in early infancy (1, 3), including NARP (neuropathy, ataxia, and retinitis pigmentosa). In diseases resulting from mutations in mitochondrial DNA (mtDNA), wild-type and mutated mtDNA coexist in patient mitochondria, a characteristic called heteroplasmy. The severity of the disease depends on the proportion of mutant alleles, with a minimal critical proportion of ∼70% for penetrance of a clinical phenotype; this property is known as the threshold effect (4, 5).
The budding yeast Saccharomyces cerevisiae serves as a good model for mitochondrial disease because (i) mitochondrial genes and function are particularly well conserved from yeast to human (1); (ii) yeast are genetically tractable; and (iii) yeast have the ability to survive either by fermentation or by respiration, where only the latter requires oxidative phosphorylation (OXPHOS). This last point is particularly beneficial because mutant strains in which ATP synthase activity is impaired can be easily maintained on fermentable media (e.g., glucose); therapeutic strategies can then be tested on media where respiration is required (e.g., glycerol, ethanol, or lactate). In addition, along with Chlamydomonas reinhardtii (6), yeast is the only eukaryote in which site-directed mutagenesis of the mitochondrial genome has been established (7). Because of the natural instability of heteroplasmy in yeast, homoplasmic populations in which 100% of mitochondria contain mutated mtDNA can be easily generated. Thus, yeast models of the five most common ATP6 mutations found in NARP patients (T8993G, T8993C, T9176G, T9176C, and T8851C) have been generated and characterized (8–11). Other patients exhibiting ATP synthase deficiency have been found to carry mutations in two nuclear genes, ATP12 (12) and TMEM70 (13), which encode proteins that are required for ATP synthase assembly. An appropriate yeast model of such disorders is the deletion mutant for the nuclear gene FMC1 that encodes a protein required at high temperatures (35–37 °C) for assembly of the F1 sector of ATP synthase (14). When the fmc1Δ mutant is grown at high temperatures, its mitochondria contain far fewer assembled ATP synthase complexes than a wild-type (WT) strain, whereas the ones that assemble are fully functional. This heterogeneity is also found in patients with decreased levels of ATP synthase due to ATP12, TMEM70, or heteroplasmic ATP6 mutations. Therefore, the fmc1Δ mutant constitutes an appropriate model of these disorders.
In this study, we establish a two-step screening assay designed to identify drugs active against inherited ATP synthase disorders modeled in yeast. In the primary screen, ∼12,000 compounds from various chemical libraries were tested for their ability to suppress the respiratory growth defect of the fmc1Δ mutant. In the secondary screen, active compounds were tested on the five yeast atp6-NARP mutants. Our screen identified chlorhexidine (CH) and oleate (OA); further experiments confirmed that they improve various respiratory phenotypes of both the fmc1Δ and NARP mutants. Dihydrolipoic acid (DHLA), which has previously been reported as active against mitochondrial encephalopathies and is currently being tested in patients (15, 16), was also active in our yeast-based method. Moreover, we show that CH, OA, and DHLA are effective in a human cybrid-based model of NARP. These results validate our yeast-based approach as a method for identifying compounds with potential to treat inherited mitochondrial diseases affecting ATP synthase.
Results
Development of a Yeast-Based Screen for Drugs That Suppress NARP Phenotypes.
The fmc1Δ mutant and the five yeast atp6-NARP mutants exhibit growth defects on glycerol-based medium at 35 and 37 °C (Fig. 1A). At 35 °C, the temperature used for the screening assay, the different mutants present growth defects of varying severity, with the atp6-NARP T9176G, T8851C, and T8993G mutants (the latter being the most frequent mutation in human) exhibiting the most severe phenotypes. Our measurements of mitochondrial ATP synthesis in these mutants (Fig. 1A Right) correlate well with their fitness on glycerol. They also confirm previous observations that defects in ATP synthesis must be severe (estimated at ∼80% or more; refs. 8 and 17) before a clear growth defect on respiratory medium can be observed in yeast. Even more significant is the fact that these observations also correlate with what is known about the relative severity of the corresponding atp6-NARP mutations in human (2, 18, 19), which likely reflects a high level of evolutionary conservation within the region of subunit a affected by these mutations. These observations constitute a preliminary validation of the use of yeast to model human inherited mitochondrial diseases affecting ATP synthase. Because the fmc1Δ strain displays an intermediate respiratory growth phenotype and is a good model of heteroplasmy, we selected it for our primary screen.
Fig. 1.
