Abstract
Burkholderia pseudomallei and Burkholderia thailandensis are related pathogens that invade a variety of cell types, replicate in the cytoplasm, and spread to nearby cells. We have investigated temporal and spatial requirements for virulence determinants in the intracellular life cycle, using genetic dissection and photothermal nanoblade delivery, which allows efficient placement of bacterium-sized cargo into the cytoplasm of mammalian cells. The conserved Bsa type III secretion system (T3SSBsa) is dispensable for invasion, but is essential for escape from primary endosomes. By nanoblade delivery of B. thailandensis we demonstrate that all subsequent events in intercellular spread occur independently of T3SSBsa activity. Although intracellular movement was essential for cell–cell spread by B. pseudomallei and B. thailandensis, neither BimA-mediated actin polymerization nor the formation of membrane protrusions containing bacteria was required for B. thailandensis. Surprisingly, the cryptic (fla2) flagellar system encoded on chromosome 2 of B. thailandensis supported rapid intracellular motility and efficient cell–cell spread. Plaque formation by both pathogens was dependent on the activity of a type VI secretion system (T6SS-1) that functions downstream from T3SSBsa-mediated endosome escape. A remarkable feature of Burkholderia is their ability to induce the formation of multinucleate giant cells (MNGCs) in multiple cell types. By infection and nanoblade delivery, we observed complete correspondence between mutant phenotypes in assays for cell fusion and plaque formation, and time-course studies showed that plaque formation represents MNGC death. Our data suggest that the primary means for intercellular spread involves cell fusion, as opposed to pseudopod engulfment and bacterial escape from double-membrane vacuoles.
The robust Gram-negative bacillus Burkholderia pseudomallei (Bp) is endemic to warm, fecund soils of tropical regions (1, 2). Infections acquired from the environment can lead to melioidosis, a serious and sometimes fatal human disease. Accumulating evidence suggests that adaptations selected in the rhizosphere are responsible for “accidental virulence” in mammals (3). Bp has a large (7.2 Mb) genome that has been shaped by extensive horizontal exchange (4). Burkholderia mallei (Bm) is a clonal descendant of Bp that has undergone genome decay and has lost the capacity for environmental survival. Bm is the agent of equine glanders and it can also cause fatal human infections (2). Resistance to antibiotics and their low infectious dose have led to the classification of Bp and Bm as biowarfare threats.
The geographic distribution of Bp overlaps with that of B. thailandensis (Bt) and their genomes are highly similar and syntenic (5). Although Bt is rarely associated with human disease and is considered relatively nonpathogenic, this assessment is not absolute. Following aerosol challenge of mice, Bt causes fulminant, lethal infections that are dependent on virulence determinants shared with Bp and Bm (2, 6). Bp, Bt, and Bm invade and replicate in a wide range of cell types and exhibit nearly identical intracellular life cycles (1, 2). Following invasion and escape from endosomes, replication in the cytoplasm is accompanied by actin-based motility and cell–cell spread, analogous to Shigella flexneri and Listeria monocytogenes (7–9). Actin motility is mediated by BimA, a polarly localized surface protein that binds actin and promotes polymerization (9). An unusual feature of infection is the induction of cell fusion and the formation of multinucleate giant cells (MNGCs) (10). For Bp and Bm this requires BimA and has been observed with multiple cell types in vitro and in tissues from patients with melioidosis (2).
Bp possesses a generous endowment of specialized export systems including three “injection” type III secretion systems (T3SS), two of which are similar to those in phytopathogenic bacteria. The third, T3SSBsa, is homologous to the Shigella Mxi-Spa and Salmonella SPI-1 T3SSs and is highly conserved in Bp, Bt, and Bm (1, 2). T3SSBsa is required for virulence in hamster and murine models of pathogenesis (2) and has been implicated in invasion of epithelial cells, escape from endosomes, intracellular survival, and evasion of autophagy (11). In addition, Bp encodes six type VI secretion systems (T6SSs) (12). Using the nomenclature of Schell et al. (13), T6SS-1 (also referred to as T6SS-5) (14) is critical for virulence in the Bt murine model of acute melioidosis and contributes to the lethality of Bm in hamsters (13). Recently, T6SS-1 mutants in Bp were shown to be capable of endosome escape in RAW264 cells but were defective in MNGC formation (15).
For intracellular pathogens, understanding the roles of virulence determinants is complicated by their involvement in temporally and spatially staged events. T3SSBsa has been proposed to be required for late events associated with cell–cell spread, but direct investigation has been difficult since mutants are defective in earlier steps in the intracellular life cycle. To address this conundrum, we have used a photothermal nanoblade to deliver live bacteria directly into the cytoplasm of mammalian cells (16). The photothermal nanoblade device uses a laser pulse to excite a thin titanium coating on the tip of a glass capillary pipette. Rapid thermal excitation of the metallic nanostructure produces an explosive nanoscale vapor bubble that creates a small incision in the cell membrane at the point of pipette contact. This incision provides a transient delivery portal through which variably sized cargo—from molecules to bacteria—can be efficiently delivered with high cell viability. We have combined the use of this technology with traditional genetic ablation techniques and infection analysis to probe virulence mechanisms participating in the intracellular life cycle of Burkholderia.
