Abstract
In mice and humans the circadian rhythm of many biochemical reactions, physiology, and behavior is generated by a transcriptional-translation feedback loop (TTFL) made up of the so-called core clock genes/proteins. The circadian system interfaces with most signaling pathways including those involved in cell proliferation and inflammation. Cryptochrome (CRY) is a core clock protein that plays an essential role in the repressive arm of the TTFL. It was recently reported that mutation of CRY in p53-null mice delayed the onset of cancer. It was therefore suggested that CRY mutation may activate p53-independent apoptosis pathways, which eliminate premalignant and malignant cells and thus delay overt tumor formation. Here we show that CRY mutation sensitizes p53 mutant and oncogenically transformed cells to tumor necrosis factor α (TNFα)-initiated apoptosis by interfacing with the NF-κB signaling pathway through the GSK3β kinase and alleviating prosurvival NF-κB signaling. These findings provide a mechanistic foundation for the delayed onset of tumorigenesis in clock-disrupted p53 mutant mice and suggest unique therapeutic strategies for treating cancers associated with p53 mutation.
Keywords: hepatocellular carcinoma, inflammatory cytokine, extrinsic apoptotic pathway
The circadian rhythm in mammalian organisms is generated by a molecular clock comprising four gene/protein groups (1–5): The CLOCK (NPAS2) and BMAL1 transactivators and the cryptochrome 1 and 2 (CRY1/2) and the period 1 and 2 (PER1/2) repressors. CLOCK (or NPAS2) and BMAL1 make heterodimers, which act on the promoters of CRY1/2 and PER1/2 and activate their transcription. The CRY and PER proteins in turn, after a time delay of ill-defined mechanism, inhibit the CLOCK-BMAL1 transactivator and hence their own transcription to close the TTFL. This core TTFL directly or indirectly affects most cellular functions, and as a result, clock disruption by core clock gene mutation is expected to have serious repercussions at the cellular and organismal levels.
We recently reported that mice of the p53−/−Cry1−/−Cry−/− (hereafter p53KOCryDKO) genotype exhibited delayed onset of spontaneous cancer relative to p53−/− (p53KO) mice (6). When analyzed for their response to genotoxic stress, it was found that p53KOCryDKO cells were more sensitive to UV-induced apoptosis than p53KO cells even though they are identical in terms of DNA repair and DNA damage checkpoint functions (6). Therefore, we ascribed the reduced clonogenic survival of p53KOCryDKO cells upon UV irradiation, relative to p53KO cells, to enhanced p53-independent apoptosis as a consequence of CRY mutation. We also suggested that the delayed onset of cancer and prolonged lifespan of mice of p53KOCryDKO genotype may have been caused by enhanced apoptosis of oncogenically transformed cells before giving rise to macroscopic tumors (6). In line with these findings we recently determined that p53KOCryDKO tumors are more responsive to oxaliplatin than p53KO tumors (7). However, UV and oxaliplatin activate the intrinsic apoptosis pathway (8) that is commonly induced by DNA damaging agents, whereas the difference in tumor incidence between the two mouse lines was observed in the absence of any external genotoxic stress. Therefore, we considered the possibility that the loss of CRY may amplify the extrinsic pathway for apoptosis to eliminate cells with potential to give rise to cancer. Here we present evidence that the circadian clock and NF-κB signaling pathway intersect and that CRY mutation enhances extrinsic apoptosis by interfering with NF-κB–mediated activation of transcription of genes required for antiapoptosis upon cytokine stimulation.
Results
Effect of the Circadian Clock on Extrinsic Apoptosis Induced by TNFα.
