Abstract
Evidence for cooperation between actin nucleators is growing. The WH2-containing nucleator Spire and the formin Cappuccino interact directly, and both are essential for assembly of an actin mesh during Drosophila oogenesis. Their interaction requires the kinase noncatalytic C-lobe domain (KIND) domain of Spire and the C-terminal tail of the formin. Here we describe the crystal structure of the KIND domain of human Spir1 alone and in complex with the tail of Fmn2, a mammalian ortholog of Cappuccino. The KIND domain is structurally similar to the C-lobe of protein kinases. The Fmn2 tail is coordinated in an acidic cleft at the base of the domain that appears to have evolved via deletion of a helix from the canonical kinase fold. Our functional analysis of Cappuccino reveals an unexpected requirement for its tail in actin assembly. In addition, we find that the KIND/tail interaction blocks nucleation by Cappuccino and promotes its displacement from filament barbed ends providing insight into possible modes of cooperation between Spire and Cappuccino.
Many processes in the eukaryotic cell depend upon the timely generation and disassembly of actin filaments. The rate-limiting step of filament formation is the creation of a stable actin nucleus. At least three different classes of proteins have evolved to accelerate this step: formins, the Arp2/3 complex, and Wiscott–Aldrich homology 2 (WH2)-domain nucleators (1). How actin nucleators from different classes cooperate to build particular actin structures is an area of intense interest and investigation. For example, the direct biochemical and genetic links between WH2-based Spir (2, 3) and the formin Cappuccino (4) suggest close mechanistic collaboration between these proteins in actin assembly (5–7).
Spire (spir) and cappuccino (capu) were first identified in screens for genes affecting embryonic pattern formation in Drosophila melanogaster (Dm) (7). They synergize to build a cytoplasmic actin mesh, and mutation of either of these genes results in loss of this structure, premature microtubule-dependent cytoplasmic streaming, and gross defects in embryonic morphology (8, 9). Mice lacking formin-2 (Fmn2; a mammalian Capu ortholog) exhibit egg failure and female hypofertility (10) due to loss of an actin-based structure during meiosis (11, 12), supporting the functional conservation of these proteins in higher eukaryotes.
Formins possess an actin-nucleating formin homology 2 (FH2) domain and an adjacent proline-rich FH1 domain (Fig. 1A). The FH2 domain remains bound to the barbed end of the actin filament as additional subunits are added, protecting growing ends from the activity of capping proteins, which would otherwise terminate elongation (13). Fmn-family formins, including Capu and orthologs Fmn1 and Fmn2 (5, 14, 15), lack obvious regulatory domains found in diaphanous-related formins (DRFs), which include a GTPase-binding domain, diaphanous inhibitory domain, and C-terminal diaphanous autoregulatory domain (DAD) (16, 17). Fmn-family formins do have short (approximately 25 aa) sequences C-terminal to the FH2 domain that do not resemble the DAD motif but are highly conserved, suggesting their functional importance (18).
Fig. 1.
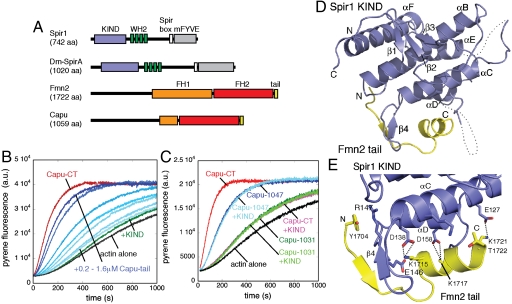
The Capu-tail is essential for Capu activity and Spir-KIND binding. (A) Schematics of Spir and Fmn2/Capu. KIND, kinase noncatalytic C-lobe domain (blue); WH2, Wiscott-Aldrich homology-2 motif (green); Spir box (white); mFYVE, modified Fab1/YOTB/Vac1/EEA1 zinc-binding domain (gray); FH1, formin homology-1 (orange); FH2, formin homology-2 (red); tail (yellow). (B) KIND inhibition of Capu-CT is competed by Capu-tail. Baseline conditions are 10 nM Capu-CT (red) plus 800 nM KIND (green). Addition of 200 nM to 1.6 μM Capu-tail (increasing with shades of blue) competes with KIND indicating that this is the major site of interaction. (C) KIND cannot inhibit truncated Capu. The last 12 or 28 residues of Capu were deleted from Capu-CT (Capu1047 and Capu1031, respectively). Further characterization of these truncations is presented in Fig. S2 D–F. Twenty nM Capu-CT, Capu1047, or Capu1031 and 2 μM Dm-Spir-KIND are added where indicated. (D) The structure of the Spir1-KIND (blue)/Fmn2-tail (yellow) complex. Unstructured loops are drawn with dashed lines. (E) Detailed view of the Spir1/Fmn2 peptide interface. Selected main chain and side chain atoms are drawn in stick representation. Hydrogen bonds are illustrated with dashed black lines.
In addition to a central cluster of four WH2 domains, Spir contains an N-terminal kinase noncatalytic C-lobe domain (KIND) (19), a modified Fab1/YOTB/Vac1/EEA1 (mFYVE) zinc-binding domain, and a C-terminal DEJL motif (Fig. 1A) (2, 20). Several spir gene family members have been identified in other organisms (3, 21), including paralogs Spir1 and Spir2 in higher eukaryotes. The clusters of four WH2 domains of Dm-Spir and human Spir1 nucleate actin in vitro (5, 15, 22).