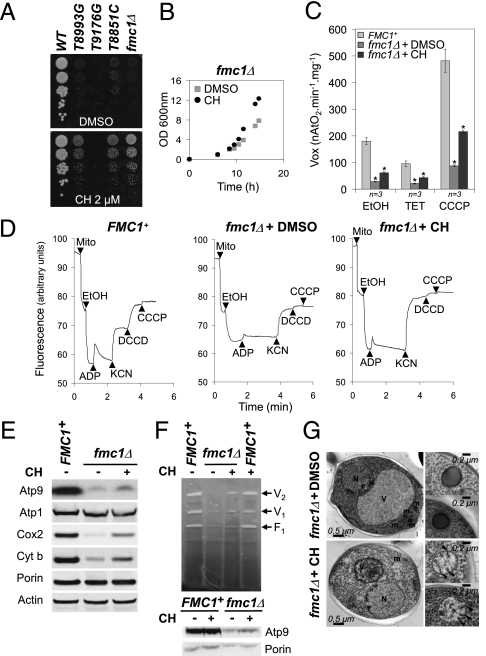
Development of a yeast-based screening assay for NARP-related diseases and identification of CH. (A) Serial dilutions of the WT strain, five NARP mutants, and the fmc1Δ strain were spotted onto glucose and glycerol plates. The plates were incubated at the indicated temperature for 3 d (glucose) or 7 d (glycerol). Rates of ATP synthesis relative to WT are indicated in the right-hand column. (B) Candidate screen. The fmc1Δ strain was spread onto rich glycerol plates. Small sterile filters were then placed on the agar surface and DHLA or OA (chemical structures depicted at Right) were added to the filters at the indicated quantities. The plates were then incubated at 35 °C for 7 d. (C) Screening assay carried out as described in B except that DMSO was added to the upper left filter (negative control, −) and DHLA to the bottom right filter (positive control, +). At the remaining positions, compounds from the chemical libraries were added, and plates were incubated for 7 d at 35 °C. (D) The improvement of fmc1Δ growth obtained with CH is shown, and its molecular structure is depicted. (E) The dose-dependent effect of CH on three yeast mutants is shown.
We next tested candidate compounds that could hold therapeutic potential for mitochondrial disease in our yeast-based assay. We selected DHLA because it has already been used as a treatment for patients presenting mitochondrial encephalopathies (15, 16). We also selected OA, a fatty acid known to induce expression of the mitochondrial Odc1p protein (20), a carrier for various Krebs cycle intermediates encoded by the ODC1 gene in yeast (21). We have previously isolated ODC1 as a multicopy suppressor of the respiratory growth defect of the fmc1Δ strain (22). By using our simple assay described below, we found that both DHLA and OA partially suppress, in a dose-dependent manner, the growth defect of the fmc1Δ strain (Fig. 1B). The fact that DHLA, a compound displaying therapeutic benefits for patients affected by mitochondrial encephalopathies, also suppresses the respiratory growth phenotype of the fmc1Δ strain further validates our yeast-based method.
Primary Screen of Chemical Libraries Using Yeast-Based Assay.
We then performed our primary screen by testing 12,000 compounds from various chemical libraries for their ability to suppress the respiratory growth defect of the fmc1Δ mutant strain. Similarly to an assay we previously developed (23, 24), fmc1Δ cells are spread on solid glycerol medium and exposed to filters spotted with the compounds. Active compounds are then identified by a halo of enhanced growth around a filter (example in Fig. 1C Right). The advantage of this method is that, in one simple experiment, it allows numerous compounds to be tested across a large range of concentrations due to diffusion of the drugs in the growth medium. This design improves the sensitivity of the screen drastically because many compounds (including OA and CH, see below) are toxic at high concentrations (Fig. 1B and C). The positive hits obtained were then tested in a secondary screen by using the yeast atp6-NARP mutants in the same experimental procedure.
Our screen used, among others, the Prestwick Chemical Library, a collection of drugs for which bioavailability and toxicity studies have already been carried out in humans; therefore, active compounds from this library can directly enter drug optimization programs. The percentage of active compounds in our primary screen was quite low (only ∼10 of 12,000 molecules tested, corresponding to <0.1%), indicating that the screening assay is specific and stringent.
Identification of CH.
Among the hits from our primary screen was CH, an antiseptic compound from the Prestwick Chemical Library (Fig. 1D). The secondary screen (Fig. 1E) showed that, in addition to its activity in the fmc1Δ strain, CH elicits a dose-dependent partial suppression of the respiratory growth defects of the atp6-NARP T8993G and T8851C strains. In contrast, CH had no visible effect on the atp6-NARP T9176G strain, which is the most severe NARP mutation in both human and yeast (Fig. 2A). The severity of this phenotype is due to an almost complete lack of incorporation of subunit a into ATP synthase (10), whereas substantial residual mitochondrial ATP synthesis is observed in the mutants that are rescued by CH (11).
Fig. 2.
CH suppresses multiple phenotypes of yeast models of ATP synthase disorders. (A) Serial dilutions of the WT strain, three NARP mutants, and the fmc1Δ strain were spotted onto glycerol plates supplemented with CH (2 μM final concentration) or the equivalent quantity of DMSO (− control). The plates were incubated at 35 °C for 7 d. (B) Growth curves of the fmc1Δ strain grown at 35 °C in liquid galactose medium supplemented with 1.25 μM CH (the optimal concentration) or DMSO. (C) Mean respiration rates of the WT and fmc1Δ strains treated in vivo with 1.25 μM CH or DMSO were determined using three different substrates as indicated. Error bars represent SD. * represents significant difference compared with untreated cells (P = 0.05, one-sided Wilcoxon rank test). (D) Energization of the mitochondrial membrane was determined by rhodamine 123 fluorescence quenching with intact mitochondria from the fmc1Δ strain treated in vivo with 1.25 μM CH (Right) or DMSO (Center), and the WT isogenic strain (FMC1+; Left). The contents were added as follows: mitochondrial proteins (Mito), ethanol (EtOH), ADP, potassium cyanide (KCN), DCCD, and CCCP. (E) SDS-PAGE and Western blot analysis of mitochondrial proteins from the mitochondria used in D. Porin was used as a loading control. (F) BN-PAGE, ATPase activity, and Western blot analysis of extracts from isolated mitochondria. V2 and V1 indicate the dimeric and monomeric forms of the F1F0 ATP synthase complex, respectively; F1 indicates the free F1 particles. (G) Ultrastructure electron micrographs of fmc1Δ cells grown at 35 °C in presence of 1 μM CH (Lower) or DMSO as control (Upper). Arrowheads indicate mitochondrial cristae.