Results
T3SSBsa Is Required for Plaque Formation and Endosome Escape but Is Dispensable for Invasion.
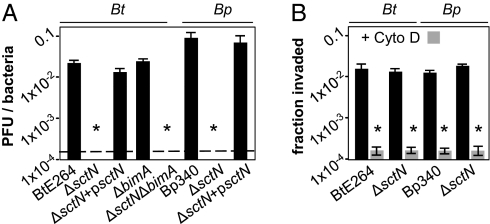
HEK293 cells are efficiently invaded by Bp and Bt and are highly amenable to photothermal nanoblade-mediated cytosolic delivery. In the experiment in Fig. 1A, HEK293 monolayers were infected with B. thailandensis E264 (17), B. pseudomallei Bp340 (18), or derivatives containing in-frame deletions in sctN, which encodes the ATPase required for activity of the Bsa T3SS. Plaque formation, a hallmark of cell–cell spread, was eliminated in ΔsctN mutants and restored by complementation in trans. These observations are consistent with previous reports that plaque formation by Bp is dependent on T3SSBsa (19).
Fig. 1.
T3SSBsa is required for endosome escape but not invasion. (A) Plaque-forming units (pfu) per bacteria 18 h after infection of HEK293 cells with BtE264, Bp340, ΔsctN mutants, or complemented derivatives (psctN). Dashed line, limit of detection. (B) Colony forming units (cfu) recovered per bacteria in 2-h invasion assays. HEK293 cells were untreated (solid bars) or treated with 2 μg/mL cytochalasin D (shaded bars) before infection. Assays were performed in triplicate and error bars represent ±SEM. *P < 0.005.
It has been suggested that T3SSBsa is required for invasion of nonphagocytic cells, a prerequisite for plaque formation (20). For Bp340 and BtE264, invasion was inhibited by cytochalasin D as expected, but it was unaffected by ΔsctN alleles (Fig. 1B). These results indicate that whereas invasion requires actin polymerization, it occurs independently of T3SSBsa activity. Analogous results were obtained using HeLa cells (Fig. S1A). To determine whether T3SSBsa facilitates endosome escape in HEK293 cells, monolayers were stained 8 h after infection for F-actin. As shown in Fig. S1B, BtE264 and Bp340 formed actin tails, indicating successful escape, whereas ΔsctN mutants and ΔbimA control strains did not. Moreover, ΔsctN mutants colocalized with the late endosomal marker LAMP1 (Fig. S1C). At later time points, only scattered bacterial debris was detected for ΔsctN mutants, suggesting they were killed and degraded due to endosomal entrapment (Fig. S1D). Although our results do not support a role for T3SSBsa in invasion, they are consistent with previous reports of its essential contribution to endosome escape (2, 21).
Intercellular Spread Following Cytoplasmic Delivery by a Photothermal Nanoblade.
The inability of ΔsctN mutants to escape from primary endosomes confounds efforts to understand the involvement of T3SSBsa in subsequent steps required for cell–cell spread. Solutions to this dilemma require the ability to bypass early events that are otherwise essential during infection. To accomplish this, we exploited the capabilities of our recently developed photothermal nanoblade to place bacteria directly into the cytoplasm of host cells (Fig. 2A) (16). Because a nanoblade has not yet been customized and approved for use with Bp in a select agent BSL-3 facility, we focused our analysis on Bt, which can be safely manipulated under BSL-2 conditions.
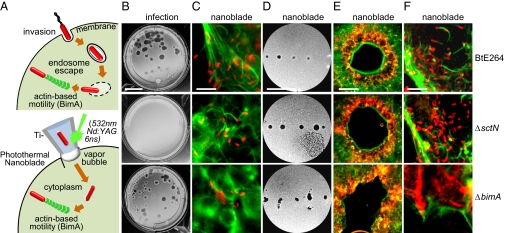
Fig. 2.
Intercellular spread following photothermal nanoblade-mediated delivery. (A) Upper, invasion by BtE264 is followed by T3SSBsa-mediated endosome escape and BimA-mediated actin polymerization in the cytoplasm. Lower, photothermal excitation of Ti-coated microcapillary pipettes using a 6-ns, 532-nm laser pulse facilitates pressurized delivery of bacteria directly into the cytosol. (B) Plaque formation on HEK293 monolayers after infection with BtE264 (Top), ΔsctN (Middle), or ΔbimA mutants (Bottom). (Scale bar, 1 cm.) (C) Bt and mutants (red) were delivered into HEK293 cells using a photothermal nanoblade and stained for actin (green) 12 h later. (Scale bar, 20 μm.) (D) Plaque formation on HEK293 monolayers following nanoblade delivery. (Scale bar, 1 cm.) (E) Plaques in D stained for bacteria (red) and actin (green). (Scale bar, 500 μm.) (F) Magnified edges of plaques in E. (Scale bar, 20 μm.)