Based on sequence homology, the KIND domain was hypothesized to adopt a structure similar to the C-lobe of protein kinases (19). It is predicted to lack kinase activity, as it is missing several key residues required for kinase activity as well as the entire N-lobe of the kinase fold. Instead, it is thought to function as a protein–protein interaction domain (5, 19). The KIND domain is detected in proteins of various functions, including the protein tyrosine phosphatase-L1 and the Ras guanine nucleotide exchange factor very-KIND (19, 23, 24). To date, there are no three-dimensional structural data available for any KIND domain. The Spir KIND domain mediates specific, high-affinity interactions with C-terminal constructs of Capu (5, 18). Recently, mammalian Spir1 and Spir2 KIND domains were reported to bind directly to the C-terminal tail, distal to the FH2 domains, of Fmn1 and Fmn2 (18). The KIND domain inhibits actin polymerization by Capu (and Fmn2), but it remains to be determined whether the nucleation and/or elongation steps of actin assembly are affected when the KIND domain binds to the tail of Capu (5).
To better understand the physical association and functional cooperation between Spir and Capu, we determined the 2.2-Å crystal structure of the human Spir1 KIND domain bound to the Fmn2 tail (identical in several species including humans). We determined that the interaction observed in this structure is critical for the regulation of actin dynamics by Spir and Capu and for their colocalization in cells. We found that Capu cannot nucleate or protect the barbed ends of actin filaments in the presence of the KIND domain but that Capu, Spir, and actin monomers form a stable complex.
Results
Dual Functions of the Capu C-Terminal Tail.
We discovered that a short polypeptide segment at the extreme C-terminus of Capu (residues 1023–1059) was necessary and sufficient for binding to the Dm-Spir KIND domain (see Fig. S1 for diagrams of constructs and Fig. S2 A–C for binding data). Similar results were reported for human Spir1 and Spir2 KIND domains, which bind to the C-terminal Formin–Spir interaction (FSI) domains of Fmn1 and Fmn2 (18). Using competition fluorescence anisotropy, we measured an equilibrium dissociation constant (Kd = 290 nM) for Dm-Spir-KIND and Capu-tail (residues 1029–1059; Table S1), which is similar to that reported for binding of human Spir1 KIND to a Fmn2 FSI peptide (Kd = 260 nM; see SI Discussion) (18).
We next tested the role of the KIND/tail interaction in Capu-mediated actin assembly. The KIND domains of Dm-Spir and human Spir1 inhibit Capu and Fmn2 stimulated actin polymerization in vitro, respectively (5). We asked whether the KIND/tail interaction was required for this inhibition using a peptide competition assay (25). Indeed, the Capu-tail peptide competed the inhibitory effect of Dm-Spir-KIND on Capu-CT (Fig. 1B). A half-maximal response was observed at approximately 1 μM of the peptide, close to the concentration of Spir-KIND present (800 nM), suggesting that the Capu-tail and Capu-CT bind Dm-Spir-KIND with similar affinities (see SI Discussion).
We also tested whether Dm-Spir-KIND could inhibit the actin assembly activity of a Capu construct lacking the tail domain. To our surprise, we discovered that Capu FH1FH2 constructs in which the tail was truncated were defective in actin assembly (Fig. 1C). In particular, Capu-1031 (residues 467–1031; Fig. S2 D and E) showed little activity above actin alone in pyrene-actin assembly assays. A construct with about half of the tail (Capu-1047) retained some actin assembly activity but much less than Capu-CT. These C-terminal truncations are unlikely to affect the structural integrity of the FH2 domain, as they are outside the predicted structural core of the domain, and both constructs elute in analytical gel filtration at the volume expected for properly folded dimers (Fig. S2F). Both truncation constructs exhibit longer lag times, suggesting that nucleation rates are decreased. The residual activity of both Capu-1047 and Capu-1031 was not affected by addition of Dm-Spir-KIND, confirming that tail binding is necessary for inhibition of Capu by Spir-KIND (Fig. 1C). We conclude that the C-terminal tail of Capu has at least two functions: It is required for efficient actin assembly and for binding to Spir. Because of its multiple roles, we use “tail” to refer to the C-terminal sequence instead of the more specific “FSI domain.”
Structure of the Spir1/Fmn2 Complex.
To better understand the interaction between Spir and Capu, we undertook structural studies of the KIND domain in complex with formin C-terminal fragments. We failed to obtain suitable crystals with Drosophila proteins or with any formin construct that included the FH2 domain, but were able to crystallize and determine the structure of the human Spir1 KIND domain in complex with the Fmn2 tail (Table S2). As expected, the architecture of the KIND domain resembles the C-terminal lobe of protein kinases (19). For ease of comparison, we label secondary structure elements of the KIND domain to correspond to those of the PAK1 protein kinase (see below). The KIND fold consists of six α-helices (αD, E, F, H, I, and J) and four β-strands (β6–β9) arranged to form a compact, globular domain (Fig. 1D). The structure spans residues 37–231 of Spir1, but no interpretable electron density is observed for the loop connecting strand β8 and helix αF (residues 104–118) or for the loop between helices αH and αI (residues 166–191). The Fmn2 tail docks in a wide groove on the base of the KIND domain with two primary sites of interaction (Fig. 1E). Residues 1704–1722 of the Fmn2 tail are visible in the structure; residues 1706–1708 form a short β-strand that interacts with β9 of the KIND domain, whereas residues 1714–1722 form an amphipathic α-helix that packs with helices αF and αH of the KIND domain. In total, Fmn2 binding buries 848 Å2 of solvent-accessible surface on Spir1. The crystal structure reveals a 1∶1 stoichiometry of binding between the KIND domain and the Fmn2 tail, and thus suggests that two molecules of Spir-KIND could bind to the formin dimer. This is indeed the case; our structural and functional analysis shows that two molecules of Spir-KIND bind to dimeric formin constructs in solution (Fig. S2 G and H and SI Discussion).