CH Rescues Multiple Mitochondrial Defects in fmc1Δ Cells.
To characterize the effects of CH, we used the fmc1Δ strain because it responds best to the drug. The fmc1Δ strain exhibits a low rate of ATP synthase assembly and low levels of respiratory complexes III and IV when grown at high temperatures (35-37 °C; ref. 25). In addition, its mitochondria are devoid of cristae and contain large inclusion bodies composed mostly of aggregated, unassembled α and β subunits of the F1 moiety of ATP synthase. Finally, its inner mitochondrial membrane energization and respiration rate are impaired. We thus set out to determine whether CH treatment could rescue these phenotypes. For growth in liquid, we selected galactose as a carbon source to allow faster growth of the mutant strains while retaining their mitochondrial activity; although galactose is a fermentable substrate, unlike glucose it does not elicit repression of mitochondrial biogenesis. In addition, the growth defect of fmc1Δ yeast as well as its rescue by CH could be clearly observed in liquid galactose medium (Fig. 2B).
Oxygen consumption rates.
Using ethanol as an electron donor, we measured the oxygen consumption levels of WT (FMC1+) and fmc1Δ whole cells grown at 35 °C in rich galactose medium with or without CH. In the absence of CH, fmc1Δ cells exhibited respiration activity six times lower than WT; treatment with CH resulted in a significant (2.2-fold; P = 0.05, SI Methods) increase in respiration (Fig. 2C). We also measured respiration in the presence of triethyltin (TET), an inhibitor of ATP synthase (basal or state 4 respiration), and the mitochondrial membrane uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP), i.e., in conditions where respiration is maximal (uncoupled state). In these conditions, treatment of fmc1Δ cells with CH also resulted in significantly improved respiration rates (2.1- and 2.5-fold, respectively; P = 0.05; Fig. 2C).
Respiratory enzyme abundance.
The respiration data suggest that CH treatment increases the amount of respiratory enzymes in fmc1Δ cells, which we confirmed by Western blot analysis (Fig. 2E). We observed a partial restoration of steady-state levels of complex III–V subunits except for Atp1p, whose abundance is unaffected by the FMC1 deletion as previously observed (14). In contrast, CH treatment did not affect the amount of respiratory enzymes in WT cells (Fig. S1).
Energization of mitochondrial membrane.
We assessed the proton pumping activity of isolated mitochondria prepared from WT (FMC1+) and fmc1Δ cells grown with or without CH using the fluorescent membrane potential (ΔΨ)-sensitive dye rhodamine 123 (26). Consistent with their low oxygen consumption and our previous results (14), mitochondria from untreated fmc1Δ cells were poorly energized with ethanol relative to WT, whereas those from fmc1Δ cells grown in the presence of CH were energized almost as efficiently as WT mitochondria (Fig. 2D). In WT mitochondria, a further addition of ADP, as expected, led to a transient decrease of fluorescence quenching, reflecting the use of ΔΨ by ATP synthase to phosphorylate the added ADP. Because of the low levels of ATP synthase in fmc1Δ mitochondria, the addition of ADP had little effect on membrane potential. In contrast, a significant ΔΨ decrease was induced by ADP addition in mitochondria isolated from CH-treated fmc1Δ cells. This observation reflects a higher level of ADP phosphorylation in mitochondria from fmc1Δ cells upon CH treatment.
ATP synthesis rates.
We measured ATP synthesis rates of mitochondria isolated from both WT and fmc1Δ cells grown with or without CH, using NADH as a respiratory substrate and an excess of external ADP. In good agreement with the partial suppression of the respiratory growth phenotype, we observed a modest but reproducible effect of CH on ATP synthesis rates in fmc1Δ cells, whereas CH had almost no effect on WT mitochondria (Table S1).
Blue native (BN)-PAGE and ATPase activity.
We evaluated the effect of CH on both assembly and activity of ATPase in isolated mitochondria from both fmc1Δ and WT cells grown with or without CH. The BN-PAGE and ATPase activity stain demonstrate that CH treatment led to a significant increase in fully assembled ATP synthase in the fmc1Δ strain (Fig. 2F).
Mitochondrial morphology.
We evaluated the effect of CH on the mitochondrial morphology of fmc1Δ cells using electron microscopy. Consistent with previous observations (25), 77% of cell sections of fmc1Δ cells grown at 35 °C display matrix-localized, electron-dense inclusion bodies consisting of ATP synthase subunits α and β. Strikingly, this proportion was reduced to 6% when fmc1Δ cells were grown in the presence of CH (Fig. 2G and Table S2). Moreover, mitochondrial cristae were clearly discernible in some of these cells, whereas they were completely absent in untreated fmc1Δ cells. The restoration of cristae is consistent with the CH-induced increase in oxygen consumption and respiratory chain subunits, because cristae allow higher amounts of respiratory enzymes to be assembled within mitochondria.
Transcription Profile of fmc1Δ Strain and Its Response to CH Treatment.
