Abstract
Protein kinase C (PKC) phosphorylation stimulates the cystic fibrosis transmembrane conductance regulator (CFTR) channel and enhances its activation by protein kinase A (PKA) through mechanisms that remain poorly understood. We have examined the effects of mutating consensus sequences for PKC phosphorylation and report here evidence for both stimulatory and inhibitory sites. Sequences were mutated in subsets and the mutants characterized by patch clamping. Activation of a 4CA mutant (S707A/S790A/T791A/S809A) by PKA was similar to that of wild-type CFTR and was enhanced by PKC, whereas responses of 3CA (T582A/T604A/S641A) and 2CA (T682A/S686A) channels to PKA were both drastically reduced (>90%). When each mutation in the 3CA and 2CA constructs was studied individually in a wild-type background, T582, T604, and S686 were found to be essential for PKA activation. Responses were restored when these three residues were reintroduced simultaneously into a 9CA mutant lacking all nine PKC consensus sequences (R6CA revertant); however, PKC phosphorylation was not required for this rescue. Nevertheless, two of the sites (T604 and S686) were phosphorylated in vitro, and PKC alone partially activated wild-type CFTR, the 4CA mutant, and the point mutants T582A and T604A, but not S686A channels, indicating that PKC does act at S686. The region encompassing S641 and T682 is inhibitory, because S641A enhanced activation by PKA, and T682A channels had 4-fold larger responses to PKC compared to wild-type channels. These results identify functionally important PKC consensus sequences on CFTR and will facilitate studies of its convergent regulation by PKC and PKA.
The cystic fibrosis transmembrane conductance regulator (CFTR) is a chloride channel that is activated by phosphorylation and gated by ATP (1–3). Mutations in CFTR lead to cystic fibrosis, an autosomal recessive disease characterized by abnormal ion transport across epithelia, viscous mucus secretions, chronic bacterial infections in the airways, and inflammation (4). CFTR has two membrane domains, each comprising six transmembrane segments, two nucleotide-binding domains (NBD1 and NBD2), and a regulatory domain (R domain) with potential sites for phosphorylation by protein kinase A (PKA) and protein kinase C (PKC) (5). Two predicted PKC sites are also situated in the distal region of NBD1, which begins near residue 433 and contains ≈200 amino acids (reviewed by ref. 3).
CFTR is phosphorylated by PKA at multiple sites (2, 6–9). Mutagenesis of the dibasic and monobasic PKA consensus sequences strongly inhibits channel activity (6, 7, 10, 11), although partial activation persists after PKA sites are removed in various combinations, and no one site seems to be essential (7, 12). CFTR is also phosphorylated by PKC at multiple sites, but their functional roles have not been studied systematically. After prolonged phosphatase-mediated rundown in excised patches, channel responses to PKA are strongly enhanced by pretreatment with PKC (13–18), suggesting PKC modulates responses to PKA in addition to causing a small activation on its own. PKC regulation may be direct because it is abolished in a mutant called 9CA (T582A/T604A/S641A/T682/S686A/S707A/S790A/T791A/S809A), which lacks all nine PKC consensus sequences between the first Walker B motif and second membrane domain (18); i.e., in the R domain and distal part of NBD1 (Fig. 1A). Eliminating consensus PKC sites strongly inhibits channel responses to PKA and abolishes partial activation by PKC alone. Because responses of the 9CA mutant to PKA stimulation in patch–clamp and iodide efflux assays resemble those of wild-type channels that had been exposed to PKC inhibitors, it was proposed that functional differences between 9CA and wild-type channels reflect, at least in part, loss of PKC phosphorylation at those predicted sites (14, 18).
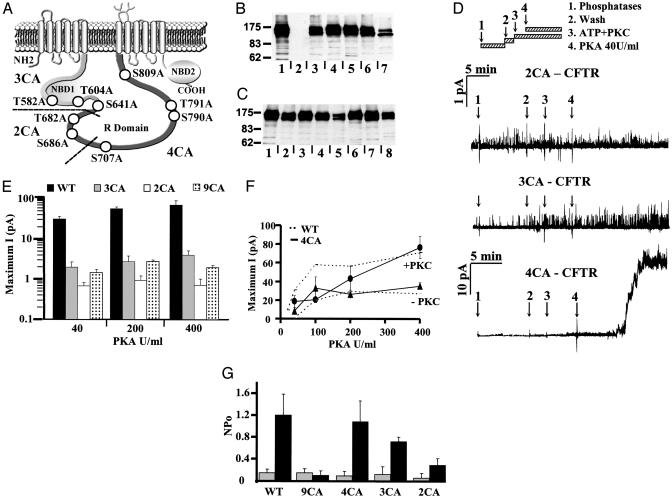
Fig. 1.