We also determined the structure of the Spir1 KIND domain alone (Table S2). Comparison of the apo-KIND structure with the KIND/tail complex reveals no dramatic structural differences; the structures superimpose with an rmsd of 0.45 Å over 153 α-carbon atoms (Fig. S4A). We do note that strand β9 and adjacent loops are poorly ordered in the apo-KIND structure. This segment interacts extensively with the Fmn2 peptide, apparently leading to its stabilization.
Evolution of a Binding Domain from the Protein Kinase Fold.
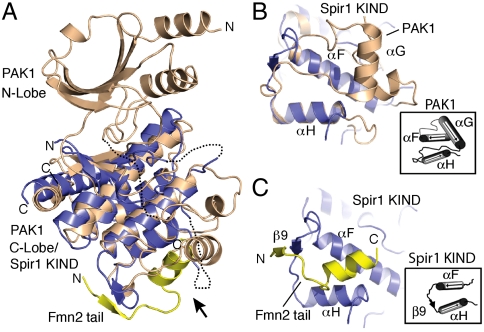
A search of the Protein Data Bank (PDB) for structural relatives of the KIND domain using the Dali structural similarity server (27) revealed similarity with the C-lobe of several hundred protein kinase structures with rmsd values in the range of 2.4–3.6 Å and sequence identity for equivalent residues from 9–22%. We compare the KIND domain with the kinase PAK1 as a representative example (Fig. 2) (19). The KIND domain superimposes with an rmsd of 2.9 Å over 134 atoms in the C-lobe of PAK1, and the superimposed residues are 19% identical (Fig. S5A). In PAK1 and other kinases, the N- and C-lobes are independent structural domains connected by a “hinge” segment. There is no equivalent of the N-lobe in the KIND domain; instead, the N-terminus of the KIND domain maps to the hinge region of the kinase fold.
Fig. 2.
Structure of the Spir1-KIND/Fmn2-tail complex and comparisons to the protein kinase fold. (A) Superposition of the Spir1-KIND/Fmn2-tail structure with the C-lobe of PAK1 protein kinase. Spir1 and Fmn2 are colored as in Fig. 1 and PAK1 is tan; PDB ID code 1F3M (26). The direction of view in panels B and C is indicated by an arrow. (B) Close-up view of the Spir1 superposition with PAK1. The Fmn2 tail has been omitted for clarity. Labels indicate secondary structural elements of the kinase fold. The organization of helices F, G, and H is shown schematically in the inset. Note the absence of an equivalent to helix G in the KIND domain, and the relationship to the binding site of the Fmn2 tail shown in panel C. (C) Close-up view of the Spir1/Fmn2 tail interaction in the same orientation as panel B. Secondary structural elements of Spir1 are labeled, and shown schematically in the Inset.
How might the binding function of the KIND domain have arisen from the protein kinase fold? The superposition with PAK1 reveals that there is no equivalent of helix αG in the KIND domain fold (Fig. 2 B and C and Fig. S5A). This secondary structure element lies at the base of the kinase C-lobe, and is conserved in the protein kinase fold. Helix αG and the loops that connect it with adjacent helices αF and αH are replaced instead by a short polypeptide segment containing strand β9, which is not present in PAK1. Interestingly, the hydrophobic “scar” left by deletion of these elements is the binding site for the Fmn2 tail. Strand β9 interacts directly with the tail peptide and the amphipathic helix in the tail peptide packs with its hydrophobic face against helices αF and αH (Fig. 2C). Thus the binding function of the KIND domain appears to have evolved by deletion of a conserved element of secondary structure.
Interactions in the Spir1-Fmn2 Interface.
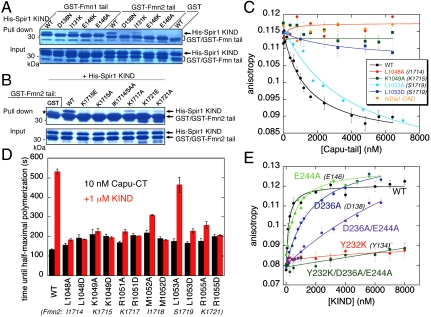
N-terminal to the mFmn2-tail β-strand, the side chain of Y1704 stacks with the guanidinium group of R147 in the KIND domain, and just C-terminal to it, residues 1709–1713 form an irregular but well-ordered loop that leads to the α-helical segment (Fig. 1E). Isoleucine residues 1714 and 1718 in the tail α-helix pack with KIND domain residues including Y134 (Fig. S4D) to form a hydrophobic core in the interface. Lysine residues extend from both sides of the tail α-helix to contact acidic residues on the KIND domain; these Spir1/Fmn2 salt-bridges include K1715 with D138, K1717 with D158, and K1721 with E127 (Fig. 1E). Mutation of D138 in the KIND domain to asparagine or alanine abrogates binding to both Fmn1 and Fmn2 tail peptides in GST-pulldown experiments (Fig. 3A). Similarly, mutation of residue K1715 in Fmn2 to either glutamic acid or alanine abrogates pulldown of the KIND domain (Fig. 3B). Mutation of tail residues K1717 or K1721 also impairs binding of the KIND domain but to a lesser extent (Fig. 3B). Additional polar interactions with the KIND domain include dual hydrogen bonds between E146 in the KIND domain and the backbone amides of H1710 and K1715 in Fmn2. Mutation of E146 in the KIND domain to either alanine or lysine also disrupts binding to both Fmn1 and Fmn2 tail peptides in vitro (Fig. 3A).