To investigate the global effects of CH on cellular function in the fmc1Δ mutant, we carried out genome-wide comparative analyses of the transcriptional responses to the FMC1 gene deletion and the addition of the drug using high-resolution tiling microarrays (ref. 27; Fig. 3A). Overall, 336 of 5,446 expressed genes [defined in SI Methods; results in Dataset S1 and available from ArrayExpress (details in SI Methods)] showed at least 1.5-fold differential expression in response to the deletion of FMC1 (Fig. 3A, triangles). We analyzed these genes according to Gene Ontology (28) and transcription factor targets (ref. 29; SI Methods; results in Dataset S2). As expected, these genes are mostly related to mitochondrial respiration; in particular, the down-regulated genes are enriched for subunits of the respiratory chain complexes (30/47; P = 6 × 10−17; P values corrected for multiple testing; SI Methods) and targets of the respiratory gene expression activator Hap4p (25/38; P = 5 × 10−24). Additionally, enrichment for targets of the transcription factors Rtg3p (5/31; P = 8 × 10−2) and Gcn4p (15/121; P = 5 × 10−3) indicate that the retrograde pathway, a transcriptional program that responds to mitochondrial dysfunction by modulating metabolism, was activated in the fmc1Δ mutant (30–32). Finally, iron homeostasis was also perturbed in the fmc1Δ strain (11/38; P = 6 × 10−7).
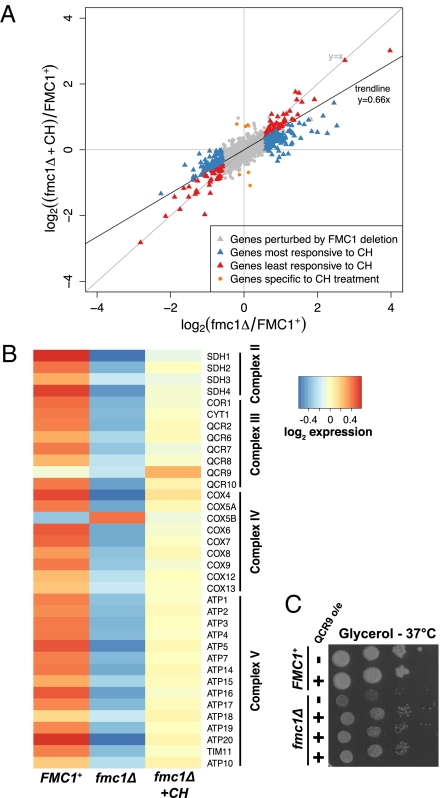
Fig. 3.
Transcription profiles of fmc1Δ, fmc1Δ + CH, and WT (FMC1+). (A) Scatterplot of log2-fold changes in gene expression of CH-treated (y axis) and untreated (x axis) fmc1Δ yeast relative to WT in rich galactose medium at 35 °C. CH treatment induces a genome-wide shift toward WT expression levels of ∼1/3 as evidenced by the trendline y = 0.66x. (B) Expression of genes encoding respiratory chain complex subunits. Log2-scale normalized expression values were centered for each gene by subtracting the mean expression level from all values per gene. Subunits show a general pattern of down-regulation in fmc1Δ yeast relative to WT and partial rescue by CH treatment. (C) Overexpression of the QCR9 gene in the fmc1Δ strain (bottom three rows, compared with the fmc1Δ strain in third row and WT strain in first two rows) results in partial recovery of growth on rich glycerol medium at a nonpermissive temperature.
The drug CH induced a partial rescue of the fmc1Δ mutant at the transcriptional level, with gene expression fold changes compared with WT reduced genome-wide by ∼1/3 (Fig. 3A; trendline y = 0.66x, where complete rescue would be ∼0 and no drug effect would be ∼1). Almost all genes perturbed in the fmc1Δ mutant responded positively to CH treatment, although to varying extents. The most responsive genes (perturbed genes whose fold change relative to WT was reduced by >1/3; Fig. 3A, blue), included all 30 respiratory chain complex subunits and 23/25 (P = 3 × 10−21) of the Hap4p targets down-regulated in the mutant; they were also enriched for genes involved in cristae formation (3/4; P = 10−3). Accordingly, signals of the retrograde response largely disappeared after CH treatment. Among the genes least responsive to CH treatment (perturbed genes whose response to the drug was lower than the genome-wide trend; Fig. 3A, red) were those involved in iron homeostasis. The small number of genes (six) specific to CH treatment (those whose treatment-induced expression was significantly different from both other conditions) and their lack of functional enrichments suggests that the drug has no major side effects unrelated to the mutant phenotype.
The transcriptional behavior of the respiratory chain subunits provides important insight into the regulatory impact of CH on the fmc1Δ mutant (Fig. 3B). Among the subunits, there was a clear pattern of down-regulation in the fmc1Δ mutant relative to WT, with CH treatment recovering expression to intermediate levels (corroborating the measurements of protein levels; Fig. 2E). The only genes that did not follow this pattern were COX5B (encoding an “anaerobic” isoform of cytochrome c oxidase subunit 5) and, more interestingly, QCR9, encoding a component of cytochrome bc1 complex, whose expression was up-regulated beyond WT levels by CH treatment. The unique response of QCR9 prompted us to investigate whether overexpression of this gene alone was sufficient to suppress the respiratory growth defect of the fmc1Δ strain. Indeed, a partial rescue was observed in fmc1Δ cells overexpressing the QCR9 gene (Fig. 3C). These findings suggest that QCR9 could be a key regulatory target for determining the cellular levels of bc1 complex (Discussion). Together, our transcription data correspond well to our biochemical data and strongly indicate that the main effect CH has on the fmc1Δ strain is improvement of its respiratory function, in which QCR9 may play a role. Notably, WT cells displayed a very limited response to CH at the transcriptional level that included neither activation of respiratory pathways nor up-regulation of QCR9 expression (Dataset S1). These results suggest that CH does not act via a general transcriptional induction of respiratory pathways; rather, it requires specific conditions to exert its beneficial effects (Discussion).