Partial PKC mutants. (A) Schematic representation of CFTR channel indicating potential PKC phosphorylation sites in NBD1 and the R domain. Mutations were made in subsets (named 3CA, 2CA, and 4CA, respectively) to identify sequences involved in CFTR regulation. (B) Western blot of BHK cells stably transfected with wild-type CFTR (lane 1), 9CA-CFTR (lane 3), R6CA-CFTR (lane 4), 4CA-CFTR (lane 5), 3CA-CFTR (lane 6), or 2CA-CFTR (lane7). Lane 2 shows control cells transfected with empty pNUT vector lacking CFTR cDNA. (C) Western blot showing BHK cells stably transfected with wild-type CFTR (lane 1), 9CA-CFTR (lane 2), R6CA-CFTR (lane 3), T582A-CFTR (lane 4), T604A (lane 5), S641A (lane 6), T682A (lane 7), or S686A (lane 8). (D) Recordings of 2CA, 3CA, and 4CA mutant channels by using the inside–out configuration [pipette potential (Vp) = +30 mV]. After excision, patches were treated with a mixture of three phosphatases to reduce basal CFTR phosphorylation, washed, then exposed to 3.78 nM PKC + 5 μM DiC8 and 1 mM ATP for 5 min followed by 40 units/ml PKA (see Materials and Methods). (E) Maximum currents [current (I), pA, measured at the plateau] for wild-type, 3CA, 2CA, and 9CA channels, plotted logarithmically at three PKA concentrations. (F) Relationship between maximum current and [PKA] for 4CA channels pretreated (circles) or not pretreated (triangles) with PKC. Values from wild-type CFTR tested under the same conditions are shown for comparison (upper and lower dashed lines, respectively). For each concentration, values are mean ±SE for n = four patches. (G) Activation by PKC alone. NPo was calculated for wild-type, 9CA, 4CA, 3CA, and 2CA channels before (gray bars) and after (dark bars) treatment with PKC + ATP for 15 min. Mean ± SE, n = six patches.
PKC phosphorylation has been demonstrated at S686 and S790 (2), although the other PKC consensus sequences are highly conserved from fish to humans and may also be phosphorylated (19). The main goal of this work was to examine the impact of altering predicted PKC sites on CFTR activation by PKA and PKC. Our approach was to narrow down the number of PKC consensus sequences potentially involved by examining channels with subsets of mutations. To identify the ones most critical for channel function, individual sites were then mutated and their importance confirmed by reintroduction into the 9CA construct mentioned above, which served as a “null” background. This yielded several surprising results: (i) three sequences were identified that are essential for activation by PKA but do not require PKC phosphorylation; (ii) at least one of those sites is nonetheless phosphorylated and needed for partial activation by PKC alone; and (iii) two other sequences are probably inhibitory, because mutating them leads to gains of function during PKC exposure. Identification of these consensus sequences clarifies PKC regulation and will enable study of more subtle aspects of CFTR regulation by multiple kinases (20, 21).
Materials and Methods
Chemicals. Type II bovine cardiac PKA catalytic subunit (PKA) and rat brain PKC II (PKC) were obtained from the laboratory of M. P. Walsh (University of Calgary, Calgary, AB, Canada). Protein phosphatase type 2 (PP2) Cα (PP2Cα) was from Upstate Biotechnology (Lake Placid, NY). PP2A catalytic subunit (PP2Aα) was from Promega. Other chemicals were from Sigma and of the highest grade available.
Cell Culture. Baby hamster kidney (BHK) cells stably expressing wild-type or mutated CFTR lacking PKC consensus sequences were plated on glass coverslips at low density 3 days before patch–clamp experiments. Cells were grown at 37°C with 5% CO2 in MEM (Life Technologies, Burlington, ON) containing 5% FBS and the selecting drug methotrexate (500 μM).
Mutagenesis. 3CA (T582A/T604A/S641A) was made by PCR mutagenesis by using wild-type CFTR cDNA as the template and the following forward (F) and reverse (R) primers: T582A (F, TACCTAGATGTTTTAGCAGAAAAAGAAATATTTGAA; R, TTCAAATATTTCTTTTTCTGCTAAAACATCTAGGTA), T604A (F, ACTAGGATTTTGGTCGCTTCTAAAATGGAACAT; R, ATGTTCCATTTTAGAAGCGACCAAAATCCTAGT), S641A (F, CTACAGCCAGACTTTGCCTCAAAACTCATGGGA; R, TCCCATGAGTTTTGAGGCAAAGTCTGGCTGTAG). 2CA (T682A/S686A) was constructed by using (F, GGACAGAAGCAAAAAAACAAGCTTTTAAAC), (R, CTGTTTAAAAGCTTGTTTTTTTGCTTCTGTCC), and wild-type CFTR cDNA as the template. Finally, pUCF2.5 3CA (S707/790/809A; ref. 7) was used as template cDNA for constructing 4CA (S707/790/791/809A) with the primers (F, GACAACAGCAGCCGCACGAAAACTC) and (R, CACTTTTCGTGCGGCTGCTGTTGTC). All constructs were confirmed by dideoxynucleotide sequencing and subcloned into pNUT-CFTR by replacement of the corresponding fragments in wild-type CFTR. Individual point mutations T582A, T604A, S641A, T682A, and S686A were subsequently introduced into wild-type CFTR by using the QuickChange (Stratagene) mutagenesis kit, which was also used to create a revertant mutant R6CA (A582T/A604T/A686S-9CA) in which three wild-type PKC consensus sequences were restored in the 9CA mutant described (18). BHK cells were transfected by calcium phosphate coprecipitation, and cell colonies stably expressing the mutants were selected by using 500 μM methotrexate as described (7, 18).