Fig. 3.
Structure-function analysis of the tail and KIND domains. (A) GST-Fmn1-tail, or GST-Fmn2-tail were tested for their ability to pull down WT or mutant His6-Spir1 KIND domain. (B) Pull-downs of His6-Spir1 KIND domain with WT or mutant GST-Fmn2-tail. A copurified translational byproduct in the GST control condition is indicated by an asterisk. Data in (A) and (B) are representative of three independent experiments. (C) Competition anisotropy of 10 nM Capu-tail-AlexaFluor488 from 600 nM Dm-Spir-KIND with unlabeled Capu-tail, WT and mutants. Point mutations of conserved residues virtually abolish binding. Data are mean ± standard deviation (SD) for eight time points. Additional data are presented in Fig. S3A. (D) Point mutants of conserved tail residues in Capu-CT prevent inhibition of actin assembly by Dm-Spir-KIND in pyrene-actin polymerization assays. Time until half-maximal polymerization for all mutants in the presence or absence of 1 μM Dm-Spir-KIND are plotted. Data are mean ± SD for three trials. See raw data in Fig. S3B. (E) Polarization anisotropy of 10 nM Capu-tail and increasing concentrations of unlabeled WT or mutant Dm-Spir-KIND. Data are presented as in panel C. See Fig. S3C for additional mutants and Table S1 for Kd values.
Structure-Function Analysis of Spir–Capu Interactions.
Guided by the crystal structure and mutagenesis of the human Spir1/Fmn2 complex, we engineered mutations in Dm-Spir and Capu designed to disrupt their interaction. In fluorescence anisotropy assays, tail peptides with mutations in well-conserved basic residues K1049, R1051, and R1055 (corresponding to K1715, K1717, and K1721 in Fmn2) and in hydrophobic residues L1048 and M1052 (I1714 and I1718 in Fmn2) were unable to compete with WT Capu-tail for binding to Dm-Spir-KIND (Fig. 3C and Fig. S3A). Consistent with the anisotropy data, these mutations in the context of Capu-CT rendered the formin insensitive to inhibition by the KIND domain in pyrene-actin assembly assays (Fig. 3D and Fig. S3B). In contrast, an L1053A mutant (S1719 in Fmn2) retained activity comparable to WT in both assays. This residue is not conserved and is expected to be relatively solvent exposed in the complex based on its counterpart at the same position in Fmn2 (S1719; Figs. S4D and S5B). We also tested mutations in Dm-Spir KIND (Table S1 and Fig. 3E and Fig. S3C). Mutations at residues E229, D236, E244, and E252 (I131, D138, E146, and E154 in Spir1) decreased the binding affinity between 4- and 15-fold (Fig. 3E, Fig. S3C, and Table S1). Whereas single mutations at acidic residues did not eliminate KIND/tail binding, mutation of Dm-Spir Y232 to lysine abolished detectable binding in both the anisotropy assay (Fig. 3E) and the Capu inhibition assay (Fig. S3D). This residue corresponds to Y134 in human Spir1, which is buried in the KIND/Fmn2-tail interface, and is conserved as tyrosine or phenylalanine among KIND domains (Fig. S5A).
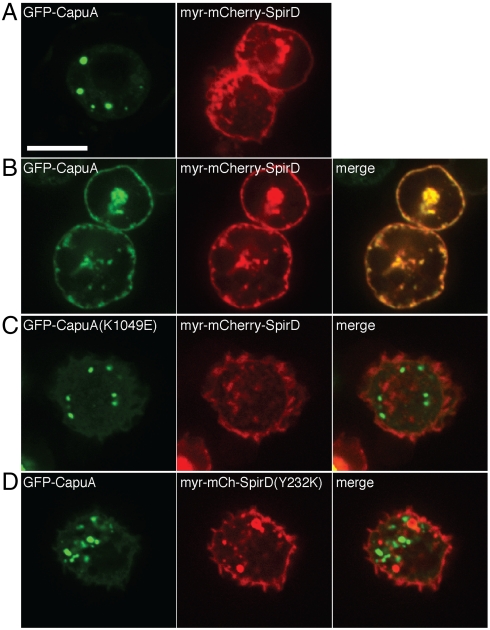
To assess importance of the KIND/tail binding interaction on full-length proteins in a cellular environment, we developed a colocalization assay in Drosophila S2 cells. Myristoylated Dm-SpirD fused to mCherry (myr-mCherry-SpirD) was membrane-localized in S2 cells (Fig. 4A). In the absence of Spir, Capu fused to green fluorescent protein (GFP–CapuA) was found in isolated puncta (Fig. 4A). Upon coexpression with myr-mCherry-SpirD, GFP-CapuA was predominantly membrane-localized (Fig. 4B). This colocalization was abolished by the K1049E mutation in the Capu tail (K1715 in Fmn2) and by the Y232K mutation in the Dm-Spir KIND domain (Fig. 4 C and D). Analysis of additional mutants is presented in Fig. S6. These defects in binding and colocalization are not simply due to gross misfolding of the KIND domain, as confirmed by circular dichroism (Fig. S7A).