Drugs Active in the Yeast-Based Assay are also Active in Human NARP Cells.
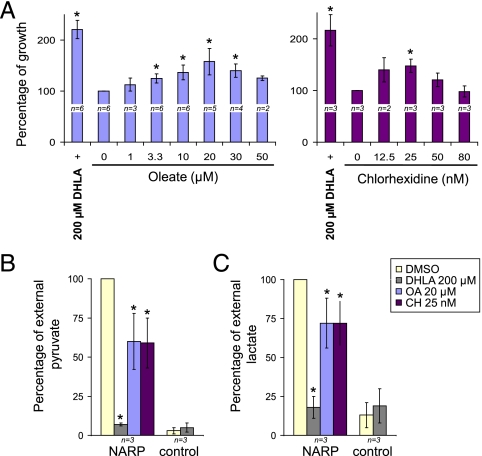
We next tested the compounds that were active in our yeast-based assay in a human cell-based model of NARP syndrome; in particular, we used a cybrid-based model nearly homoplasmic for the NARP T8993G mutation (33). To encourage the cells to rely on OXPHOS rather than glycolysis, we used glucose-deprived medium (34), in which the NARP cybrids exhibit a significant growth defect (33). Strikingly, DHLA, CH, and OA all significantly increased the growth rate of NARP cybrids (Fig. 4A). In agreement with its effects in mitochondrial encephalopathy patients, treatment with 200 μM DHLA increased the survival of NARP cybrid cells by 2.2-fold, recovering their survival to nearly the rate measured in control cybrids (Fig. 4A). Remarkably, CH (Fig. 4A Right) and OA (Fig. 4A Left) treatments resulted in a clear dose-dependent improvement of NARP cybrid survival (the maximal increases were 1.5- and 1.6- fold, respectively). As in yeast, however, CH did not affect growth of WT cybrids (Fig. S2). The improved survival of the NARP cybrids suggests that DHLA, OA, and CH improve OXPHOS production of ATP in these cells.
Fig. 4.
DHLA, CH, and OA are active in a human cybrid-based model of NARP syndrome. NARP and control cybrids were grown in glucose-deprived medium with DHLA, CH, and OA at the indicated final concentrations or with the equivalent quantity of DMSO as a control (0). (A) Percentage of growth after 3 d in glucose-deprived medium containing OA or CH at the indicated concentrations, compared with treatment with DHLA at 200 μM final concentration (+), a compound being tested in clinical trials for the treatment of mitochondrial encephalopathies. * represents conditions with growth significantly different from in DMSO (P < 0.05; Methods). (B) Mean percentage of external pyruvate in NARP cybrids treated with DHLA, CH, OA (left-hand side of graph) compared with control cybrids (right-hand side of graph). Error bars represent SD. * represents significant difference compared with the DMSO condition (P = 0.01). (C) Mean percentage of external lactate as in B.
As one of the main symptoms of NARP-related mitochondrial disorders is lactic acidosis, which is most likely due to a shift toward glycolytic metabolism, it would be particularly valuable if these compounds could reduce glycolysis. Indeed, CH, OA, and DHLA effected a reduction in extracellular levels of the glycolytic byproducts pyruvate and lactate in NARP cybrids (Fig. 4B and C), comparable to their amelioration of growth: DHLA reduced external pyruvate and lactate to the levels observed in control cybrids, and OA and CH each reduced pyruvate by ∼40% and lactate by ∼30% (P = 0.01; Methods). These observations demonstrate that DHLA, OA, and CH reduce glycolysis in NARP cybrids, indicating that they induce a metabolic shift toward OXPHOS.
Discussion
Our results validate the use of a yeast-based assay to identify compounds potentially active in the treatment of inherited mitochondrial diseases caused by ATP synthase deficiency. Firstly, there is a strong correlation between the severity of mutations in patients and the respiratory defects caused by the homologous mutations in yeast. Secondly, DHLA, a compound previously found to be active against mitochondrial encephalopathies in humans, is also active in yeast. Thirdly, CH and OA, two compounds identified by our yeast-based assay, are also active in human cybrids derived from NARP patients.
>CH has remarkable suppressor activity in our yeast model (fmc1Δ) of ATP synthase assembly disorders, with a substantial (more than twofold) increase in respiration due to higher amounts of respiratory complexes III and IV. Consistent with this result, the fmc1Δ mutant recovered an effective capacity to energize the inner mitochondrial membrane upon CH treatment. Another striking effect of CH is the restoration of mitochondrial cristae and elimination of matrix-localized inclusion bodies formed by aggregation of α and β ATP synthase subunits. In agreement with these suppressive effects, CH partially restored the expression of most components of the OXPHOS pathway perturbed in the fmc1Δ mutant. The transcription data, which provide insight into the cellular response to CH treatment, tell us that the strongest beneficial effect of CH is at the level of the respiratory chain. The fact that CH did not exert this effect on WT cells indicates that (i) the transcriptional response of fmc1Δ cells to CH is more likely a downstream rather than a direct effect of treatment, and (ii) CH requires specific conditions to improve respiratory function, including but not limited to ATP synthase deficiency (otherwise it would have rescued all of the atp6-NARP mutants).