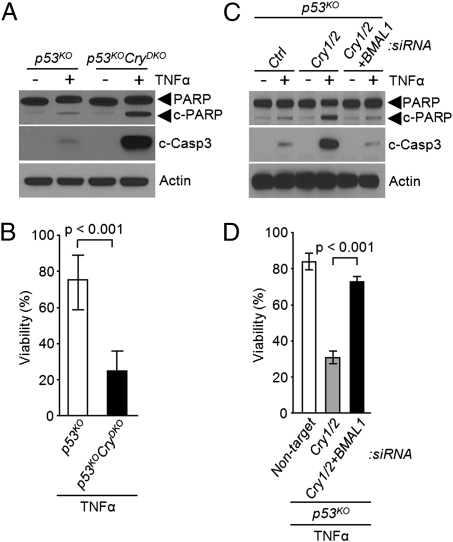
To find out whether CRY mutation sensitizes cells to the extrinsic apoptosis pathway, we treated p53KO and p53KOCryDKO dermal fibroblasts (6) with tumor necrosis factor α (TNFα), which is one of the key inflammatory cytokines known to activate the extrinsic apoptosis pathway (9–11). As apparent from Fig. 1A, the cytokine elicits remarkably enhanced apoptotic response in p53KOCryDKO cells as measured by the extent of proteolytic cleavage of poly ADP ribose polymerase (PARP) and caspase 3. Importantly, this increased sensitivity to apoptosis is reflected in cell viability as measured by clonogenesis assay (Fig. 1B), suggesting that the enhanced apoptosis may play a role in preferential elimination of CRY-null cancer cells exposed to cytokines. Next, we wished to determine whether the enhanced sensitivity to TNFα-induced apoptosis and the associated reduction in clonogenic survival were the phenotypic manifestation of CRY mutation independent of its role in the core circadian clock circuitry or a reflection of its role in the molecular clock. To this end, either CRY1/2 alone or both CRY1/2 (negative arm) and BMAL1 (positive arm) were down-regulated in p53KO cells and then the cells were treated with TNFα and analyzed for apoptosis and clonogenicity. As seen in Fig. 1C, depletion of CRY1/2 by siRNA in p53KOCryDKO cells has essentially the same consequence as null mutations of these genes in p53KOCryDKO cells (Fig. 1A) in terms of TNFα-induced apoptosis and clonogenic survival (Fig. 1D). Importantly, depletion of BMAL1 as well negates the enhanced apoptosis and clonogenic lethality promoted by CRY1/2 down-regulation (Fig. 1 C and D). Thus, we conclude that in p53KO cells the core circadian clock strongly influences the extent of TNFα-induced apoptosis.
Fig. 1.
Regulation of cytokine-initiated apoptosis by the circadian clock. (A and B) Effect of cryptochrome. (A) Immunoblots showing the apoptotic index probed with the indicated antibodies. Dermal fibroblasts of the indicated genotypes were treated with TNFα (50 ng/mL) for 48 h and then cell lysates were analyzed by immunoblotting. Actin was examined as a loading control. (B) Clonogenic cell survival assay showing the viable cells remaining after treatment with TNFα (50 ng/mL) for 48 h as a percentage of viable untreated cells. Results represent the mean of three independent experiments (±SD). (C and D) Contribution of BMAL1. (C) Apoptotic index. p53KO cells were transfected with the indicated siRNAs and then incubated with TNFα (50 ng/mL) for 48 h, followed by lysis and immunoblotting. (D) Clonogenic cell survival assay was used to determine cell viability of p53KO cells transfected with the indicated siRNAs and then treated with TNFα for 48 h, diluted, and plated. (±SD, n = 3).
Role of NF-κB in Linking the Circadian Clock to Extrinsic Apoptosis.
Next, we wished to determine the point at which the clock interfaces with the TNFα-initiated apoptosis pathway. It is well established that the NF-κB transcription factor plays a critical role in immune responses stimulated by inflammatory cytokines such as TNFα and IL-1β (12, 13). In addition, NF-κB has also been shown to have a role in cellular transformation and tumorigenesis by up-regulating the expression of some antiapoptosis genes upon inflammatory cytokine stimulation (14, 15). The effect of NF-κB on tumorigenesis, however, is strongly modulated by the p53 tumor suppressor, which can antagonize NF-κB activity (16–18). In p53-null background, the NF-κB signaling pathway has been shown to promote cell proliferation through NF-κB–mediated transcription of antiapoptotic factors and thus promote tumor growth (18). Conversely, inhibition of NF-κB signaling in a p53-null tumor model resulted in robust cell death by apoptosis (18), suggesting a potential approach for selectively eliminating p53-null cancer cells by interfering with NF-κB signaling.