Fig. 4.
Requirements for colocalization of Capu and Spir in Drosophila S2 cells. GFP-CapuA and myr-mCherry-SpirD are expressed either (A) individually or (B–D) together in S2 cells. (A) The myristoylation sequence fused to mCherry-SpirD drives it to membranes. GFP-CapuA is concentrated in bright puncta when expressed alone. (B) When coexpressed with myr-mCherry-SpirD, WT CapuA colocalizes with SpirD at the plasma membrane. (C) The Capu mutant K1049E (K1715 in Fmn2) fails to colocalize with SpirD. (D) The SpirD mutant Y232K (Y134 in Spir1) fails to colocalize with CapuA. Scale bar is 10 μm. Analysis of additional mutants is shown in Fig. S6.
Spir-KIND Promotes Dissociation of Capu From the Filament Barbed End.
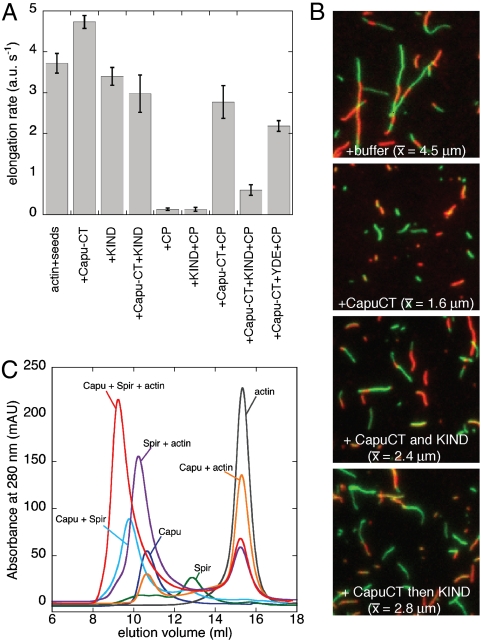
In bulk polymerization assays, the Spir KIND domain inhibits Capu-mediated polymerization (5). Nucleation is inhibited, as demonstrated by increased lag times; however, it is not clear whether Spir-KIND affects elongation rates of Capu or even whether Capu is able to bind the barbed end of growing filaments when bound to Spir-KIND. To test the effect of Spir-KIND on Capu-bound barbed ends, we measured elongation by monitoring pyrene-actin polymerization from preformed seeds in the presence of 0.5 μM monomeric actin, which is below the critical concentration of the pointed end (28). We found that Capu alone does not cause a decrease in the elongation rate in the absence of profilin, an unusual property among formins (Fig. 5A (+Capu-CT), Fig. S7F, and SI Discussion) (29, 30). Like other formins (13, 31, 32), Capu protects the growing barbed end from capping protein [Fig. 5A (+Capu-CT+CP)]. To determine if Capu can processively associate with the barbed end in the presence of Dm-Spir-KIND, we added Capu-CT, Dm-Spir-KIND, and capping protein to the elongation assay. The presence of Dm-Spir-KIND dramatically reduces the ability of Capu-CT to protect the barbed end from capping protein, whether Capu-CT and Dm-Spir-KIND were mixed before addition to actin or Dm-Spir-KIND was added after elongation assays were initiated (Fig. 5A (+Capu-CT+KIND+CP) and Fig. S7E). The Dm-Spir-KIND “YDE” mutant (Y232K/D236A/E244A) has a reduced ability to inhibit Capu protection of the barbed end, confirming that the effect of Spir-KIND requires its interaction with the Capu-tail [Fig. 5A (+Capu-CT+YDE+CP)].
Fig. 5.
Interactions between Spir, Capu, and actin. (A) Actin filament assembly from preformed seeds is measured in the absence or presence of Capu-CT, WT or mutant Dm-Spir-KIND, and capping protein (CP) as indicated. The Spir-KIND YDE mutant has a significantly reduced effect on protection by Capu-CT compared to WT KIND (p < 0.0001, unpaired Student’s t test). Protein concentrations are 0.5 μM Mg-G-Actin (20% pyrene labeled), 0.25 μM F-actin seeds (approximately 50 pM barbed ends), 0.375 nM capping protein, 50 nM Capu-CT, 5 μM Dm-Spir-KIND (WT or YDE). Data are mean ± SD for at least three independent trials (see Fig. S7D for representative raw data). (B) Actin filaments stabilized with either AlexaFluor488 (green) or AlexaFluor647 (red) labeled phalloidin were imaged 30 min after shearing. When 10 nM Capu-CT was added after shearing, few reannealing events are detected. Addition of KIND and Capu-CT gave an intermediate result regardless of the order of addition. (C) Gel filtration of Capu-CT, Spir-NT, and actin demonstrates that these three proteins form a stable complex. See Fig. S7C for SDS-PAGE analysis of peak fractions.