The expression pattern of QCR9, which encodes a subunit (Qcr9p) of complex III, suggests that this gene plays a role in the rescue mediated by CH. The possibility that increased Qcr9p synthesis alone could result in a higher abundance of complex III is in line with previous studies showing that such a regulatory mechanism is commonly used in the expression of mitochondrial and chloroplastic energy-transducing enzymes (35–37). Because complexes III and IV are mainly present together in the form of supercomplexes (38), increasing complex III abundance may allow more complex IV to be incorporated into mitochondria. QCR9 overexpression does indeed improve respiratory growth of the fmc1Δ mutant, but to a lesser extent than CH treatment; CH treatment also effected a partial increase in ATP synthase assembly (Fig. 2 E and F). It is therefore possible that CH works by increasing both the number of ATP synthase complexes and the efficiency with which they are used (via the stronger proton-motive force produced by complexes III/IV, whose increase in quantity may be mediated by up-regulation of QCR9). The combination resulted in a modest but significant improvement of ATP production in fmc1Δ cells, leading to restoration of respiratory growth. The modest improvement of ATP production by CH has significant therapeutic potential when considering the threshold phenomenon: Small increases in ATP production can be sufficient to restore a healthy state (4, 5). Further investigations into how exactly CH improves the abundance of respiratory chain complexes and ATP synthase are needed to more thoroughly characterize the therapeutic potential of this drug. In addition, evidence of CH displaying detrimental effects at high concentrations (39) will be important to consider as CH continues to be developed as a therapeutic.
In addition to providing a simple and powerful screening assay for identifying drugs active against ATP synthase disorders, the system presented here constitutes a proof of principle that yeast can be used as a pharmacological model for the study of mitochondrial diseases. The drugs identified can be used in various reverse screening strategies (40) to identify their intracellular targets, potentially revealing novel cellular mechanisms involved in disease pathologies (41, 42). In addition, the use of multiple atp6-NARP mutants in our secondary screen, which resulted in candidate compounds with allele-specific efficacies, holds promise for the development of personalized therapeutics for mitochondrial diseases.
As yeast models of inherited mitochondrial disorders continue to be developed, we believe the screening approach presented here will continue to yield promising chemical therapeutics and insights into disease mechanisms (1, 43).
Methods
Yeast Strains and Culture Medium.
The S. cerevisiae strains used and their genotypes are listed in Table S3. For details on growth procedures, see SI Methods.
Yeast-Based Drug Screening Assay.
This assay was adapted from an existing test (23, 24). Two hundred forty microliters of exponentially growing cell cultures, adjusted to an OD600 of 0.2, was spread homogeneously with sterile glass beads (∼3 mm diameter) on a square Petri dish (12 cm × 12cm) containing YPAGly solid medium. Sterile filters (similar to those used for antibiograms) were placed on the agar surface, and 2.5 μl of individual compounds from the various chemical libraries were applied to each filter in addition to DMSO, the vehicle, as a negative control, and a DHLA solution in DMSO as a positive control. Plates were then incubated at 35 °C for 7 d and scanned using a Snap Scan1212 (Agfa). For information on compounds screened, see SI Methods.
Isolation of Yeast Mitochondria and Subsequent Experiments.
Mitochondria were prepared by the enzymatic method as described (44) from cells grown for 7–8 generations in YPAGal medium at 35 °C in the presence of CH or DMSO. For details on the experiments with isolated mitochondria, see SI Methods.
Ultrastructural Studies.
Please see SI Methods for details.
Transcription Profiling.
Two biological replicates of strains MC1 and MC6 were cultured in YPAGal + DMSO/CH at 35 °C and harvested; total RNA was isolated and reverse-transcribed into cDNA, which was hybridized to whole-genome tiling arrays. For more details, see SI Methods.
Statistical Analysis of Transcription Profiles.
Raw tiling array data were processed to provide normalized intensity values for each probe in each hybridization. The expression level of each transcript was estimated by the median value of the probe intensities of the transcript across both arrays per strain and condition (27). For more details, see SI Methods.
Human Cell Lines and Culture Conditions.
The cybrid cell lines JCP213 (control) and JCP239 (NARP T8993G) (33) were cultivated in high-glucose DMEM; growth measurements were performed in glucose-deprived DMEM supplemented with CH, DHLA, OA, or DMSO. For each treatment condition, four wells were used. After 3 d of incubation with the drugs, cell proliferation was estimated by using Neutral Red staining (45) and also by cell counting with an Adam cell counter. The cells were then assayed for lactate and pyruvate by using kits from DiaSys-Poles. For more details, see SI Methods.
Supplementary Material
Acknowledgments
Part of this work was initiated in the Centre National de la Recherche Scientifique laboratory of L. Meijer, whom we thank for his continuous support and friendship. We also thank P. Lehn, Y. Bizais, and M.-F. Giraud for their warm welcome, encouragements, and helpful scientific discussions; and C. Voisset for critical reading of the manuscript. This work was supported by grants from the following organizations: Agence Nationale de la Recherche (ANR) “Maladies Rares” from the French government, Association Française Contre les Myopathies (AFM; to M.B. and J.-P.d.R.), the “Conseil Régional de la Région Aquitaine” (to J.-P.d.R.), and the National Institutes of Health/Deutsche Forschungsgemeinschaft (to L.M.S.). E.C., R.K., and N.E. were supported by postdoctoral fellowships from ANR and AFM. The study was supported technically by the European Molecular Biology Laboratory Genomics Core Facility.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101478108/-/DCSupplemental.