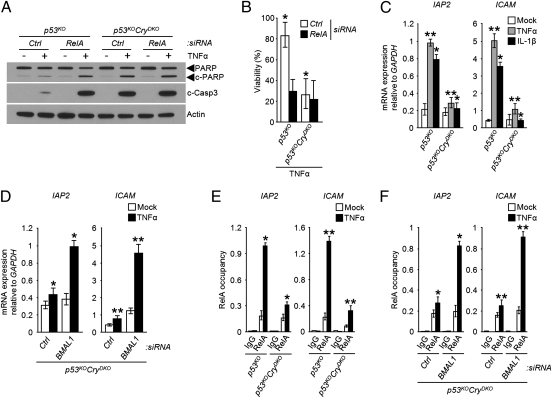
To analyze the role of NF-κB in TNFα-induced apoptosis, the RelA subunit of NF-κB heterodimer (RelA-p50) was down-regulated in p53KO and p53KOCryDKO cells by siRNA; the cells were then treated with TNFα and probed for apoptosis, using PARP and caspase 3 cleavages as readout (Fig. 2). We found that whereas knockdown of RelA in p53KOCryDKO cells, which have heightened apoptotic response to the cytokine already, has no further effect on the level of apoptosis and cell viability, down-regulation of RelA in the in p53KO cells enhanced the extent of apoptosis and lethality to a level comparable to that seen in p53KOCryDKO cells (Fig. 2 A and B). These results support the view that NF-κB plays an important role in the suppression of TNFα-initiated apoptosis in p53-null cells and suggest that the TNFα–NF-κB–apoptosis signaling pathway is controlled by the circadian clock. It is known that NF-κB activates the transcription of antiapoptotic genes such as IAPs to suppress TNFα-initiated apoptosis in the tumor cells (14, 15, 19). Hence, to assess the effect of CRY on this signaling axis, we treated cells with TNFα or IL-1β to activate the NF-κB pathway (12, 20) and measured the transcriptional activation of NF-κB target genes IAP2 and ICAM. As expected, both cytokines induce transcription of these genes in p53KO cells (Fig. 2C). Importantly, in the p53KOCryDKO background, the cytokine-initiated transcriptional activation of NF-κB target genes was abolished (Fig. 2C). Moreover, the similar effects of CRY mutation on NF-κB activation by both TNFα and IL-1β (Fig. 2C) indicate that CRY affects the NF-κB signaling pathway in general. Furthermore, the effect of CRY mutation on TNFα-stimulated transcription of IAP2 and ICAM is reversed by down-regulating BMAL1 (Fig. 2D), indicating that the observed effect of CRY mutation on the cytokine–NF-κB signaling axis is not due to a nonclock function of CRY, but is a manifestation of interfacing of the core circadian clock with the NF-κB signaling pathway.
Fig. 2.
Regulation of NF-κB activity by the circadian clock. (A and B) Apoptosis and clonogenic death. (A) Apoptosis. Immunoblots of extracts from p53KO and p53KOCryDKO cells transfected with the indicated siRNAs and then treated with TNFα (50 ng/mL) for 48 h. (B) Viability. Cells were transfected with siRNAs as indicated and then treated with TNFα (50 ng/mL, 48 h), diluted, and plated. Survival was determined by clonogenic assay (±SD, n = 3). *P < 0.005. (C and D) Transcription. Regulation of target genes of NF-κB by the clock. Quantitative real-time PCR analysis following TNFα (50 ng/mL, 1 h) or IL-1β (30 ng/mL, 1.5 h) of the indicated cell lines without (C), and with (D) siRNA transfections as indicated.*P < 0.005, **P < 0.001. (E and F) Promoter binding. Regulation by the clock of the occupancy of the promoters of NF-κB target genes was probed by ChIP with RelA antibodies following TNFα treatment of p53KO and p53KOCryDKO cells (E) and of the p53KOCryDKO cells transfected with the indicated siRNAs (F). RelA occupancy of the promoters relative to the untreated control is shown. Data represent means ± SD (n = 3). *P < 0.005, **P < 0.001.
This conclusion was further confirmed by analyzing the occupancy of the IAP2 and ICAM promoters by NF-κB using the chromatin immunoprecipitation (ChIP) assay with antibodies against the RelA subunit of the NF-κB heterodimer. As seen in Fig. 2E, TNFα stimulates binding of NF-κB to the promoters of both IAP2 and ICAM in p53KO cells and this stimulation is nullified by CRY mutation. Furthermore, in agreement with the transcription data in Fig. 2C, down-regulation of BMAL1 in p53KOCryDKO cells restores the TNFα-induced occupancy of NF-κB in the promoters of its target genes (Fig. 2F). Thus, we conclude that the core circadian clock activity regulates the NF-κB pathway and hence apoptosis in response to cytokine stimulation in p53-impaired cells, with the negative arm of the clock (CRY) inhibiting and the positive arm (BMAL1) promoting apoptosis in this genetic background.
Role of GSK3β in Linking the Circadian Clock to the Proapoptotic NF-κB Signaling.