In Fig. 2, wild-type (WT) BtE264 or mutant derivatives were introduced into HEK293 cells by infection or by photothermal nanoblade delivery. Plaque formation following infection was absolutely dependent on T3SSBsa activity (Fig. 2B). When ΔsctN mutants were delivered into the cytosol using the nanoblade, they divided and polymerized actin as well as wild-type bacteria (Fig. 2C), demonstrating that actin motility functions independently of T3SSBsa and does not require passage through the endosomal environment. We were surprised, however, to find that ΔsctN mutants were capable of forming plaques following nanoblade delivery that were indistinguishable in size and morphology from those formed by WT Bt following infection or nanoblade delivery (Fig. 2 D–F). The ability to bypass early events in the intracellular life cycle allows us to conclude that the only requirement for T3SSBsa in cell–cell spread and plaque formation is for escape from primary endosomes following invasion. It had been assumed that for cell–cell spread to occur, T3SSBsa would be required to lyse double-membrane secondary vacuoles formed during penetration of adjacent cells (1). Our observations indicate that either some other factor(s) performs this function, or intercellular spread occurs by an entirely different mechanism.
Two Distinct Motility Systems Facilitate Plaque Formation.
A deletion mutant in bimA was included as a control for plaque formation in photothermal delivery experiments. As shown in Fig. S1B and Fig. 2C, the ΔbimA allele eliminates actin polymerization as previously described (22), and it also eliminates the formation of membrane protrusions containing Burkholderia at their tips. Although we assumed that actin motility would be required as a driving force for cell–cell spread, this notion was clearly incorrect. Following infection or nanoblade delivery, ΔbimA mutants formed plaques that were similar in size and morphology to those of their WT parent (Fig. 2 B, D, and E). BimA-mediated actin motility and the formation of membrane protrusions are therefore dispensable for cell–cell spread by B. thailandensis.
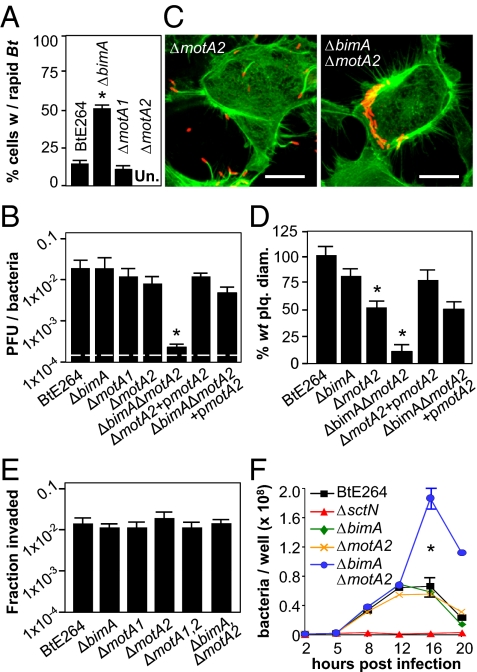
Intrigued by these results, we examined live infected cells by microscopy and discovered that Bt exhibits remarkably rapid intracellular motility. Bacteria move at speeds of >20 μm/s and often reverse course and change direction (Fig. S2A and Movies S1, S2, and S3). Because rapid intracellular motility was independent of BimA (Fig. 3A), we explored the possibility that flagellar motility was involved. Deletion of motA1, a motor component locus in the chromosome 1 flagellar biosynthesis gene cluster (fla1), eliminated swarming in soft agar but had no effect on intracellular motility or plaque formation (Fig. S2, Fig. 3 A and B, and Table S1). Previous studies identified potential chemotaxis and flagellar loci on chromosome 2 in Bt (23), and on closer inspection we found a full complement of flagellar structural and regulatory genes that could encode a second, functional motility system (fla2; Fig. S3). Deletion of motA2 from the fla2 flagellar cluster had no effect on swarming in agar or actin polymerization following invasion (Fig. S2B and Fig. 3C), but it eliminated rapid intracellular motility (Fig. 3A), as did deletion of fliC2, which is predicted to encode flagellin (Table S1). Although significant differences were not observed in plaquing efficiency (Fig. 3B), ΔmotA2 mutants formed plaques that were smaller than those produced by WT or ΔbimA strains (Fig. 3D and Table S1). When motA2 was deleted from a ΔbimA background, plaque formation was almost completely abolished, and the defect was reversed by complementation with motA2 (Fig. 3B).