Patch Clamping. Macroscopic CFTR currents were recorded from inside–out patches with the pipette potential held at + 30 mV (i.e., membrane potential, Vm = -30 mV) and inverted for purposes of illustration. Initial bath and pipette solutions contained 150 mM NaCl, 2 mM MgCl2, and 10 mM N-Tris[hydroxymethyl]methyl-2-aminoethanesulfonic acid (pH 7.2). Macroscopic current flowing through the patch was recorded as described (18), sampled at 500 Hz and filtered at 125 Hz. Maximum currents were measured at the plateau by using PCLAMP software. Number of channels × open probability (NPo) was calculated by using DRSCAN software (22) before and during exposure to PKC + ATP for 15 min. Excised patches were pretreated with a mixture of 95 units/ml alkaline phosphatase, 2 units/ml PP2Aα, 1 unit/ml PP2Cα for 10 min to minimize basal phosphorylation and standardize conditions before adding exogenous kinases. Phosphatases were removed by perfusing the recording chamber for 3 min (>8 chamber volumes) before adding kinases. Experiments were carried out at room temperature (≈20°C).
Immunoblotting. BHK cells stably expressing wild-type or mutated CFTR were washed twice with ice-cold PBS, harvested by scraping, and lysed on ice for 30 min in RIPA buffer supplemented with protease inhibitor mixture as described (18). The lysate was centrifuged and an aliquot of supernatant assayed for protein by using bicinchoninic acid. After 7.5% SDS/PAGE, proteins were transferred to a poly(vinylidene difluoride) membrane and probed by using the monoclonal anti-CFTR antibodies M3A7 or 450 (23, 24). The secondary antibody (goat anti-mouse conjugated to peroxidase; Jackson ImmunoResearch) was detected by chemiluminescence (Amersham Pharmacia Biotech). Expression of the mutants was estimated by densitometry of scanned Western blots by using IMAGEJ software (provided by Wayne Rasband, National Institutes of Health, Bethesda) and the density of band C (mature complex-glycosylated CFTR, Mr ≈175,000) was expressed as a percentage of wild type.
Phosphopeptide Mapping of PKC Sites on a Cytoplasmic Domain Fusion Protein. A fusion protein containing NBD1, R domain, NBD2, and maltose-binding protein containing CFTR residues 417–832 and 1208–1431 joined by a linker of 11 amino acids was expressed in Escherichia coli and purified on amylase resin according to the manufacturer's instructions. It was phosphorylated in a reaction buffer containing 50 mM NaCl, 2 mM MgCl2, 25 mM Tris (pH 7.2), 100 μM Na2ATP, 10 μCi [γ-32P]ATP, 2.5 μg of phosphatidylserine, 2.5 mM b-glycerophosphate, 0.1 mM CaCl2, 0.1 mM DTT, and 20 ng of PKC for 30 min at 27°C. Gel slices containing radiolabeled proteins were excised from SDS/PAGE gels, washed with four changes of 10% acetic acid/50% methanol followed by four changes of 50% methanol, and dried under vacuum. The dried gel slices were resuspended in 150 μl of 50 mM NH4HCO3 (pH 8.0), diced into small fragments, and digested overnight with 20 μg of N-Tosyl-l-phenylalanine chloromethyl ketone–trypsin at 37°C. Fresh trypsin (20 μg) was added 1–2 h before the end of the digestion period. Gel pieces were removed by centrifugation, and the supernatant containing digested peptides was collected and dried under vacuum, resuspended in water and dried, resuspended in 1% ammonium bicarbonate (pH 8.9), and loaded onto a 20 × 20-cm thin-layer cellulose sheet (EM Science). Peptides were resolved in the first dimension by electrophoresis (2 h at 250 V in 1% bicarbonate, pH 8.9) and in the second dimension by chromatography [butanol/acetic acid/pyridine/water (75:15:50:60)]. Separated peptides were visualized by using a Molecular Dynamics PhosphorImager. Phosphorylated CFTR residues were identified by comigration of phosphorylated synthetic peptides (Sheldon Biotechnology Centre, McGill University). The synthetic peptides were radiolabeled by using PKC and [γ-32P] ATP and then digested with trypsin and resolved as described above. The peptides (>85% purity) used in this study were as follows (/denotes trypsin cut site, radiolabeled peptide sequences are shown in capital letters, and predicted PKC-phosphorylated residues are underlined): T604,/lmank/tr/ILVTSK/mehlk/; T682-(S686A), FSLEGDA PVSW TETK/k/qafk/; S686-(T682A), teak/k/QSFK/qtgefgek/; S707,/NSILNPINSIR/k/fsivqk/; S790, ihr/k/TTASTR/k/vsla.
Statistics and Models. Results are reported as the means ± SEM, n = number of observations. Differences were assessed by using the unpaired Student's t test, with P < 0.05 considered significant. The locations of functionally important PKC consensus sequences in the distal end of NBD1 were predicted by using a dimeric NBD1/NBD2 model developed by Structural GenomiX, San Diego, in collaboration with Cystic Fibrosis Foundation Therapeutics (Bethesda).
