Abstract
Lutein, a dihydroxy xanthophyll, is the most abundant carotenoid in plant photosynthetic tissues and plays crucial structural and functional roles in the light-harvesting complexes. Carotenoid β-and ε-hydroxylases catalyze the formation of lutein from α-carotene (β,ε-carotene). In contrast to the well studied β-hydroxylases that have been cloned and characterized from many organisms, the ε-hydroxylase has only been genetically defined by the lut1 mutation in Arabidopsis. We have isolated the LUT1 gene by positional cloning and found that, in contrast to all known carotenoid hydroxylases, which are the nonheme diiron monooxygenases, LUT1 encodes a cytochrome P450-type monooxygenase, CYP97C1. Introduction of a null mutant allele of LUT1, lut1-3, into the β-hydroxylase 1/β-hydroxylase 2 (b1 b2) double-mutant background, in which both Arabidopsis β-hydroxylases are disrupted, yielded a genotype (lut1-3 b1 b2) in which all three known carotenoid hydroxylase activities are eliminated. Surprisingly, hydroxylated β-rings were still produced in lut1-3 b1 b2, suggesting that a fourth unknown carotenoid β-hydroxylase exists in vivo that is structurally unrelated to β-hydroxylase 1 or 2. A second chloroplast-targeted member of the CYP97 family, CYP97A3, is 49% identical to LUT1 and hypothesized as a likely candidate for this additional β-ring hydroxylation activity. Overall, LUT1 defines a class of carotenoid hydroxylases that has evolved independently from and uses a different mechanism than nonheme diiron β-hydroxylases.
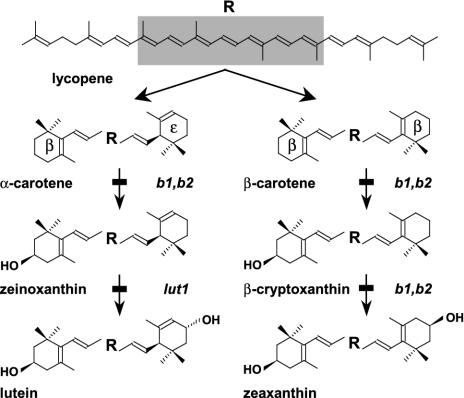
Carotenoids are terpenoid compounds that perform a variety of critical roles in photosystem structure, light harvesting, and photoprotection. Lutein (3R,3′R-β,ε-carotene-3,3′-diol), is the most abundant carotenoid in all plant photosynthetic tissues, in which it plays an important role in light-harvesting complex II assembly and function. Zeaxanthin (3R,3′R-β,β-carotene-3,3′-diol) is a structural isomer of lutein and is a critical component of nonphotochemical quenching (1, 2). The synthesis of lutein and zeaxanthin involves cyclization of lycopene to form α- and β-carotene, respectively, followed by the introduction of hydroxyl groups onto the ionone rings by a class of enzymes known as carotenoid hydroxylases (Fig. 1). β-Hydroxylases add hydroxyl groups to carbon 3 (C-3) of β-rings, whereas hydroxylation of C-3 on ε-rings is carried out by ε-hydroxylases. Two β-ring hydroxylations of β-carotene yield zeaxanthin, whereas one β-ring and one ε-ring hydroxylation of α-carotene yield lutein (Fig. 1).
Fig. 1.
Biosynthetic steps leading to lutein and zeaxanthin from lycopene. Carotenoid ring hydroxylations are key reactions for the biosynthesis of lutein and zeaxanthin. The steps blocked by the b1 (β-hydroxylase 1), b2 (β-hydroxylase 2), and lut1 (ε-hydroxylase) mutations are indicated.