References
- 1.Kucharczyk R, et al. Mitochondrial ATP synthase disorders: Molecular mechanisms and the quest for curative therapeutic approaches. Biochim Biophys Acta. 2009;1793:186–199. doi: 10.1016/j.bbamcr.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Houstek J, et al. Mitochondrial diseases and genetic defects of ATP synthase. Biochim Biophys Acta. 2006;1757:1400–1405. doi: 10.1016/j.bbabio.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Tuppen HA, Blakely EL, Turnbull DM, Taylor RW. Mitochondrial DNA mutations and human disease. Biochim Biophys Acta. 2010;1797:113–128. doi: 10.1016/j.bbabio.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Letellier T, Heinrich R, Malgat M, Mazat JP. The kinetic basis of threshold effects observed in mitochondrial diseases: A systemic approach. Biochem J. 1994;302:171–174. doi: 10.1042/bj3020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rossignol R, et al. Mitochondrial threshold effects. Biochem J. 2003;370:751–762. doi: 10.1042/BJ20021594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Remacle C, Cardol P, Coosemans N, Gaisne M, Bonnefoy N. High-efficiency biolistic transformation of Chlamydomonas mitochondria can be used to insert mutations in complex I genes. Proc Natl Acad Sci USA. 2006;103:4771–4776. doi: 10.1073/pnas.0509501103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnefoy N, Fox TD. Genetic transformation of Saccharomyces cerevisiae mitochondria. Methods Cell Biol. 2001;65:381–396. doi: 10.1016/s0091-679x(01)65022-2. [DOI] [PubMed] [Google Scholar]
- 8.Kucharczyk R, et al. Consequences of the pathogenic T9176C mutation of human mitochondrial DNA on yeast mitochondrial ATP synthase. BBA Bioenergetics. 2010;1797:1105–1112. doi: 10.1016/j.bbabio.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kucharczyk R, Rak M, di Rago JP. Biochemical consequences in yeast of the human mitochondrial DNA 8993T>C mutation in the ATPase6 gene found in NARP/MILS patients. Biochim Biophys Acta. 2009;1793:817–824. doi: 10.1016/j.bbamcr.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 10.Kucharczyk R, Salin B, di Rago JP. Introducing the human Leigh syndrome mutation T9176G into Saccharomyces cerevisiae mitochondrial DNA leads to severe defects in the incorporation of Atp6p into the ATP synthase and in the mitochondrial morphology. Hum Mol Genet. 2009;18:2889–2898. doi: 10.1093/hmg/ddp226. [DOI] [PubMed] [Google Scholar]
- 11.Rak M, et al. A yeast model of the neurogenic ataxia retinitis pigmentosa (NARP) T8993G mutation in the mitochondrial ATP synthase-6 gene. J Biol Chem. 2007;282:34039–34047. doi: 10.1074/jbc.M703053200. [DOI] [PubMed] [Google Scholar]
- 12.De Meirleir L, et al. Respiratory chain complex V deficiency due to a mutation in the assembly gene ATP12. J Med Genet. 2004;41:120–124. doi: 10.1136/jmg.2003.012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cízková A, et al. TMEM70 mutations cause isolated ATP synthase deficiency and neonatal mitochondrial encephalocardiomyopathy. Nat Genet. 2008;40:1288–1290. doi: 10.1038/ng.246. [DOI] [PubMed] [Google Scholar]
- 14.Lefebvre-Legendre L, et al. Identification of a nuclear gene (FMC1) required for the assembly/stability of yeast mitochondrial F(1)-ATPase in heat stress conditions. J Biol Chem. 2001;276:6789–6796. doi: 10.1074/jbc.M009557200. [DOI] [PubMed] [Google Scholar]
- 15.DiMauro S, Hirano M, Schon EA. Approaches to the treatment of mitochondrial diseases. Muscle Nerve. 2006;34:265–283. doi: 10.1002/mus.20598. [DOI] [PubMed] [Google Scholar]
- 16.Mattiazzi M, et al. The mtDNA T8993G (NARP) mutation results in an impairment of oxidative phosphorylation that can be improved by antioxidants. Hum Mol Genet. 2004;13:869–879. doi: 10.1093/hmg/ddh103. [DOI] [PubMed] [Google Scholar]
- 17.Mukhopadhyay A, Uh M, Mueller DM. Level of ATP synthase activity required for yeast Saccharomyces cerevisiae to grow on glycerol media. FEBS Lett. 1994;343:160–164. doi: 10.1016/0014-5793(94)80310-2. [DOI] [PubMed] [Google Scholar]
- 18.Baracca A, et al. Biochemical phenotypes associated with the mitochondrial ATP6 gene mutations at nt8993. Biochim Biophys Acta. 2007;1767:913–919. doi: 10.1016/j.bbabio.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Vazquez-Memije ME, et al. Cellular and functional analysis of four mutations located in the mitochondrial ATPase6 gene. J Cell Biochem. 2009;106:878–886. doi: 10.1002/jcb.22055. [DOI] [PubMed] [Google Scholar]
- 20.Tibbetts AS, Sun Y, Lyon NA, Ghrist AC, Trotter PJ. Yeast mitochondrial oxodicarboxylate transporters are important for growth on oleic acid. Arch Biochem Biophys. 2002;406:96–104. doi: 10.1016/s0003-9861(02)00419-8. [DOI] [PubMed] [Google Scholar]
- 21.Fiermonte G, et al. Identification of the human mitochondrial oxodicarboxylate carrier. Bacterial expression, reconstitution, functional characterization, tissue distribution, and chromosomal location. J Biol Chem. 2001;276:8225–8230. doi: 10.1074/jbc.M009607200. [DOI] [PubMed] [Google Scholar]