Results
Identifying Subsets of PKC Consensus Sequences That Are Essential for Activation by PKA. We reported previously that CFTR channel responses to PKA are strongly enhanced by pretreatment with PKC. A mutant lacking all nine PKC consensus sequences between the first Walker B motif and distal R domain (9CA, T582A/T604A/S641A/T682A/S686A/S707A/S790A/T791A/S809A) was functional, but responses to PKA were drastically reduced (≤5% of wild-type CFTR) and were not enhanced by PKC pretreatment (18). To identify the residues required for activation, functional studies were carried out by using mutants containing subsets of two to four mutations from the 9CA construct. The responsiveness of 3CA (T582A/T604A/S641A), 2CA (T682A/S686A), and 4CA (S707A/S790A/T791A/S809A) channels to PKA and PKC was compared with those of wild-type and 9CA channels, the latter serving as an end point (see Fig. 1A). Because mutations in the proximal R domain (but surprisingly not distal NBD1) caused partial misprocessing, cell variants were chosen to match channel expression as closely as possible. This mutant-CFTR expression level was determined in each stable cell line by Western blotting (Fig. 1 B and C). Based on densitometry of complex-glycosylated band C (apparent Mr ≈175 kDa), expression was highest for 4CA (76.43 ± 13.3% of wild-type CFTR) and somewhat lower for the others: 3CA (43.8 ± 5.2%), 2CA (19.1 ± 1%), and 9CA (36.2 ± 10.2%) (Fig. 1B). These differences were taken into account when interpreting functional responses to PKA and PKC; however, it was clear that such decreases in expression could not explain the dramatic loss of channel activity.
Inside–out patches containing 2CA, 3CA, or 4CA channels were studied by using a protocol described previously (18). Briefly, excised patches were incubated with alkaline phosphatase + PP2Cα + PP2Aα catalytic subunit for 10 min to reduce basal phosphorylation; phosphatases were then rinsed from the chamber, and ATP + PKC was added, followed 5 min later by 40 units/ml PKA (Fig. 1D). In marked contrast to wild-type channels (and despite PKC pretreatment), 2CA and 3CA channels were only slightly responsive to PKA, reminiscent of 9CA channels lacking all nine PKC consensus sequences (18). Maximal 3CA and 2CA currents evoked by PKC + PKA 40–400 units/ml are summarized in Fig. 1E and compared with wild-type and 9CA responses (note logarithmic scale). On the other hand, 4CA (S707A/S790A/T791A/S809A) channels had robust responses to PKA (Fig. 1F). Activations of 4CA (solid line) and wild-type channels (dashed lines, reported previously by ref. 18) were similar when the bath contained high (200–400 units/ml) PKA, and PKC pretreatment more than doubled the maximum currents for both wild-type and 4CA channels. 4CA channels were less responsive at low PKA concentrations (40 and 100 units/ml), perhaps reflecting loss of the confirmed PKC site, S790 (ref. 2 and this paper; see below). Stimulation of 4CA channels by PKC alone was also similar to that of wild-type CFTR (P > 0.217; n = 6; Fig. 1G), whereas stimulation of 3CA (P < 0.045; n = 4) and 2CA channels (P < 0.002; n = 4) was reduced although still ≈5-fold above baseline. Because PKA activation of both 3CA and 2CA channels was nearly abolished, at least two of the sequences mutated in 9CA are needed for normal responses. 4CA channels were activated by PKC similarly to wild-type channels; therefore, we focused our attention on the five sequences that are mutated in the 3CA and 2CA constructs.
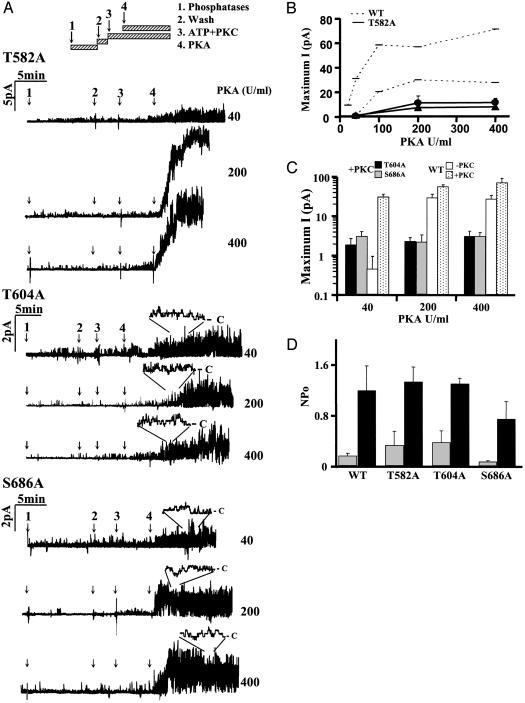
Identifying Individual Sequences Required for Channel Activation by PKA. To evaluate individual sequences, mutations present in 3CA and 2CA were introduced individually into wild-type CFTR. Densitometry of Western blots (Fig. 1C) revealed that most mutants had moderately reduced expression compared to wild-type CFTR (T682A 80.2%, S641A 79.2%, T582A 70.5%, S686A 45.4%, and T604A 24.6%; P < 0.05, n = four to six blots of each mutant). PKA and PKC activation of T582A, T604A, S641A, T682A, and S686A channels was assessed by using the same protocol as in Fig. 1. Two single mutants (S641A and T682A) were activated by PKA, whereas the other three (T582A, T604A, and S686A) had only small responses (reduced by >90%; Fig. 2 A–C; note logarithmic scale of the ordinate in Fig. 2C), indicating that among the mutations in 2CA and 3CA, T582A, T604A, and S686A are essential for CFTR activation. T582A was partially responsive to 200–400 units/ml PKA (≈26% compared to wild-type CFTR) (Fig. 2 A and B), whereas activations of T604A and S686A were nearly abolished at all PKA concentrations tested. Maximum currents mediated by T604A and S686A channels are summarized in Fig. 2C for comparison with wild-type channels. The current carried by T604A channels was only 4% that of wild-type channels (3.12 ± 1.1 pA; n = 12 patches). S686A channels appeared more active than T604A channels, but this difference was small enough to be explained by their higher expression (compare Fig. 1C).