Fig. 3.
fla2-mediated flagellar motility facilitates plaque formation. (A) Fraction of HEK293 cells containing rapidly motile bacteria 8 h after infection with BtE264 and derivatives. Approximately 300 cells were monitored per strain. (B) Plaque-forming efficiency on HEK293 cell monolayers 18 h after infection. (C) Representative infected cells stained for bacteria (red) and actin (green). (D) Plaque diameters 24 h after infection of HEK293 cell monolayers. (E) HEK293 cell invasion efficiencies by BtE264 or mutant strains 2 h postinfection. (F) Time course of intracellular replication in HEK293 cells. All assays were performed in triplicate and error bars represent ±SEM. *P < 0.005; Un, undetectable.
These observations demonstrate that MotA2-dependent flagellar motility can drive intercellular spread independently of BimA-mediated actin polymerization, and at least one of the two motility systems must be active for plaque formation. Although flagellar motility does not affect invasion or endosome escape (Fig. 3E and Table S1), an interesting phenotype is observed in intracellular growth assays. As shown in Fig. 3F, WT, ΔbimA, and ΔmotA2 strains multiply, plateau at 12 h, and decrease in numbers due to cell disruption and exposure to extracellular antibiotics. In contrast, ΔbimAΔmotA2 mutants continue to multiply and reach significantly higher levels, suggesting a relationship between intracellular movement and cell death (see below).
T6SS-1 Facilitates Plaque Formation.
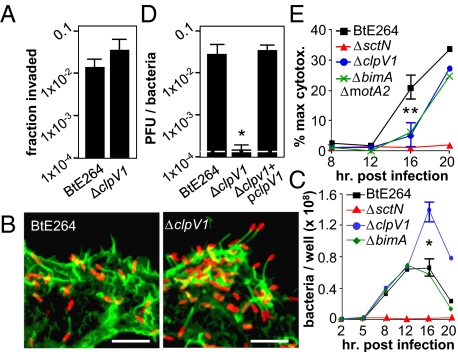
Of the multiple T6SSs encoded by Burkholderia species, T6SS-1 has been repeatedly linked to host–pathogen interactions (12–15, 19). These correlations with virulence led us to investigate the role of T6SS-1 following infection and nanoblade delivery. T6SS-1 was inactivated by deletion of the clpV1 ATPase (24). ΔclpV1 mutants were fully invasive, escaped from endosomes, replicated in the cytoplasm, polymerized actin, and displayed rapid intracellular motility similar to WT (Fig. 4 A–C, Movie S3, and Table S1). In contrast, plaque formation following infection (Fig. 4D) or nanoblade delivery (Table S1) was reduced to near background levels, demonstrating that clpV1-dependent T6SS activity is crucial for intercellular spread.
Fig. 4.
T6SS-1 is critical for efficient intercellular spread. (A) Invasion efficiencies by BtE264 or ΔclpV1 mutants 2 h postinfection in HEK293 cells. (B) HEK293 cells were infected and stained for bacteria (red) and actin (green) 8 h postinfection. (Scale bar, 20 μm.) (C) Time course of intracellular replication in HEK293 cells. (D) Plaque-forming efficiency on HEK293 cell monolayers 18 h after infection. (E) Cytotoxicity assays in HEK293 cells. All assays in A and C–E were performed in triplicate and error bars represent ±SEM. **P < 0.05; *P < 0.005.
Motility and T6SS-1 mutants exhibited similar phenotypes. In both cases major defects in plaque formation are observed, and as shown in Fig. 4C, ΔclpV1 mutants replicate to higher numbers in intracellular growth assays, similar to the ΔbimAΔmotA2 double mutant (Fig. 3F). As shown in Fig. 4E, the kinetics of cell death are significantly delayed following infection by ΔclpV1 mutants, suggesting that increased bacterial numbers reflect prolonged cell survival. In considering how these and earlier observations might be related, a closer look at the process of plaque formation itself is revealing.
Intercellular Spread and Plaque Formation Occur Through Cell Fusion.
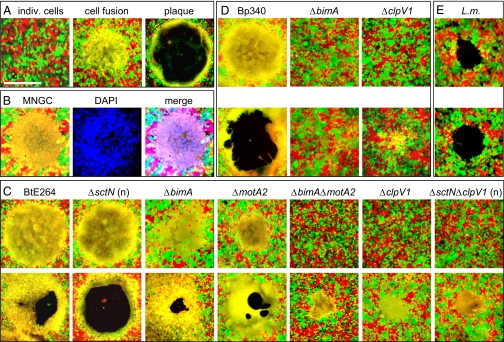
Bp, Bm, and Bt induce cell fusion and multinucleate giant cell (MNGC) formation with remarkable efficiency in a variety of cell types (2, 10, 25). In observing cell monolayers following nanoblade delivery, we often noticed the appearance of MNGCs in areas that developed to become plaques. To examine the process more closely, we constructed HEK293 cell lines that constitutively express green fluorescent protein (GFP) or monomeric strawberry red fluorescent protein (RFP) by stable transduction with recombinant lentivirus. In Fig. 5A, GFP- and RFP-expressing cells were seeded at a 1:1 ratio and Bt or mutant derivatives were introduced by infection or nanoblade delivery. The progression of events leading to plaque formation is readily apparent; individual cells (red or green) initially fuse to form one or more MNGCs (yellow), which later lyse to form a clear zone in the monolayer surrounded by a ring of fused cells (i.e., a plaque). A MNGC containing numerous DAPI-stained nuclei before lysis is shown in Fig. 5B.