The inability of the Spir/Capu complex to protect filament barbed ends from capping protein indicates that binding of the KIND domain to Capu diminishes its affinity for the barbed end. To further explore this topic, we tested the effect of Capu versus the Dm-Spir-KIND/Capu complex on reannealing of sheared actin filaments (Fig. 5B). As expected based on structural data (33) and demonstrated for other formins (34), addition of Capu-CT alone markedly decreased filament annealing (average length = 1.6 μm, compared with 4.5 μm in the absence of Capu). Addition of the KIND domain diminished this effect, but did not completely reverse it (average length = 2.4 μm when added simultaneously, or 2.8 μm when added sequentially; see SI Discussion). Taken together, our data indicate that Spir-KIND binding promotes dissociation of Capu from the barbed end of actin filaments, and that mutations that block the KIND/tail interaction diminish this effect. We therefore conclude that Spir-KIND inhibition of Capu-CT-mediated polymerization is solely the result of the KIND domain blocking nucleation.
A Stable Spir–Capu–Actin Complex.
To gain further insight into the Spir/Capu complex and its interaction with actin, we asked whether the three proteins form a stable complex under nonpolymerizing conditions. First, we examined pairwise combinations of these proteins by gel filtration and found that Spir-NT and Capu-CT coelute as expected (Fig. 5C and Fig. S7C). Capu-CT does not coelute with actin in these conditions. Spir-NT does coelute with actin in a single peak, as reported for human Spir1 (22). However, an actin peak was also detected despite the fact that actin was subsaturating. The presence of unbound actin suggests that Dm-Spir-NT does not bind actin monomers with the high degree of cooperativity observed for human Spir1 (22). When Spir, Capu, and actin were gel filtered, all three components eluted in a single peak, indicating that they form a stable complex (Fig. 5C). Actin does not elute with a Dm-Spir-KIND/Capu-CT complex; actin does elute with Spir-NT/Capu-CT (I706A) [a mutation that blocks nucleation (5, 35)] (Fig. S7B). Together these data show that Spir-WH2 domains are necessary and sufficient to stabilize actin in the complex.
Discussion
The work presented here defines the structural relationship between the KIND domain and the C-lobe of protein kinases. Whereas the C-lobe of protein kinases is a frequent site of interaction with substrates, inhibitors, and other binding partners, (36) the formin/KIND interaction appears to be structurally unique. Indeed, the Spir KIND domain binds the Fmn2 tail in a cleft at the base of the domain that is created by deletion of a helix from the kinase fold (helix αG). The absence of this helix in KIND domains was not previously appreciated due to inaccuracies in primary sequence alignments between kinases and KIND domains (19). The resulting cleft is well-conserved in the KIND domain of Spir family members, indicating that the observed binding interaction is conserved as well. However, the variability in amino acid residues predicted to line the cleft in other KIND domains outside the Spir family makes it difficult to predict whether its binding function is general among all KIND domains.
Our structure-function data indicate that the contacts observed in the crystal structure are essential to the interaction between these two proteins in solution and in cells. Unexpectedly, we find that the C-terminal tail of Capu is required for efficient actin assembly. The mechanistic basis for this effect remains to be elucidated, but one possibility is enhancement of nucleation via participation in binding of actin monomers, as has recently been described for the DAD domain of mDia1 and other DRFs (37). The Capu tail does not contain a DAD, but it is notable that it is enriched in basic residues, similar to the nucleation-crticial region of mDia1 DAD (37). Although our gel filtration data indicate that Capu does not bind monomers at physiological salt concentrations in vitro, our preliminary data do show weak binding of the Capu tail to actin at lower salt concentrations. Even weak association between the Capu tail and actin could affect nucleation. Our finding that the Capu/Spir complex binds actin monomers in a stable complex may provide insight into the nucleus formed by the collaboration. Further study of this ternary complex will be required to determine how the actin associates with Spir and Capu and whether this complex contains a nascent filament.
Our functional studies indicate that binding of Spir to the Capu tail promotes dissociation of the formin from the actin filament barbed end. Biochemically, this effect is similar to that reported for the action of Bud14 on the yeast formin Bnr1 (38). However, these apparently inhibitory biochemical activities of Spir on the formin are at odds with the genetic data, which indicate that both Spir and Capu play a positive role in assembling the actin meshwork during oogenesis. This apparent paradox is resolved if Spir and Capu dissociate in the process of filament assembly, perhaps in a “handoff” of a Spir-nucleated filament to the formin, which would protect the elongating barbed end via its processive capping activity (39). Further efforts are required to determine if and how the Spir/Capu interaction is broken.
Methods
Protein Constructs, Purification, and Assays.
Proteins were bacterially expressed and purified by standard protocols as described in SI Methods. Constructs are depicted in Fig. S1. Procedures for mutagenesis, GST pull-down assays, filament annealing, analytical gel filtration, S2 cell culture, and imaging are described in SI Methods.
Crystallization and Structure Determination.