Fig. 2.
Activity of CFTR channels with mutations at single PKC consensus sequences. (A) Recordings of T582A, T604A, and S686A channels by using the inside–out configuration [pipette potential (Vp) = +30 mV]. Several PKA concentrations were tested by using the same protocol as in Fig. 1. (B) Maximum current [current (I)], pA, measured at the plateau) for T582A mutant recorded with (•) or without (▴) PKC pretreatment. Values for wild-type CFTR in the absence (upper dashed line) or presence (lower dashed line) of PKC are shown for comparison. (C) Maximum current plotted logarithmically for T604A (dark bars) and S686A (gray bars), for comparison with wild-type CFTR (white and dotted bars represent PKA alone and PKC + PKA activity, respectively). At each concentration, the values shown are means ± SE for n = four patches. (D) Activation by PKC alone. NPo calculated before (gray bars) and after (dark bars) exposure to PKC + ATP for 15 min. Mean ± SE, n = six patches.
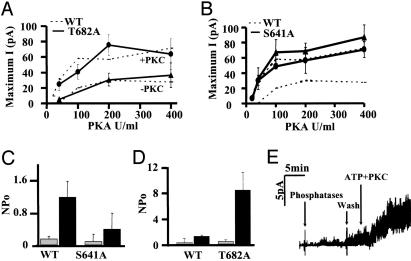
Responses to PKA and PKC Can Be Partially Dissociated, and Some PKC Sites Are Inhibitory. PKA responses of T582A, T604A, and S686A were not enhanced by PKC pretreatment, therefore we expected their stimulation by PKC alone to be similarly impaired; however, this was observed only for S686A (NPo = 0.53 ± 0.2 vs. 1.2 ± 0.38; n = 6, P = 0.015). T582A and T604A resembled wild-type channels with respect to their activation by PKC alone (Fig. 2D). The roles of S641 and T682 were also examined by using channels with single mutations. Responsiveness of T682A to PKA was similar to wild-type (Fig. 3A). On the other hand, replacing serine 641 with alanine enhanced channel activation at all PKA concentrations and eliminated the dependence on PKC (Fig. 3B). These two mutations also had different effects on the response to PKC alone. S641A dramatically reduced activation by PKC alone (Fig. 3C), whereas T682A enhanced PKC activation by >4-fold (Fig. 3 D and E). This stronger activation by PKC alone is consistent with PKC phosphorylation at T682 having an inhibitory action. Thus in wild-type CFTR, PKC phosphorylation of S641 and T682 may exert inhibitory effects on channel activation by PKA and PKC, respectively.
Fig. 3.
Activation of T682A and S641A channels. Maximum current (measured at the plateau) vs. PKA concentration for (A) T682A or (B) S641A channels pretreated (circles) or not pretreated (triangles) with PKC. Values obtained for wild-type channels under the same conditions are shown for comparison (dashed lines). Values are mean ± SE for n = four patches. (C) Activation by PKC alone. NPo of wild-type and S641A channels before (gray bars) and after (dark bars) addition of PKC for 15 min. Values are means ± SE for n = six patches. (D)NPo of wild-type and T682A channels measured before (gray bars) and after (dark bars) addition of PKC + ATP for 15 min. (E) Patch–clamp recording of T682A channels by using the inside–out configuration [pipette potential (Vp) = +30 mV] activated by PKC + ATP after preincubation with phosphatases as described in Materials and Methods.