Fig. 5.
Intercellular spread and plaque formation occur through cell fusion. (A) Progression of events leading to plaque formation. (Left) HEK293-RFP (red) and -GFP (green) cells immediately after infection. Twelve hours later, a MNGC is formed (yellow, Center), which undergoes lysis and forms a plaque at 24 h (Right). (Scale bar, 500 μm.) (B) A MNGC (Left) was stained with DAPI (blue, Center). (Right) Triple-color merged image. (C) MNGC and plaque formation by BtE264 and mutants on HEK293 RFP + GFP monolayers 12 h (Upper) or 24 h (Lower) following infection or nanoblade delivery (“n”, ΔsctN, ΔsctNΔclpV1). (D) MNGC and plaque formation 12 h (Upper) and 24 h (Lower) after infection with Bp340 and mutants. (E) Plaque formation 56 h following infection with L. monocytogenes 10403S. The slight appearance of yellow at the edge of plaques is due to physical overlap of red and green cells and does not indicate cell fusion. Images are representative of multiple independent experiments.
Fig. 5C shows early (12 h, Upper) and late (24 h, Lower) time points in cell fusion-plaque assays with BtE264 or mutant strains. Although ΔsctN mutants failed to form MNGCs or plaques following infection due to endosomal entrapment, both events occurred normally after nanoblade delivery, demonstrating that T3SSBsa is not required for MNGC formation. In BtE264, deletion of bimA or motA2 individually had little effect, whereas a ΔbimAΔmotA2 double mutant was greatly delayed in MNGC formation. A similar phenotype was observed with the ΔclpV1 strain. A ΔsctNΔclpV1 mutant was incapable of MNGC formation following infection, but resembled the ΔclpV1 single mutant after nanoblade delivery, showing that T6SS-1 affects cell–cell spread downstream of T3SSBsa-mediated endosomal escape (Fig. 5C and Table S1). Fig. 5D shows that plaque formation by B. pseudomallei also occurs through a process that involves MNGC formation and lysis and is dependent on clpV1. Because Bp340 lacks fla2 (Fig. S3 and Discussion), this process required bimA. On the basis of these and other results, we propose that cell fusion is the central mechanism for cell–cell spread by B. thailandensis and B. pseudomallei and that plaque formation occurs through a process that is dependent on the formation of MNGCs. This result differs fundamentally from intercellular spread by Listeria monocytogenes, which requires actin-based motility, engulfment of bacterial protrusions by adjacent cells, and escape from double-membrane vacuoles (8). Indeed, as shown in Fig. 5E, plaque formation by Listeria occurs in the absence of detectable cell fusion.
Discussion
Our results are incorporated into a model for the Burkholderia intracellular life cycle shown in Fig. S4. This model is based on observations with HEK293 cells and may not take into account factors that are specifically required for survival and replication in professional phagocytes.
Invasion.
Invasion of HEK293 cells by Bp or Bt requires host-cell actin polymerization but not the activity of T3SSBsa, contrasting with a clear requirement for the Mxi-Spa and SPI-1 T3SSs in invasion by Shigella (7) and Salmonella (26), respectively, and with expectations for Burkholderia based on analyses of BopE, a T3SSBsa substrate homologous to Salmonella SopE and SopE2 (20). In an experiment often cited as supporting a role for T3SSBsa in invasion (20), insertion mutations in bipD (a T3SSBsa translocon gene) or bopE conferred modest decreases in invasion (35–40%) 6 h after infection of HeLa cells. The discrepancy between these data and ours is likely due to the use of a late time point where endosomally trapped mutants loose viability (6 h in ref. 20 vs. 2 h in Fig. 1), giving the appearance of an invasion defect. A similar conclusion was previously reported by Haraga et al., using BtE264 and HeLa cells (21). Their study and ours support the conclusion that for Bp and Bt, invasion of nonphagocytic cells can occur by mechanisms that are independent of T3SSBsa. Virulence determinants that mediate invasion await discovery.
Bsa T3SS.
T3SSBsa is required for escape from endosomes following invasion of HEK293 cells by Bp or Bt, consistent with numerous prior studies with a variety of cell types (1, 2). Cytosolic delivery of Bt using a photothermal nanoblade allowed us to bypass endosome escape and directly examine the role of T3SSBsa in downstream events: actin polymerization, cell–cell spread, and MNGC formation. In all cases the results were remarkably clear; plaque formation following infection was absolutely dependent on T3SSBsa, whereas plaque formation following nanoblade delivery was independent of its activity. The same was true for MNGC formation. We also show that replication and actin polymerization occur normally when ΔsctN mutants are placed directly into the cytosol.