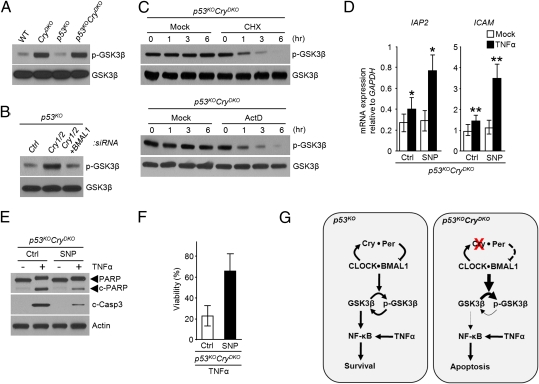
The data presented so far establish a link between the NF-κB activity and the circadian clock but does not provide an insight into the nature of this association. We considered GSK3β kinase as a potential candidate to link the two systems because GSK3β is known to activate NF-κB (21, 22) and, recently, it was reported that the phosphorylated (inactive) form of GSK3β exhibited a circadian rhythm in the mouse liver with the inactive GSK3β-P in phase with BMAL1 and antiphase with CRY1 (23). Hence, we wished to determine how the interfacing of the molecular clock with NF-κB through GSK3β would impact the cytokine-initiated apoptosis in p53-null cells with a functional or disrupted circadian clock. As apparent in Fig. 3A, in CRY-null cells the level of phosphorylated GSK3β is elevated without measureable change in the overall level of GSK3β and without a noticeable impact of the p53 status of the cell. Moreover, this effect appears to be the consequence of the core clock function of CRY because it is abolished by down-regulation of BMAL1 in CRY-null cells (Fig. 3B).
Fig. 3.
Role of GSK3β in circadian clock control of NF-κB activity. (A) Immunoblots comparing the levels of phospho-GSK3β in wild-type (WT), CryDKO, p53KO, and p53KOCryDKO cells. Total GSK3 was examined as a loading control. (B) Immunoblot analysis of phospho-GSK3β levels in p53KO cells transfected with the indicated siRNAs. (C) Decay kinetics of phospho-GSK3β. Shown are immunoblots of extracts of p53KOCryDKO cells treated with 20 μg/mL of cycloheximide (CHX) or 5 μg/mL of actinomycin D (ActD) for the indicated times. Immunoblot of total GSK3β is included as a loading control. (D) Effect of GSK3β activity on NF-κB target gene expression in p53KOCryDKO cells. The cells were preincubated with sodium nitroprusside (SNP), which activates GSK3β through dephosphorylation, and then treated with TNFα where indicated, and IAP2 and ICAM transcription levels were determined by quantitative real-time PCR. The data are normalized to GAPDH levels (±SD, n = 3). *P < 0.005, **P < 0.001. (E and F) Effect of GSK3β activity on TNFα-induced apoptosis (E), and clonogenic survival (F) in p53KOCryDKO cells without and with SNP treatment to control the phospho-GSK3β levels. (G) Model for the regulation of cytokine initiated NF-κB antiapoptotic function through the clock-controlled GSK3β activity in p53-deficient tumor cells and the consequence of cryptochrome inactivation.
To gain some insight into the mechanism by which the circadian TTFL controls the activity of GSK3β, we treated p53KOCryDKO cells with either the protein synthesis inhibitor cycloheximide or the transcription inhibitor actinomycin D and examined the levels of GSK3β and phospho-GSK3β over a 6-h period. We found that, whereas there was no significant change in GSK3β levels over the course of the experiment, the phospho-GSK3β levels decayed with a half-life of ∼1 h when either translation or transcription was inhibited (Fig. 3C). As a relatively short half-life of a protein or of its activity is essential for a robust circadian rhythmicity, this finding supports the report of strong circadian oscillation of GSK3β activity without a measurable change in the total GSK3β level in the mouse liver. Thus, we conclude that GSK3β activity is a bona fide clock-controlled function.
To examine the contribution of clock-controlled GSK3 activity on the cytokine-mediated NF-κB activation and the subsequent apoptotic response, we treated the p53KOCryDKO cells with sodium nitroprusside (SNP), which is known to activate GSK3β through dephosphorylation (24–26) (Fig. S1) and analyzed the transcription of IAP2 and ICAM (Fig. 3D), apoptotic proteolysis of PARP and caspase 3 (Fig. 3E), and cell viability (Fig. 3F) in response to TNFα. The data reveal that SNP-mediated accumulation of dephosphorylated GSK3β leads to enhanced transcription of IAP2 and ICAM (Fig. 3D), decreased apoptotic response to TNFα (Fig. 3E), and reduced clonogenic lethality of TNFα (Fig. 3F). Taken together, the data in Fig. 3 lead to the conclusion that the circadian clock, through the intermediating of GSK3β, gates the cytokine-mediated antiapoptotic function of NF-κB. Fig. 3G summarizes the mechanistic findings of our study: In p53 mutant cells with a functional clock, inflammatory cytokines such as TNFα activate the transcription of antiapoptotic genes in a manner gated by the circadian clock through GSK3β phosphorylation. In contrast, the CRY-null mutation in p53 dysfunctional cells leads to hyperphosphorylation and inactivation of GSK3β and failure to phosphorylate and activate the antiapoptotic effect of NF-κB, making cells more sensitive to TNFα-induced apoptosis.