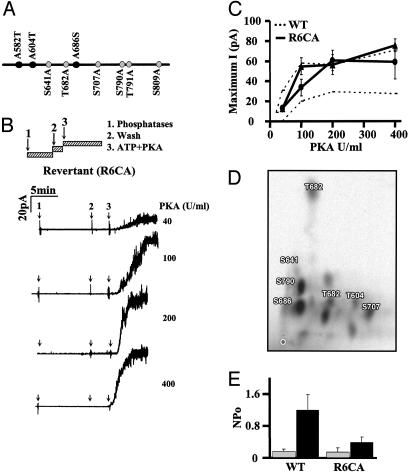
Mutagenizing PKC Consensus Sequences Has PKC-Dependent and -Independent Effects. Results obtained after point mutations indicate that T582, T604, and S686 are necessary for CFTR activation by PKA. To investigate whether they are also sufficient, they were reintroduced simultaneously into the 9CA construct lacking all nine PKC consensus sequences to generate a revertant mutant R6CA (A582T/A604T/A686S-9CA; Fig. 4A). Mature R6CA protein was expressed at approximately the same level as wild-type CFTR (91.1% ± 18.1, mean ± SE, n = 8; P = 0.267; Fig. 1 B and C). R6CA channels had robust responses to PKA after PKC pretreatment (Fig. 4C). Similar currents were obtained even without PKC pretreatment, suggesting that those three sequences are not essential as PKC phosphorylation sites per se. Phosphopeptide mapping was performed to see whether they might nevertheless be phosphorylated. T604 and S686 were in vitro phosphorylated by PKC by using a fusion protein that included NBD1, R domain, and NBD2; however, phosphorylation of T582 was not detected (Fig. 4D). R6CA channels were more active than wild-type channels, consistent with the absence of the inhibitory sequence at S641 (Fig. 4 B and C). Interestingly, reintroducing wild-type sequences at positions 582, 604, and 686 did not restore activation by PKC alone (Fig. 4E). This result, when combined with the activation of T582A and T604A mutants by PKC alone, argues that S686 and S641 (Fig. 3C) are both necessary for PKC activation. Thus S686 is required for the channel to be competent for gating (as reflected by responsiveness to PKA) and is also a site of regulatory PKC phosphorylation.
Fig. 4.
Characterization of the 9CA revertant R6CA. (A) R6CA-CFTR. Schematic of the PKC consensus sequences in NBD1 and R domain that were mutated in 9CA (18). The black circles show those sites restored in the revertant; gray circles indicate those left as alanines. (B) Recordings of R6CA mutant channels made by using the inside–out configuration [pipette potential (Vp) = +30 mV]. After excision, patches were treated with a mixture of three phosphatases (see Materials and Methods) for 10 min to remove basal CFTR phosphorylation and washed for 3 min, then 1 mM ATP + PKA was added at the concentrations indicated. (C) Relationship between maximum current (measured at the plateau) and PKA concentration, for R6CA channels pretreated (•), or not pretreated (▴) with PKC. For comparison, values for wild-type CFTR are shown under the same conditions (upper and lower dashed lines, respectively). Mean ± SE for n = four patches at each point. (D) Phosphopeptide mapping of fusion protein (NBD1-R domain-NBD2-maltose-binding protein) after in vitro phosphorylation by PKC. Proteins radiolabeled by PKC and [γ-32P] ATP were digested with trypsin and separated in the vertical direction by electrophoresis, and in the horizontal direction by chromatography; ○ marks the origin. Eleven major and several minor spots are visible in this representative map. The two major spots were identified as S686 and S790 by comigration of PKC-labeled synthetic peptides as described in Materials and Methods. Other spots identified by this method include T604, S641, T682, and S707; other spots remain to be identified. (E) Activation by PKC alone. NPo for wild-type CFTR and R6CA was calculated before (gray bars) and after (dark bars) exposure to PKC + ATP for 15 min. Mean ± SE, n = six patches.
Discussion
In a previous study, we characterized a CFTR mutant lacking all nine PKC consensus sequences in the R domain and distal part of NBD1 (9CA; ref. 18). Cells expressing 9CA channels had cpt-cAMP-stimulated iodide effluxes that were delayed relative to those from cells expressing wild-type CFTR. A similar delay was observed when wild-type cells were exposed to the PKC antagonist chelerythrine. Moreover, when PKC was added to excised patches (after phosphatase pretreatment), it caused a partial activation of wild-type channels and enhanced PKA-dependent activation but had no effect on the 9CA mutant. These results indicated that direct phosphorylation of CFTR at one or more of the sites mutated in 9CA is required for activation by PKC alone and for responsiveness to PKA.
In this work, we have analyzed the functional roles of individual PKC consensus sequences in regulating CFTR and found that the NBD1 residues T582 and T604, and S686 in the R domain, are all essential for CFTR activation by PKA; however, this did not involve PKC phosphorylation. Thus T582, T604, and S686 may be needed to maintain the channel in a state that is competent for gating. Nevertheless, S686 is needed for channel stimulation by PKC alone, therefore its phosphorylation does have functional consequences. Moreover, PKC phosphorylation of both T604 and S686 was demonstrated in vitro by using a previously undescribed fusion protein; therefore, these sites may have both conformational and phosphorylation-dependent actions. Phosphorylation of S641, as shown in Fig. 4D, may also contribute to partial activation by PKC alone. Data obtained with the partial mutant 4CA (S707A/S790A/T791A/S809A) suggest that a more distal PKC site enhances responses to low PKA activity, probably the confirmed PKC phosphorylation site S790 (ref. 2 and this work). Reintroducing T582, T604, and S686 simultaneously into the 9CA mutant (R6CA, revertant) restored channel activation by PKA but not by PKC alone.