Crystals of the human Spir1 KIND domain (residues 20–237) in complex with the Fmn2 tail (residues 1700–1722) were obtained at 4 °C in 0.1 M MES (pH 6.0), 5 mM Tris(2-carboxyethyl)phosphine (TCEP), 200 mM NaCl and 28% (vol/vol) PEG 400. The structure was determined from a tetragonal crystal form (space group P43212) by multiple isomorphous replacement using mercury and platinum heavy-atom derivatives. This structure was refined to an Rcryst of 0.194 (Rfree = 0.228) with data extending to 2.2 Å resolution. Apo-Spir1 KIND domain was crystallized in 0.1 M Na-Hepes (pH 7.5), 5 mM TCEP, 150 mM NaCl, and 20% PEG 3350. The unliganded crystals belonged to space group P21 and contained four molecules in the asymmetric unit. The apo-structure was refined to an Rcryst of 26.1% (Rfree = 31.5%) at 3.2 Å resolution. See SI Methods and Table S2 for further details.
Fluorescence Anisotropy and Actin Assembly Experiments.
Anisotropy of Capu-tail-AlexaFluor488 was measured by fluorometry (PTI, Inc.). All assays were carried out at 25 °C in 10 mM Na-Hepes pH 7.0, 1 mM EGTA, 1 mM TCEP, 150 mM KCl, 1 mM MgCl2, and 1 mg/mL BSA. The fluorophore was excited by plane-polarized light at 488 nm and emission was measured at 520 nm at angles parallel and perpendicular to the angle of incidence. Data were fit as described previously (5).
Pyrene actin assembly assays were carried out essentially as described (40). Briefly, 4 μM Acanthamoeba castellani actin (5% pyrene labeled) was incubated for 2 min at 25 °C with 200 μM EGTA and 50 μM MgCl2 to convert Ca-actin to Mg-actin. Polymerization was initiated by adding polymerization buffer (final concentration: 10 mM Na-Hepes pH 7.0, 1 mM EGTA, 50 mM KCl, 1 mM MgCl2) to the Mg-G-actin. Additional components, such as Capu-CT, Capu-tail and KIND were combined in the polymerization buffer before addition to Mg-G-actin. See SI Methods for a description of elongation assays.
Supplementary Material
Acknowledgments.
We thank H. K. Song for helpful discussions and for support through the World Class University Program of the National Research Foundation of Korea (Award R33-10108). We thank the NE-CAT beamline staff at the Advanced Photon source for assitance with data collection; the NE-CAT is supported by NCRR award RR-15301. This work was supported in part by NIH Grant GM071834 (M.J.E.), grants from the Burroughs-Wellcome Fund (Career Award in the Biomedical Sciences), and March of Dimes Foundation Grant #5-FY10-81 (M.E.Q.), and NIH NRSA postdoctoral fellowship F32GM087857 (C.L.V.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 3R7G (Spir1-KIND/Fmn2 complex) and 3RBW (unliganded KIND domain)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105703108/-/DCSupplemental.
References
- 1.Campellone KG, Welch MD. A nucleator arms race: Cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Otto IM, et al. The p150-Spir protein provides a link between c-Jun N-terminal kinase function and actin reorganization. Curr Biol. 2000;10:345–348. doi: 10.1016/s0960-9822(00)00388-2. [DOI] [PubMed] [Google Scholar]
- 3.Wellington A, et al. Spire contains actin binding domains and is related to ascidian posterior end mark-5. Development. 1999;126:5267–5274. doi: 10.1242/dev.126.23.5267. [DOI] [PubMed] [Google Scholar]
- 4.Emmons S, et al. Cappuccino, a Drosophila maternal effect gene required for polarity of the egg and embryo, is related to the vertebrate limb deformity locus. Genes Dev. 1995;9:2482–2494. doi: 10.1101/gad.9.20.2482. [DOI] [PubMed] [Google Scholar]
- 5.Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, Kerkhoff E. Regulatory interactions between two actin nucleators, Spire and Cappuccino. J Cell Biol. 2007;179:117–128. doi: 10.1083/jcb.200706196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosales-Nieves AE, et al. Coordination of microtubule and microfilament dynamics by Drosophila Rho1, Spire and Cappuccino. Nat Cell Biol. 2006;8:367–376. doi: 10.1038/ncb1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manseau LJ, Schüpbach T. Cappuccino and spire: Two unique maternal-effect loci required for both the anteroposterior and dorsoventral patterns of the Drosophila embryo. Genes Dev. 1989;3:1437–1452. doi: 10.1101/gad.3.9.1437. [DOI] [PubMed] [Google Scholar]
- 8.Dahlgaard K, Raposo AASF, Niccoli T, Johnston DS. Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev Cell. 2007;13:539–553. doi: 10.1016/j.devcel.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Theurkauf WE. Premature microtubule-dependent cytoplasmic streaming in cappuccino and spire mutant oocytes. Science. 1994;265:2093–2096. doi: 10.1126/science.8091233. [DOI] [PubMed] [Google Scholar]
- 10.Leader B, et al. Formin-2, polyploidy, hypofertility and positioning of the meiotic spindle in mouse oocytes. Nat Cell Biol. 2002;4:921–928. doi: 10.1038/ncb880. [DOI] [PubMed] [Google Scholar]
- 11.Dumont J, et al. Formin-2 is required for spindle migration and for the late steps of cytokinesis in mouse oocytes. Dev Biol. 2007;301:254–265. doi: 10.1016/j.ydbio.2006.08.044. [DOI] [PubMed] [Google Scholar]