These observations have several implications; first, they allow us to conclude that in our system, the only role for T3SSBsa is to facilitate escape from primary endosomes of initially infected cells. Next, our results help explain a perplexing lack of phenotypes associated with known or putative effectors. In contrast to obvious requirements for the Bsa T3SS in vitro and in vivo (2), to our knowledge no effectors have been definitively shown to be required for invasion, replication in nonphagocytic cells, cell–cell spread, MNGC formation, or virulence in animals. BopA, a suspected T3SSBsa substrate, is reported to facilitate survival and evasion of autophagy in phagocytic cells (11), but bopA mutants are not significantly attenuated in mice (2). The precise mechanism of T3SS-mediated endosome escape is unknown for any intracellular pathogen; however, it could conceivably be a function of translocon insertion in the endosomal membrane and occur in the absence of additional effectors. Finally, there is an instructive contrast between the roles of the Burkholderia Bsa and Shigella Mxi-Spa T3SSs. The Shigella system is essential for both endosome escape and escape from double-membrane vacuoles formed during the process of cell–cell spread (7). For Burkholderia, our results support a model for intracellular spread that obviates the need for membrane lysis after the primary endosome has been breached.
Intracellular Motility.
Polarized, unidirectional actin polymerization is a hallmark of cell–cell spread and plays an essential role for Shigella, Listeria, and other intracellular pathogens (9). The discovery that Bt remains fully capable of plaque formation in the absence of BimA was quite unexpected. Even more surprising was the observation that a predicted flagellar system on chromosome 2 (fla2) can compensate for the lack of actin motility and drive intercellular spread and MNGC formation.
The Bt and Bp fla1 flagellar gene clusters on chromosome 1 are highly conserved. fla1 encodes polar flagella, which in Bp have been implicated in invasion of epithelial cells and virulence in animal models (2). In Bt, mutation of motA1 (fla1) or motA2 (fla2) individually or in combination had no effect on invasion. Although this is a first characterization of dual flagellar motility systems in an intracellular pathogen, their occurrence and functions have been described in Vibrio and Aeromonas spp., where they facilitate motility in response to different environmental signals (27). Not surprisingly, fla1 and fla2 in Bt were observed to function under different conditions; deletion of motA1 eliminated swarming in soft agar but had no effect on motility following infection. Conversely, deletion of motA2 had no effect in soft agar but eliminated rapid intracellular motility.
Our sequence analysis suggests that fla2 encodes lateral flagella (Fig. S3B), but this prediction awaits direct confirmation. It is also unknown how the system is regulated or whether intracellular bacteria are simultaneously capable of BimA-mediated actin polymerization and fla2-dependent motility. Because flagellin monomers are known to activate assembly of the NLRC4 inflammasome, resulting in cytokine production and inflammatory cell death (28), the use of flagella for intra- and intercellular motility is surprising. It is presently unknown whether fla2 flagellin induces inflammasome-dependent cytoplasmic responses or whether mechanisms to overcome them exist. Perhaps the most important question involves the relevance of our observations with Bt to pathogenesis in Bp and Bm. A recent analysis of the global population structure of Burkholderia species pathogenic for mammals predicts an Australian origin for Bp, with a single introduction event leading to the expansion of Southeast Asian isolates (SEA Bp) and Bm (29). Interestingly, the fla2 gene cluster is absent in SEA Bp isolates such as Bp340, which is dependent on BimA for plaque formation (Fig. 5D), but it is highly conserved in sequenced genomes from Australian strains (Fig. S3) (23). The potential role of the fla2 locus in pathogenesis by Australian Bp remains to be investigated.
T6SS-1.
T6SSs are widely distributed among pathogenic and nonpathogenic Gram-negative species; they have broad roles in survival and fitness and have been linked to virulence in numerous pathogens, including Bp, Bm, and Bt (2, 14). In our analysis, T6SS-1 was found to facilitate intercellular spread following infection or photothermal delivery. Consistent with recent reports (14, 15, 19), deletion of clpV1 resulted in a defect in plaque formation in HEK293 cells and a delay in the formation of MNGCs. ΔclpV1 mutants exhibited robust actin-polymerization and flagellar-mediated motility inside cells, and genetic dissection using the photothermal nanoblade established that T6SS-1 functions downstream of invasion and T3SSBsa-mediated endosome escape. Concomitant with decreased efficiency of MNGC formation, we observed an increase in cell survival following infection with ΔclpV1 mutants and an accumulation of intracellular bacteria. These data are consistent with the hypothesis that T6SS-1 participates in events that can alternatively facilitate intercellular spread by fusing cell membranes or kill cells by compromising their integrity.
Cell Fusion and Intercellular Spread.