Cryptochrome, TNFα, and Hepatocellular Carcinoma (HCC).
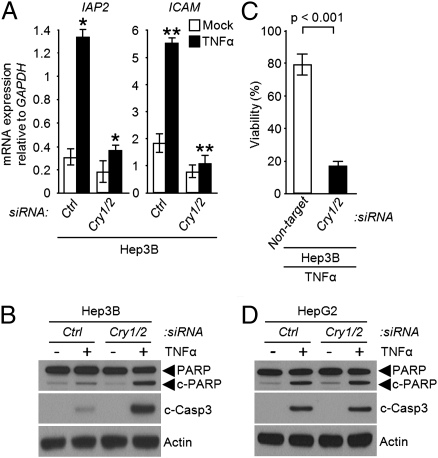
Recent studies have revealed that TNFα supports the growth of HCC, a prototype of inflammation-associated cancer (27), through activation of NF-κB–dependent expression of antiapoptotic genes (15). In addition, the loss of p53 functionality has been shown to be closely correlated with the severity of HCC (28, 29). Hence, to test for clock disruption as a potential therapeutic approach for HCC we investigated the effect of CRY down-regulation on cytokine-induced apoptosis in HCC cell lines without and with p53 functionality. In the p53 mutant Hep3B HCC cell line, down-regulation of CRY significantly reduced the transcription of NF-κB–dependent genes upon TNFα stimulation (Fig. 4A), potentiated TNFα-induced apoptosis (Fig. 4B) and loss of viability (Fig. 4C). In contrast, the down-regulation of CRY in the HepG2 HCC cell line with wild-type p53 does not affect the level of TNFα-induced apoptosis (Fig. 4D), presumably because in this genetic background the contribution of p53 to the apoptotic response to the cytokine is so robust that further enhancement by CRY down-regulation cannot be detected.
Fig. 4.
The clock regulates NF-κB-dependent apoptosis in p53-deficient hepatocellular carcinoma. (A–C) Hep3B (p53−/−) cells. The cells were transfected with the indicated siRNAs, followed by TNFα treatment and were then analyzed for expression of NF-κB controlled antiapoptotic genes by quantitative real-time PCR (A), for apoptosis by immunoblotting for PARP and cleaved caspase 3 (B), and for clonogenic survival (C).*P < 0.005, **P < 0.001 (±SD, n = 3). (D) HepG2 (p53+/+) cells. Immunoblots of extracts from siRNA transfected cells following treatment with TNFα (50 ng/mL, 48 h).
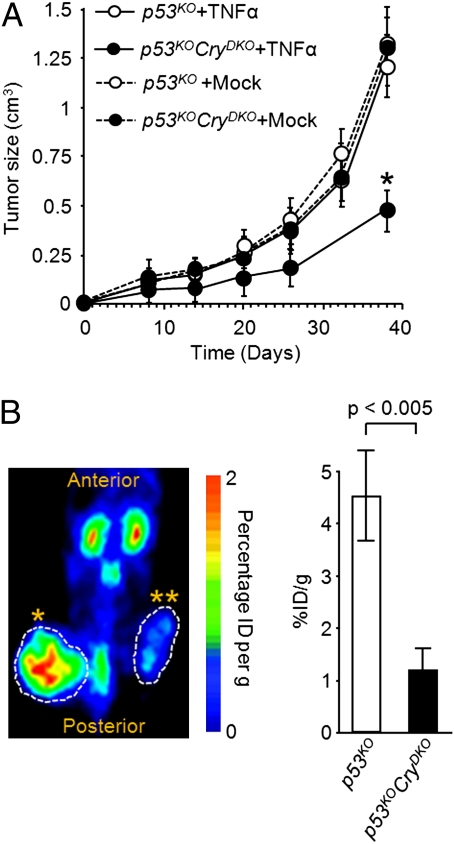
As a further proof of principle that CRY inhibition or disruption can be used as a therapeutic modality in p53 mutant tumors we performed the following experiment: Immunodeficient mice were s.c. injected with untreated or TNFα-treated p53KO cells on the left flank and untreated or TNFα-treated p53KOCryDKO cells on the right flank, and tumor growth and metabolism were monitored (Fig. 5). As seen in Fig. 5A, the tumors arising from cells of the p53KO and p53KOCryDKO genotypes grow at the same rate in the absence of TNFα treatment. However, the TNFα-treated p53KOCryDKO tumors grow slower (Fig. 5A) and have reduced metabolic activity (Fig. 5B) compared with their p53KO counterparts. We note, however, the limitation of this assay in that TNFα-treated cells were used for tumorigenesis rather than administering TNFα systemically after tumors had reached a certain size. However, systemic treatment of tumors with TNFα in this experimental system is not feasible because the doses necessary to achieve effective concentrations in the tumor are toxic to the mice. Nevertheless, these data raise the possibility that with appropriate cytokine delivery regimens CRY inhibition may be used as an adjuvant to treatment of certain tumors with cytokines.