Interestingly, mutagenizing S641 enabled full PKA stimulation without PKC pretreatment, whereas removal of T682 significantly enhanced activation by PKC without affecting PKA-dependent activation. These gains of function after mutagenesis suggest these sites are normally inhibitory. This has a precedent, in that phosphorylation of two dibasic PKA sites (S737, S768) is inhibitory based on the increase in channel activity observed when they are mutated (refs. 25 and 26, but see ref. 27). PKC phosphorylated T604, S641, T682, S686, S707, and S790 in vitro in a fusion protein containing NBD1, R domain, NBD2, and maltose-binding protein, with S686 and S790 giving the strongest signals. Although mutagenesis of consensus sequences can have actions that are independent of phosphorylation, at least mutagenesis assures that phosphorylation is removed from a particular site. We found previously that prolonged exposure to three phosphatases in vitro removed only ≈70% of the basal phosphorylation from CFTR that had been metabolically labeled in vivo by using 32PO4 (18). The functional effects of the various mutants examined in this study are summarized in Fig. 5.
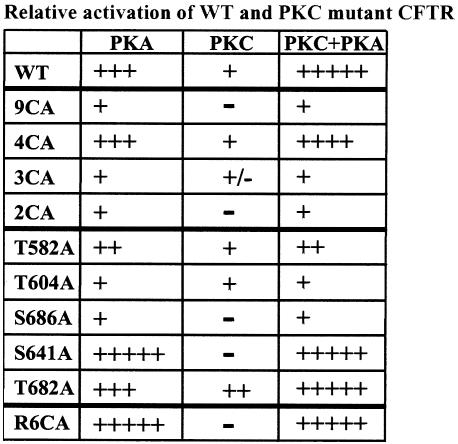
Fig. 5.
Summary of CFTR activation by PKA and/or PKC. Relative activation is shown as weak (+) to strong (+++++). - indicates no response.
The present results are consistent with previous structural and electrophysiological studies. The functionally important residues T582, T604, S641, and S686 are located in a region of the protein that is highly conserved among species (28). The inhibitory site T682 is present only in mammalian CFTRs, therefore the absence of this site may contribute to the stronger activation of amphibian CFTR by PKC. T582 and T604 are located in NBD1 near the R domain, and S641 is in a highly conserved region called RD1, which is sensitive to mutagenesis (29). Recent NBD structures from bacterial ABC transporters suggest they may function as dimers, with the LSGGQ motif (ABC transporters signature) from one NBD facing the ATP-binding site of the other NBD (reviewed by ref. 30). We examined the predicted locations of T582, T604, and S641 in a homology model developed for the NBDs of CFTR based on other ABC transporters (M. Sauder, Structural GenomiX, personal communication). According to the model, both T604 and T582 lie near the interface between the NBDs, with T604 near the ATP-binding pocket of NBD1 and T582 near the signature LSGGQ motif. For their positions, see Fig. 6, which is published as supporting information on the PNAS web site. Substitution and/or phosphorylation of these residues (and to a lesser extent S641, which is also predicted to be near the interface) may exert their effects by altering interactions between the domains. The functional revertant mutant described here should allow the roles of PKC phosphorylation at other sites to be established.
Supplementary Material
Acknowledgments
We thank J. M. Sauder (Structural GenomiX, Incorporated) for providing models of the NBDs; F. Chappe for technical assistance; and M.-A. Wioland for helpful discussions and critical reading of the manuscript. This work was supported by grants from the Canadian Cystic Fibrosis Foundation (CCFF) and Canadian Institutes of Health Research (CIHR) (to J.W.H.) and from the National Institutes of Health (National Institute of Diabetes and Digestive and Kidney Diseases) (to J.R.R.). V.C. and L.D.H. are CCFF postdoctoral fellows. J.W.H. is a senior scientist of the CIHR.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CFTR, cystic fibrosis transmembrane conductance regulator; NBD, nucleotide-binding domain; BHK, baby hamster kidney; R domain, regulatory domain; PP2, protein phosphatase type 2; NPo, number of channels × open probability.
References
- 1.Sheppard, D. N. & Welsh, M. J. (1999) Physiol. Rev. 79, S23-S45. [DOI] [PubMed] [Google Scholar]
- 2.Picciotto, M. R., Cohn, J. A., Bertuzzi, G., Greengard, P. & Nairn, A. C. (1992) J. Biol. Chem. 267, 12742-12752. [PubMed] [Google Scholar]
- 3.Hanrahan, J. W., Gentzsch, M. & Riordan, J. R. (2003) in ABC Proteins: From Bacteria to Man, eds. Holland, B., Higgins, C. F., Kuchler, K. & Cole, S. P. C. (Elsevier, New York), pp. 589-618.