Burkholderia efficiently induce MNGC formation in both phagocytic and nonphagocytic cells (10). We propose that cell fusion represents the primary path for intercellular spread and plaque formation by Bt and Bp (Fig. S4), and the same is likely to hold true for Bm. These observations are consistent with the results of time course experiments showing that the formation of MNGCs and their eventual lysis give rise to the open cores of plaques, and with the lack of a requirement for T3SSBsa in cell–cell spread following cytosolic delivery of bacteria using our photothermal nanoblade. Whereas Bt T3SSBsa mutants are incapable of endosome escape following infection, Bp T3SSBsa mutants are reported to exhibit delayed escape that eventually leads to MNGC formation (30), consistent with our proposal that cell–cell spread occurs independently of T3SSBsa activity for both Bt and Bp. Furthermore, mutations that eliminate intracellular motility or inactivate T6SS-1 have analogous effects on MNGC formation and cell–cell spread following photothermal delivery or infection. Our model also explains the lack of published reports demonstrating double-membrane vacuoles following engulfment of protrusions with Burkholderia at their tips, as observed with L. monocytogenes, S. flexneri, and other pathogens with similar lifestyles (7, 8). Although membrane protrusions are readily formed by wild-type Bt, they are not observed with ΔbimA mutants. The ability of ΔbimA strains that retain fla2 motility to efficiently form plaques shows that membrane protrusions are not required for intercellular spread.
Our results, along with a recent report from Stevens et al. (22), demonstrate a clear link between motility and the efficiency of cell fusion by intracellular Burkholderia. We hypothesize that flagellar and/or actin-mediated motility increases the frequency of contact between bacteria and host cell membranes and that contact is prerequisite for membrane fusion through a process facilitated by T6SS-1. It is tempting to speculate that a bacterially encoded fusogenic factor is involved in this unique mechanism of cell–cell spread.
Materials and Methods
Detailed experimental procedures are found in SI Materials and Methods. BtE264 (17) and Bp340 (SEA Bp 1026b ΔamrRAB-oprA) (18) mutants were constructed using allelic exchange as described (31). Nanoblade delivery was performed as described in SI Materials and Methods and in ref. 16.
Supplementary Material
Acknowledgments
We thank the University of California (Los Angeles) Vector Core for lentiviral reagents and Mary Burtnick at the University of South Alabama for Bt antiserum. This work was supported by the Pacific Southwest Regional Center of Excellence in Biodefense and Emerging Infectious Diseases (U54 A1065359) (to J.F.M.), the National Science Foundation [Chemical, Bioengineering, Environmental, and Transport Systems (CBET) 0853500 and Electrical, Communications, and Cyber Systems (ECCS) 0901154] (to P.-Y.C.), a University of California Discovery Biotechnology Award (178517) (to P.-Y.C.), the National Institutes of Health Roadmap for Medical Research Nanomedicine Initiative (PN2EY018228) (to M.A.T.), and an Innovator Award from the Broad Stem Cell Research Center at University of California, Los Angeles (to M.A.T.). C.T.F. is supported by a National Institutes of Health Ruth L. Kirschstein National Research Service Award (GM07185), the Warsaw Microbiology Fellowship, and the University of California (Los Angeles) Dissertation Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107183108/-/DCSupplemental.
References
- 1.Wiersinga WJ, van der Poll T, White NJ, Day NP, Peacock SJ. Melioidosis: Insights into the pathogenicity of Burkholderia pseudomallei. Nat Rev Microbiol. 2006;4:272–282. doi: 10.1038/nrmicro1385. [DOI] [PubMed] [Google Scholar]
- 2.Galyov EE, Brett PJ, DeShazer D. Molecular insights into Burkholderia pseudomallei and Burkholderia mallei pathogenesis. Annu Rev Microbiol. 2010;64:495–517. doi: 10.1146/annurev.micro.112408.134030. [DOI] [PubMed] [Google Scholar]
- 3.Nandi T, et al. A genomic survey of positive selection in Burkholderia pseudomallei provides insights into the evolution of accidental virulence. PLoS Pathog. 2010;6:e1000845. doi: 10.1371/journal.ppat.1000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holden MT, et al. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci USA. 2004;101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim HS, et al. Bacterial genome adaptation to niches: Divergence of the potential virulence genes in three Burkholderia species of different survival strategies. BMC Genomics. 2005;6:174. doi: 10.1186/1471-2164-6-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.West TE, Frevert CW, Liggitt HD, Skerrett SJ. Inhalation of Burkholderia thailandensis results in lethal necrotizing pneumonia in mice: A surrogate model for pneumonic melioidosis. Trans R Soc Trop Med Hyg. 2008;102(Suppl 1):S119–S126. doi: 10.1016/S0035-9203(08)70028-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ray K, Marteyn B, Sansonetti PJ, Tang CM. Life on the inside: The intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol. 2009;7:333–340. doi: 10.1038/nrmicro2112. [DOI] [PubMed] [Google Scholar]