Fig. 5.
Modulation of antitumorigenic effect of TNFα by cryptochrome. (A) Tumor growth. NOD/SCID mice were s.c. injected with untreated or TNFα-treated p53KO cells on the left flank and untreated or TNFα-treated p53KOCryDKO cells on the right flank, and tumor growth was monitored for 40 d. Error bars represent means ± SD (n = 10). *P < 0.001. (B) Tumor metabolism. MicroPET scan recorded 1 h after [18F]-FDG injection on day 38 after xenograft injection. The NOD/SCID mice were injected with TNFα-treated CryDKO cells in the left flank (*) and p53KOCryDKO cells in the right flank (**). (Right) Quantification of [18F]-FDG signal calculated as the ratio of percentage of injected dose (% ID) at the region of interest (ROI, circle) to the % ID at a background region. Error bars represent means ± SD (n = 4). The MicroPET scan (Left) is a dorsal view of a representative mouse.
Conclusion
This study was undertaken, in part, to understand the molecular basis for the delayed onset of cancer in p53KOCryDKO mice relative to p53KO mice. We believe that our data in this paper have provided a plausible explanation: The clock disruption through CRY mutation sensitizes the transformed cells to apoptosis by inflammatory cytokines such as TNFα, which is known to be elevated in CRY mutant mice (30). Our findings also link two major signaling pathways, the circadian clock, which introduces a temporal variable into many cellular and physiological functions, and the NF-κB pathway, which is a key nodal focus in the inflammatory response at a cellular and organismal level. Finally, our data suggest potential therapeutic approaches for treating p53 mutation-associated cancers by targeting the clock in general and cryptochrome in particular.
Materials and Methods
Establishment of Dermal Fibroblast Cell Lines.
Wild-type and p53KO mice of C57BL/6J background were obtained from The Jackson Laboratory. Cryptochrome mutants in C57BL/6J background, p53KO and p53KOCryDKO were generated in our laboratory as described previously (6, 31). The mice were handled in accordance with the guidelines of the National Institutes of Health and the University of North Carolina School of Medicine. Dermal fibroblasts were isolated using skin biopsy as described previously (6).
Cell Lines, siRNA, and Reagents.
Hep3B and HepG2 cells were obtained from American Type Culture Collection. These cells and dermal fibroblasts isolated in our laboratory were grown in Dulbecco's Modified Eagle Medium (Gibco), supplemented with 10% FBS (Gibco) and 100 units/mL penicillin and 100 μg/mL streptomycin (Gibco). DharmaFECT (Dharmacon) reagent was used according to the manufacturer's instruction for transcfection of ON-TARGET plus SMARTpool siRNA duplexes obtained from Dharmacon [mCry1 (L-040485-01-0005), mCry2 (L-040486-00-0005), mBMAL1 (L-040483-01-0005), mRelA (L-040776-00-0005), mGSK3β (L-041080-00-0005), mCyclophilin-B (D-001820-20-05), hCry1 (L-015421-00-0005), hCry2 (L-014151-01-0005), hCyclophilin-B (D-001820-10-05), or nontargeting siRNA (D-001810-10-05) as a control]. For Fig. 3C, cells were treated with 20 μg/mL cycloheximide (Sigma) or 5 μg/mL actinomycin D (Sigma) for the indicated times before analysis by immunoblotting. For Fig. 3 D–F, cells were treated with 2 mM SNP (Calbiochem) for 20 min before treatment of TNFα (50 ng/mL). The specificity of the antibodies and siRNAs used are shown in Fig. S2.
Immunoblotting.
Protein levels from whole cell lysate were determined by immunoblotting. Antibodies used in this study include those for RelA, Actin, Cyclophilin B, and IgG control (Santa Cruz Biotechnology), cleaved-caspase3, PARP1, GSK3β, phospho-GSK3β Ser9 (Cell Signaling Technology), IgM (Thermo Scientific), and Cry1 and Cry2 antibodies (31).