Based on the stereospecific introduction of C-3 hydroxyl groups and the requirement for molecular oxygen, carotenoid hydroxylation reactions were predicted to be catalyzed by mixed-function oxygenases such as the cytochrome P450 enzymes (3–5). However, β-hydroxylases have been cloned from a variety of photosynthetic and nonphotosynthetic bacteria, green algae, and plants (6) and in all three phyla encode nonheme diiron proteins that have a fundamentally different hydroxylation reaction mechanism than heme-binding cytochrome P450 enzymes (7). Biochemical analysis and mutagenesis of pepper (Capsicum annum) β-hydroxylases have confirmed that the enzymes require iron, ferredoxin, and ferredoxin oxidoreductase for activity and that all 10 of the conserved iron-coordinating histidines are required for activity (8). The Arabidopsis genome encodes two nonheme diiron β-hydroxylases (β-hydroxylases 1 and 2), and although both efficiently hydroxylate β-rings, they function poorly with ε-ring-containing substrates in vitro (9, 10).
Early isotope-labeling studies have shown that carotenoid hydroxylation reactions are stereospecific (3, 4). The chirality of the hydroxylated ε-ring C-3 is opposite to that of the hydroxylated β-ring C-3. This difference in product chirality was an initial suggestion that two distinct hydroxylases are needed for β-and ε-ring hydroxylations and may partially explain why β-hydroxylases function poorly with ε-ring-containing substrates in vitro. Mutational studies in Arabidopsis have provided genetic evidence for the existence of a distinct ε-ring-specific hydroxylase (11). Mutation of the LUT1 locus in Arabidopsis decreased the production of lutein by 80–95% (dependent on plant age) and resulted in accumulation of the monohydroxy precursor zeinoxanthin, a classic phenotype for a mutation affecting a biosynthetic enzyme. ε-Ring hydroxylation was specifically blocked in lut1, and production of β-carotene-derived xanthophylls was increased. From these data, it was proposed that LUT1 encodes a function specific for ε-ring hydroxylation (11).
The interactions and functional redundancies of the three known carotenoid hydroxylases in Arabidopsis (β-hydroxylases 1 and 2 and LUT1) have been studied in vivo by isolating mutations disrupting each gene and generating multiple hydroxylase-deficient mutant genotypes (12). In the β-hydroxylase 1/β-hydroxylase 2 double-null mutant (b1 b2), in which both known β-hydroxylases were eliminated, hydroxylated β-ring groups were still synthesized at significant levels (75% of wild type), indicating that an additional β-ring hydroxylation activity exists in vivo. The ethyl methane sulfonate (EMS)-derived lut1-2 mutation was introduced into the b1 b2 background to address whether this additional β-hydroxylase activity might be a secondary function of the ε-hydroxylase or be due to a third unrelated β-hydroxylase. Hydroxylated β-ring groups were reduced further to 60% of wild-type levels in the lut1-2 b1 b2 triple mutant (12), suggesting that LUT1 is capable of β-ring hydroxylation in vivo. However, a caveat of this experiment is that LUT1 activity may not have been completely eliminated in the EMS-derived lut1-2 mutant, and we could not resolve whether the remaining β-ring hydroxylation in lut1-2 b1 b2 was caused by residual LUT1 activity or the presence of a third unrelated β-hydroxylase. Cloning of the LUT1 locus and generation of a null ε-hydroxylase mutant are required to further understanding of in vivo carotenoid hydroxylase activity and for applying molecular genetic approaches to study carotenoid hydroxylase functions in vivo.
Prior attempts to clone an ε-ring-specific hydroxylase by sequence-based similarity to β-hydroxylases in Arabidopsis were not successful and only identified the β-hydroxylase 2 gene (10). A thorough search of the fully sequenced Arabidopsis genome also failed to identify any additional genes bearing significant similarity to β-hydroxylases from plants, cyanobacteria, and nonphotosynthetic bacteria (10). These results suggested that the ε-hydroxylase defines a structurally distinct carotenoid hydroxylase family. We report here identification of the LUT1 locus by positional cloning and show that LUT1 indeed defines a previously uncharacterized class of carotenoid hydroxylases in nature.