- 4.Pilewski, J. M. & Frizzell, R. A. (1999) Physiol. Rev. 79, S215-S255. [DOI] [PubMed] [Google Scholar]
- 5.Riordan, J. R., Rommens, J. M., Kerem, B.-S., Alon, N., Rozmahel, R., Grzelczak, Z., Zielenski, J., Lok, S., Plavsic, N., Chou, J.-L., et al. (1989) Science 245, 1066-1073. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, S. H., Rich, D. P., Marshall, J., Gregory, R. J., Welsh, M. J. & Smith, A. E. (1991) Cell 66, 1027-1036. [DOI] [PubMed] [Google Scholar]
- 7.Chang, X.-B., Tabcharani, J. A., Hou, Y.-X., Jensen, T. J., Kartner, N., Alon, N., Hanrahan, J. W. & Riordan, J. R. (1993) J. Biol. Chem. 268, 11304-11311. [PubMed] [Google Scholar]
- 8.Gadsby, D. C. & Nairn, A. C. (1999) Physiol. Rev. 79, S77-S107. [DOI] [PubMed] [Google Scholar]
- 9.Frizzell, R. A., Field, M. & Schultz, S. G. (1979) Am. J. Physiol. 236, F1-F8. [DOI] [PubMed] [Google Scholar]
- 10.Rich, D. P., Berger, H. A., Cheng, S. H., Travis, S. M., Saxena, M., Smith, A. E. & Welsh, M. J. (1993) J. Biol. Chem. 268, 20259-20267. [PubMed] [Google Scholar]
- 11.Seibert, F. S., Tabcharani, J. A., Chang, X.-B., Dulhanty, A. M., Mathews, C. J., Hanrahan, J. W. & Riordan, J. R. (1995) J. Biol. Chem. 270, 2158-2162. [DOI] [PubMed] [Google Scholar]
- 12.Ostedgaard, L. S., Baldursson, O. & Welsh, M. J. (2001) J. Biol. Chem. 276, 7689-7692. [DOI] [PubMed] [Google Scholar]
- 13.Tabcharani, J. A., Chang, X.-B., Riordan, J. R. & Hanrahan, J. W. (1991) Nature 352, 628-631. [DOI] [PubMed] [Google Scholar]
- 14.Jia, Y., Mathews, C. J. & Hanrahan, J. W. (1997) J. Biol. Chem. 272, 4978-4984. [DOI] [PubMed] [Google Scholar]
- 15.Middleton, L. M. & Harvey, R. D. (1998) Am. J. Physiol. 275, C293-C302. [DOI] [PubMed] [Google Scholar]
- 16.Yamazaki, J., Britton, F., Collier, M. L., Horowitz, B. & Hume, J. R. (1999) Biophys. J. 76, 1972-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Button, B., Reuss, L. & Altenberg, G. A. (2001) J. Gen. Physiol. 117, 457-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chappe, V., Hinkson, D. A. R., Zhu, T., Chang, X.-B., Riordan, J. R. & Hanrahan, J. W. (2003) J. Physiol. 548, 39-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahan, D., Evagelidis, A., Hanrahan, J. W., Hinkson, D. A. R., Jia, Y., Luo, J. & Zhu, T. (2001) Pflügers Arch. 443, S92-S96. [DOI] [PubMed] [Google Scholar]
- 20.Li, M., West, J. W., Numann, R., Murphy, B. J., Scheuer, T. & Catterall, W. A. (1993) Science 261, 1439-1442. [DOI] [PubMed] [Google Scholar]
- 21.Cohen, P. (2002) Trends Biochem. Sci. 25, 596-601. [DOI] [PubMed] [Google Scholar]
- 22.Hanrahan, J. W., Kone, Z., Mathews, C. J., Luo, J., Jia, Y. & Linsdell, P. (1998) Methods Enzymol. 293, 169-194. [DOI] [PubMed] [Google Scholar]
- 23.Kartner, N. & Riordan, J. R. (1998) Methods Enzymol. 292, 629-652. [DOI] [PubMed] [Google Scholar]
- 24.Jensen, T. J., Bone, B. G. Jennings M. A., Aleksandrov, L. A., Mengos, A., Gentzch, M., Chang, X. B. & Riordan, J. R. (2001) Pediatr. Pulmon. S20, 179. [Google Scholar]
- 25.Wilkinson, D. J., Strong, T. V., Mansoura, M. E., Wood, D. L., Smith, S. S., Collins, F. S. & Dawson, D. C. (1997) Am. J. Physiol. 273, L127-L133. [DOI] [PubMed] [Google Scholar]
- 26.Baldursson, O., Berger, H. A. & Welsh, M. J. (2000) Am. J. Physiol. 279, L835-L841. [DOI] [PubMed] [Google Scholar]
- 27.Winter, M. C. & Welsh, M. J. (1997) Nature 389, 294-296. [DOI] [PubMed] [Google Scholar]
- 28.Chen, J.-M., Scotet, V. & Ferec, C. (2003) Mol. Gen. Metabol. 71, 245-249. [DOI] [PubMed] [Google Scholar]
- 29.Pasyk, E. A., Morin, X. K., Zeman, P., Garami, E., Galley, K., Huan, L. J., Wang, Y. & Bear, C. E. (1998) J. Biol. Chem. 273, 31759-31764. [DOI] [PubMed] [Google Scholar]
- 30.Kerr, I. D. (2002) Biochim. Biophys. Acta 1561, 47-64. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.