- 22.Schwimmer C, et al. Increasing mitochondrial substrate-level phosphorylation can rescue respiratory growth of an ATP synthase-deficient yeast. J Biol Chem. 2005;280:30751–30759. doi: 10.1074/jbc.M501831200. [DOI] [PubMed] [Google Scholar]
- 23.Bach S, et al. Isolation of drugs active against mammalian prions using a yeast-based screening assay. Nat Biotechnol. 2003;21:1075–1081. doi: 10.1038/nbt855. [DOI] [PubMed] [Google Scholar]
- 24.Bach S, et al. A yeast-based assay to isolate drugs active against mammalian prions. Methods. 2006;39:72–77. doi: 10.1016/j.ymeth.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Lefebvre-Legendre L, et al. Failure to assemble the alpha 3 beta 3 subcomplex of the ATP synthase leads to accumulation of the alpha and beta subunits within inclusion bodies and the loss of mitochondrial cristae in Saccharomyces cerevisiae. J Biol Chem. 2005;280:18386–18392. doi: 10.1074/jbc.M410789200. [DOI] [PubMed] [Google Scholar]
- 26.Emaus RK, Grunwald R, Lemasters JJ. Rhodamine 123 as a probe of transmembrane potential in isolated rat-liver mitochondria: Spectral and metabolic properties. Biochim Biophys Acta. 1986;850:436–448. doi: 10.1016/0005-2728(86)90112-x. [DOI] [PubMed] [Google Scholar]
- 27.David L, et al. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashburner M, et al. The Gene Ontology Consortium. Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacIsaac KD, et al. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crespo JL, Powers T, Fowler B, Hall MN. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc Natl Acad Sci USA. 2002;99:6784–6789. doi: 10.1073/pnas.102687599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Epstein CB, et al. Genome-wide responses to mitochondrial dysfunction. Mol Biol Cell. 2001;12:297–308. doi: 10.1091/mbc.12.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guaragnella N, Butow RA. ATO3 encoding a putative outward ammonium transporter is an RTG-independent retrograde responsive gene regulated by GCN4 and the Ssy1-Ptr3-Ssy5 amino acid sensor system. J Biol Chem. 2003;278:45882–45887. doi: 10.1074/jbc.M309301200. [DOI] [PubMed] [Google Scholar]
- 33.Manfredi G, et al. Oligomycin induces a decrease in the cellular content of a pathogenic mutation in the human mitochondrial ATPase 6 gene. J Biol Chem. 1999;274:9386–9391. doi: 10.1074/jbc.274.14.9386. [DOI] [PubMed] [Google Scholar]
- 34.Weber TP, Widger WR, Kohn H. The Mg2+ requirements for rho transcription termination factor: Catalysis and bicyclomycin inhibition. Biochemistry. 2002;41:12377–12383. doi: 10.1021/bi020420w. [DOI] [PubMed] [Google Scholar]
- 35.Barrientos A, Zambrano A, Tzagoloff A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 2004;23:3472–3482. doi: 10.1038/sj.emboj.7600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kramarova TV, et al. Mitochondrial ATP synthase levels in brown adipose tissue are governed by the c-Fo subunit P1 isoform. FASEB J. 2008;22:55–63. doi: 10.1096/fj.07-8581com. [DOI] [PubMed] [Google Scholar]
- 37.Wollman FA, Minai L, Nechushtai R. The biogenesis and assembly of photosynthetic proteins in thylakoid membranes1. Biochim Biophys Acta. 1999;1411:21–85. doi: 10.1016/s0005-2728(99)00043-2. [DOI] [PubMed] [Google Scholar]
- 38.Schägger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christensen F, Bleeg HS, Jensen JE. The effect of chlorhexidine on some biochemical parameters of rat liver mitochondria. Acta Pharmacol Toxicol (Copenh) 1975;36:1–12. doi: 10.1111/j.1600-0773.1975.tb00766.x. [DOI] [PubMed] [Google Scholar]
- 40.Tribouillard D, et al. Antiprion drugs as chemical tools to uncover mechanisms of prion propagation. Prion. 2007;1:48–52. doi: 10.4161/pri.1.1.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tribouillard-Tanvier D, et al. Protein folding activity of ribosomal RNA is a selective target of two unrelated antiprion drugs. PLoS ONE. 2008;3:e2174. doi: 10.1371/journal.pone.0002174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voisset C, Thuret JY, Tribouillard-Tanvier D, Saupe SJ, Blondel M. Tools for the study of ribosome-borne protein folding activity. Biotechnol J. 2008;3:1033–1040. doi: 10.1002/biot.200800134. [DOI] [PubMed] [Google Scholar]
- 43.Schwimmer C, et al. Yeast models of human mitochondrial diseases: from molecular mechanisms to drug screening. Biotechnol J. 2006;1:270–281. doi: 10.1002/biot.200500053. [DOI] [PubMed] [Google Scholar]
- 44.Guérin B, Labbe P, Somlo M. Preparation of yeast mitochondria (Saccharomyces cerevisiae) with good P/O and respiratory control ratios. Methods Enzymol. 1979;55:149–159. doi: 10.1016/0076-6879(79)55021-6. [DOI] [PubMed] [Google Scholar]
- 45.Auré K, et al. Impact on oxidative phosphorylation of immortalization with the telomerase gene. Neuromuscul Disord. 2007;17:368–375. doi: 10.1016/j.nmd.2007.01.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.