- 8.Tilney LG, Portnoy DA. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J Cell Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stevens JM, Galyov EE, Stevens MP. Actin-dependent movement of bacterial pathogens. Nat Rev Microbiol. 2006;4:91–101. doi: 10.1038/nrmicro1320. [DOI] [PubMed] [Google Scholar]
- 10.Kespichayawattana W, Rattanachetkul S, Wanun T, Utaisincharoen P, Sirisinha S. Burkholderia pseudomallei induces cell fusion and actin-associated membrane protrusion: A possible mechanism for cell-to-cell spreading. Infect Immun. 2000;68:5377–5384. doi: 10.1128/iai.68.9.5377-5384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong L, et al. The Burkholderia pseudomallei type III secretion system and BopA are required for evasion of LC3-associated phagocytosis. PLoS ONE. 2011;6:e17852. doi: 10.1371/journal.pone.0017852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pukatzki S, et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schell MA, et al. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol Microbiol. 2007;64:1466–1485. doi: 10.1111/j.1365-2958.2007.05734.x. [DOI] [PubMed] [Google Scholar]
- 14.Schwarz S, et al. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 2010;6:e1001068 doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burtnick MN, et al. The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect Immun. 2011;79:1512–1525. doi: 10.1128/IAI.01218-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu TH, et al. Photothermal nanoblade for large cargo delivery into mammalian cells. Anal Chem. 2011;83:1321–1327. doi: 10.1021/ac102532w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brett PJ, DeShazer D, Woods DE. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int J Syst Bacteriol. 1998;48:317–320. doi: 10.1099/00207713-48-1-317. [DOI] [PubMed] [Google Scholar]
- 18.Mima T, Schweizer HP. The BpeAB-OprB efflux pump of Burkholderia pseudomallei 1026b does not play a role in quorum sensing, virulence factor production, or extrusion of aminoglycosides but is a broad-spectrum drug efflux system. Antimicrob Agents Chemother. 2010;54:3113–3120. doi: 10.1128/AAC.01803-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pilatz S, et al. Identification of Burkholderia pseudomallei genes required for the intracellular life cycle and in vivo virulence. Infect Immun. 2006;74:3576–3586. doi: 10.1128/IAI.01262-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stevens MP, et al. A Burkholderia pseudomallei type III secreted protein, BopE, facilitates bacterial invasion of epithelial cells and exhibits guanine nucleotide exchange factor activity. J Bacteriol. 2003;185:4992–4996. doi: 10.1128/JB.185.16.4992-4996.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haraga A, West TE, Brittnacher MJ, Skerrett SJ, Miller SI. Burkholderia thailandensis as a model system for the study of the virulence-associated type III secretion system of Burkholderia pseudomallei. Infect Immun. 2008;76:5402–5411. doi: 10.1128/IAI.00626-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sitthidet C, et al. Actin-based motility of Burkholderia thailandensis requires a central acidic domain of BimA that recruits and activates the cellular Arp2/3 complex. J Bacteriol. 2010;192:5249–5252. doi: 10.1128/JB.00608-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuanyok A, et al. A horizontal gene transfer event defines two distinct groups within Burkholderia pseudomallei that have dissimilar geographic distributions. J Bacteriol. 2007;189:9044–9049. doi: 10.1128/JB.01264-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shalom G, Shaw JG, Thomas MS. In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology. 2007;153:2689–2699. doi: 10.1099/mic.0.2007/006585-0. [DOI] [PubMed] [Google Scholar]
- 25.Boddey JA, et al. The bacterial gene lfpA influences the potent induction of calcitonin receptor and osteoclast-related genes in Burkholderia pseudomallei-induced TRAP-positive multinucleated giant cells. Cell Microbiol. 2007;9:514–531. doi: 10.1111/j.1462-5822.2006.00807.x. [DOI] [PubMed] [Google Scholar]
- 26.Patel JC, Galán JE. Manipulation of the host actin cytoskeleton by Salmonella—all in the name of entry. Curr Opin Microbiol. 2005;8:10–15. doi: 10.1016/j.mib.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 27.McCarter LL. Dual flagellar systems enable motility under different circumstances. J Mol Microbiol Biotechnol. 2004;7:18–29. doi: 10.1159/000077866. [DOI] [PubMed] [Google Scholar]
- 28.Miao EA, et al. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol. 2010;11:1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearson T, et al. Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer. BMC Biol. 2009;7:78. doi: 10.1186/1741-7007-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burtnick MN, et al. Burkholderia pseudomallei type III secretion system mutants exhibit delayed vacuolar escape phenotypes in RAW 264.7 murine macrophages. Infect Immun. 2008;76:2991–3000. doi: 10.1128/IAI.00263-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrett AR, et al. Genetic tools for allelic replacement in Burkholderia species. Appl Environ Microbiol. 2008;74:4498–4508. doi: 10.1128/AEM.00531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.