- 12.Schuh M, Ellenberg J. A new model for asymmetric spindle positioning in mouse oocytes. Curr Biol. 2008;18:1986–1992. doi: 10.1016/j.cub.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 13.Zigmond SH, et al. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr Biol. 2003;13:1820–1823. doi: 10.1016/j.cub.2003.09.057. [DOI] [PubMed] [Google Scholar]
- 14.Kobielak A, Pasolli HA, Fuchs E. Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nat Cell Biol. 2004;6:21–30. doi: 10.1038/ncb1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila Spire is an actin nucleation factor. Nature. 2005;433:382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- 16.Alberts AS. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J Biol Chem. 2001;276:2824–2830. doi: 10.1074/jbc.M006205200. [DOI] [PubMed] [Google Scholar]
- 17.Wallar BJ, et al. The basic region of the diaphanous-autoregulatory domain (DAD) is required for autoregulatory interactions with the diaphanous-related formin inhibitory domain. J Biol Chem. 2006;281:4300–4307. doi: 10.1074/jbc.M510277200. [DOI] [PubMed] [Google Scholar]
- 18.Pechlivanis M, Samol A, Kerkhoff E. Identification of a short Spir interaction sequence at the C-terminal end of formin subgroup proteins. J Biol Chem. 2009;284:25324–25333. doi: 10.1074/jbc.M109.030320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciccarelli FD, Bork P, Kerkhoff E. The KIND module: A putative signalling domain evolved from the C lobe of the protein kinase fold. Trends Biochem Sci. 2003;28:349–352. doi: 10.1016/S0968-0004(03)00116-6. [DOI] [PubMed] [Google Scholar]
- 20.Kerkhoff E, et al. The Spir actin organizers are involved in vesicle transport processes. Curr Biol. 2001;11:1963–1968. doi: 10.1016/s0960-9822(01)00602-9. [DOI] [PubMed] [Google Scholar]
- 21.Le Goff C, et al. pEg6, a spire family member, is a maternal gene encoding a vegetally localized mRNA in Xenopus embryos. Biol Cell. 2006;98:697–708. doi: 10.1042/BC20050095. [DOI] [PubMed] [Google Scholar]
- 22.Bosch M, et al. Analysis of the function of Spire in actin assembly and its synergy with formin and profilin. Mol Cell. 2007;28:555–568. doi: 10.1016/j.molcel.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 23.Huang J, Furuya A, Furuichi T. Very-KIND, a KIND domain containing RasGEF, controls dendrite growth by linking Ras small GTPases and MAP2. J Cell Biol. 2007;179:539–552. doi: 10.1083/jcb.200702036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mees A, et al. Very-KIND is a novel nervous system specific guanine nucleotide exchange factor for Ras GTPases. Gene Expr Patterns. 2005;6:79–85. doi: 10.1016/j.modgep.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 25.Li F, Higgs HN. Dissecting requirements for auto-inhibition of actin nucleation by the formin, mDia1. J Biol Chem. 2005;280:6986–6992. doi: 10.1074/jbc.M411605200. [DOI] [PubMed] [Google Scholar]
- 26.Lei M, et al. Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell. 2000;102:387–397. doi: 10.1016/s0092-8674(00)00043-x. [DOI] [PubMed] [Google Scholar]
- 27.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pollard TD. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J Cell Biol. 1986;103:2747–2754. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 30.Pring M, Evangelista M, Boone C, Yang C, Zigmond SH. Mechanism of formin-induced nucleation of actin filaments. Biochemistry. 2003;42:486–496. doi: 10.1021/bi026520j. [DOI] [PubMed] [Google Scholar]
- 31.Harris ES, Li F, Higgs HN. The mouse formin, FRLalpha, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J Biol Chem. 2004;279:20076–20087. doi: 10.1074/jbc.M312718200. [DOI] [PubMed] [Google Scholar]
- 32.Kovar DR, Wu JQ, Pollard TD. Profilin-mediated competition between capping protein and formin Cdc12p during cytokinesis in fission yeast. Mol Biol Cell. 2005;16:2313–2324. doi: 10.1091/mbc.E04-09-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otomo T, et al. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- 34.Kovar DR, Kuhn JR, Tichy AL, Pollard TD. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J Cell Biol. 2003;161:875–887. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Y, et al. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–723. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- 36.Goldsmith EJ, Akella R, Min X, Zhou T, Humphreys JM. Substrate and docking interactions in serine/threonine protein kinases. Chem Rev. 2007;107:5065–5081. doi: 10.1021/cr068221w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gould CJ, et al. The formin DAD domain plays dual roles in autoinhibition and actin nucleation. Curr Biol. 2011;21:384–390. doi: 10.1016/j.cub.2011.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chesarone M, Gould CJ, Moseley JB, Goode BL. Displacement of formins from growing barbed ends by bud14 is critical for actin cable architecture and function. Dev Cell. 2009;16:292–302. doi: 10.1016/j.devcel.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quinlan ME, Kerkhoff E. Actin nucleation: bacteria get in-Spired. Nat Cell Biol. 2008;10:13–15. doi: 10.1038/ncb0108-13. [DOI] [PubMed] [Google Scholar]
- 40.Zuchero JB. In vitro actin assembly assays and purification from Acanthamoeba. Methods Mol Biol. 2007;370:213–226. doi: 10.1007/978-1-59745-353-0_15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.