Clonogenic Cell Survival Assay.
The clonogenic survival assay was done as described previously (32, 33) with some modifications. Cells were seeded at low density to ensure the formation of 200 colonies per six-well plate (Corning). Following plating, cells were kept in growth medium for 10–14 h and treated with a single dose of TNFα (50 ng/mL). Readily visible colonies (over 50 cells per colony) formed after a 9- to 10-d incubation were fixed and counted. The number of observed colonies divided by the number of plated cells was expressed as cell viability.
mRNA Measurement by Quantitative PCR.
RNA was extracted using RNeasy mini kit (Qiagen) and cDNA was generated using ImProm-II reverse transcription system (Promega). PCR was performed in a 7900HT fast real-time PCR system (Applied Biosystems) using the 2× sybr green master mix (Applied Biosystems) with the primers listed in Table S1.
ChIP Assay.
Cells treated with TNFα (50 ng/mL) for 30 min were collected and washed with PBS. ChIP was performed by manufacturer's instruction (Sigma) with some exceptions. Protein–DNA complexes incubated with either an anti-RelA antibody (Santa Cruz Biotechnology) or an equivalent IgG control (Cell Signaling Technology) were precipitated using protein A/G-conjugated agarose beads (Calbiochem). Protein–DNA crosslink was resolved and followed by quantitative real-time PCR (qPCR) using sybr green master mix (Applied Biosystems). For detection of RelA binding onto the IAP2 and ICAM1 promoters, the following primers were used for PCR (for IAP2, 5′ TGTAAGAGCCAAGAGCCTTC and 3′ ACTTCCTCCTCACCCTTGGT; for ICAM1, 5′ CCAGATCTTGAGTGTTGGAA and 3′ CAGCTTGTCCTCCTGTCTCC).
Tumor Xenograft Model.
Cells were treated ex vivo for 4 h with TNFα (50 ng/mL), harvested, and resuspended for injection. A total of 2 × 106 cells mixed with an equal volume of matrigel (BD Biosciences) were inoculated s.c. into both flanks of 6-wk-old female immunodeficient NOD/SCID mice. The NOD/SCID mice were bred and kept under defined-flora pathogen-free conditions at the Animal Facility of the Division of Experimental Radiation Oncology, University of North Carolina. Tumor volume and body weight were recorded every 3 d. Tumor size was measured with a caliper in three mutually perpendicular diameters (a, b, and c) and the volume was calculated as V = (π/6) × a × b × c.
MicroPET Imaging.
Whole-body images of the biodistribution of 18F-2-deoxyglucose ([18F]-FDG) tracers in mice was obtained by micropositron emission tomography (microPET) scans. Mice were kept warm, under gas anesthesia (2% isoflourane), and injected with [18F]-FDG intraperitoneally. A 1-h interval for uptake was allowed between probe administration and microPET scanning. Data were acquired using a Siemens Preclinical Solutions microPET Focus 220 instrument. MicroPET data were acquired for 10 min and were reconstructed using statistical maximum a posteriori probability (MAP) algorithms into multiple frames. The spatial resolution of microPET is ∼1.5-mm, 0.4-mm voxel size. Three dimensional regions of interest (ROIs) were drawn using AMIDE software (Andreas Loening, Stanford University, Stanford, CA). Color scale is proportional to tissue concentration with red being the highest and lower values in yellow, green, and blue.
Statistical Analysis.
Values are shown as mean ± SD calculated using a two-tailed t test.
Supplementary Material
Acknowledgments
We appreciate critical reading and useful comments of the manuscript by our laboratory members, in particular T. H. Kang, C. P. Selby, L. A. Lindsey-Boltz, and S. Gaddameedhi. We are grateful to A. Baldwin, J. W. Park, H. M. Lee, T. W. Kim, T. Y. Eom, T. Y. Kim, and S. H. Lee for critical comments and helpful discussions of the manuscript. We acknowledge the services of the Mouse Facility Center and Biomedical Research Imaging Center at University of North Carolina, Chapel Hill. We appreciate experimental assistance and helpful discussions for microPET analysis from S. H. Kim and J. H. Lee. This work was supported by National Institutes of Health Grants GM31082 and GM32833.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108125108/-/DCSupplemental.
References
- 1.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 2.Hastings MH, Reddy AB, Maywood ES. A clockwork web: Circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahar S, Sassone-Corsi P. Metabolism and cancer: The circadian clock connection. Nat Rev Cancer. 2009;9:886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 5.Vitaterna MH, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozturk N, Lee JH, Gaddameedhi S, Sancar A. Loss of cryptochrome reduces cancer risk in p53 mutant mice. Proc Natl Acad Sci USA. 2009;106:2841–2846. doi: 10.1073/pnas.0813028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JH, Sancar A. Circadian clock disruption improves the efficacy of chemotherapy through p73-mediated apoptosis. Proc Natl Acad Sci USA. 2011;108:10668–10672. doi: 10.1073/pnas.1106284108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hotchkiss RS, Strasser A, McDunn JE, Swanson PE. Cell death. N Engl J Med. 2009;361:1570–1583. doi: 10.1056/NEJMra0901217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 10.Ashkenazi A. Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat Rev Cancer. 2002;2:420–430. doi: 10.1038/nrc821. [DOI] [PubMed] [Google Scholar]
- 11.Ashkenazi A, Dixit VM. Death receptors: Signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 12.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 13.Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 14.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 15.Pikarsky E, et al. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–466. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- 16.Kawauchi K, Araki K, Tobiume K, Tanaka N. Loss of p53 enhances catalytic activity of IKKbeta through O-linked beta-N-acetyl glucosamine modification. Proc Natl Acad Sci USA. 2009;106:3431–3436. doi: 10.1073/pnas.0813210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawauchi K, Araki K, Tobiume K, Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat Cell Biol. 2008;10:611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- 18.Meylan E, et al. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–107. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin AS., Jr NF-kappaB antiapoptosis: Induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 1998;281:1680–1683. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- 20.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 21.Hoeflich KP, et al. Requirement for glycogen synthase kinase-3beta in cell survival and NF-kappaB activation. Nature. 2000;406:86–90. doi: 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- 22.Wilson W, 3rd, Baldwin AS. Maintenance of constitutive IkappaB kinase activity by glycogen synthase kinase-3alpha/beta in pancreatic cancer. Cancer Res. 2008;68:8156–8163. doi: 10.1158/0008-5472.CAN-08-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iitaka C, Miyazaki K, Akaike T, Ishida N. A role for glycogen synthase kinase-3beta in the mammalian circadian clock. J Biol Chem. 2005;280:29397–29402. doi: 10.1074/jbc.M503526200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang YJ, Xu YF, Liu YH, Yin J, Wang JZ. Nitric oxide induces tau hyperphosphorylation via glycogen synthase kinase-3beta activation. FEBS Lett. 2005;579:6230–6236. doi: 10.1016/j.febslet.2005.09.095. [DOI] [PubMed] [Google Scholar]
- 25.Urbanska K, et al. Inhibition of IGF-I receptor in anchorage-independence attenuates GSK-3beta constitutive phosphorylation and compromises growth and survival of medulloblastoma cell lines. Oncogene. 2007;26:2308–2317. doi: 10.1038/sj.onc.1210018. [DOI] [PubMed] [Google Scholar]
- 26.Baryawno N, et al. Small-molecule inhibitors of phosphatidylinositol 3-kinase/Akt signaling inhibit Wnt/beta-catenin pathway cross-talk and suppress medulloblastoma growth. Cancer Res. 2010;70:266–276. doi: 10.1158/0008-5472.CAN-09-0578. [DOI] [PubMed] [Google Scholar]
- 27.Nakamoto Y, Guidotti LG, Kuhlen CV, Fowler P, Chisari FV. Immune pathogenesis of hepatocellular carcinoma. J Exp Med. 1998;188:341–350. doi: 10.1084/jem.188.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staib F, Hussain SP, Hofseth LJ, Wang XW, Harris CC. TP53 and liver carcinogenesis. Hum Mutat. 2003;21:201–216. doi: 10.1002/humu.10176. [DOI] [PubMed] [Google Scholar]
- 29.Xue W, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hashiramoto A, et al. Mammalian clock gene Cryptochrome regulates arthritis via proinflammatory cytokine TNF-alpha. J Immunol. 2010;184:1560–1565. doi: 10.4049/jimmunol.0903284. [DOI] [PubMed] [Google Scholar]
- 31.Gauger MA, Sancar A. Cryptochrome, circadian cycle, cell cycle checkpoints, and cancer. Cancer Res. 2005;65:6828–6834. doi: 10.1158/0008-5472.CAN-05-1119. [DOI] [PubMed] [Google Scholar]
- 32.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 33.Gaddameedhi S, et al. Similar nucleotide excision repair capacity in melanocytes and melanoma cells. Cancer Res. 2010;70:4922–4930. doi: 10.1158/0008-5472.CAN-10-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.