Materials and Methods
Positional Cloning of LUT1. Homozygous lut1-1 (ecotype Columbia) was crossed to wild-type Landsberg erecta. F2 progeny homozygous for the lut1 mutation were identified by a TLC screening method. Briefly, carotenoid samples were extracted as described (10), resuspended in ethyl acetate, spotted on a silica TLC plate (J. T. Baker), and developed in 90:10 (v/v) hexane/isopropanol. F2 plants homozygous for lut1 contain a characteristic extra yellow band caused by accumulation of zeinoxanthin.
Genomic DNA from homozygous lut1 F2 plants was isolated by using the DNAzol reagent following manufacturer instructions (Invitrogen). PCRs were performed with 1 μl of genomic DNA in a 20-μl reaction mixture. The PCR program was 94°C for 3 min, 60 cycles of 94°C for 15 s, 50–60°C (the annealing temperature was optimized for each specific pair of primers) for 30 s, 72°C for 30 s, and finally 72°C for 10 min. A portion of the PCR product then was separated on a 3% agarose gel. lut1 had been mapped previously to 67 ± 3 centimorgans on chromosome 3 (10). Additional simple sequence length polymorphism markers for fine-mapping in this interval were designed based on the insertions/deletions information obtained from the Monsanto web site (www.arabidopsis.org/Cereon).
Cosmid Screening and Complementation of lut1. An Arabidopsis cosmid library (13) was screened, and cosmids carrying the At3g53130 gene were identified. For complementation of the lut1 mutation, a 4.2-kb restriction fragment containing the At3g53130 gene was subcloned into the pMLBART vector (14). Homozygous lut1 plants were transformed with Agrobacterium tumefaciens strain GV3101 containing pMLBART-At3g53130 by using the floral dip method (15). Basta-resistant T1 transformants were selected, and the carotenoid composition of leaf tissue was analyzed by HPLC (10).
Isolation of T-DNA Knockout Mutants in At3g53130 and Generation of a Carotenoid Hydroxylase Triple-Knockout Mutant Line. At3g53130-specific primers (forward, 5′-CTTCCTCTTCTTACTCTTCTCTCTTCACT-3′; reverse, 5′-AAGAACGATGGATGTTATAGACTGAAATC-3′) were sent to the University of Wisconsin Arabidopsis T-DNA (portion of the tumor-inducing plasmid that is transferred to plant cells) knockout facility to identify knockout mutants of the LUT1 gene. A single knockout line, designated lut1-3, was identified and isolated as described (www.biotech.wisc.edu/Arabidopsis). To generate a hydroxylase triple-knockout mutant line, homozygous lut1-3 and b1 b2 plants were crossed. Putative lut1-3b1b2 triple mutants were identified from the segregating F2 population by HPLC, and their genotypes were confirmed by PCR as described (12).
TaqMan Real-Time PCR Assay. LUT1 mRNA levels were quantified by TaqMan real-time PCR by using elongation factor 1α mRNA levels for normalization (12). The LUT1 TaqMan probe and primers are: 5′-CCGTCTCGCTGCTGGTCCTCG-3′ (TaqMan probe), 5′-GGATGAATGAGTACGGACCCAT-3′ (forward primer), and 5′-GGGTCGCTCACAATTACGAAA-3′ (reverse primer). The relative quantity of the transcripts was calculated by using the comparative threshold cycle (CT) method (16).
Phylogenetic Analysis of LUT1 Homologs. Full-length protein sequences of putative LUT1 homologs from Arabidopsis thaliana, Glycine max, Oryza sativa, and Pisum sativum were obtained from GenBank: CYP97A3 (accession no. AAL08302), CYP97B1 (accession no. CA A89260), CYP97B2 (accession no. AAB94586), CYP97B3 (accession no. CAB10290), CYP97C1 (accession no. A A M13903), CYP97C2 (accession no. AAK20054), and CYP86A8 (accession no. CAC47665). Rice CYP97A4 and CYP97B4 sequences were obtained from the cytochrome P450 web site (http://drnelson.utmem.edu/CytochromeP450.html).
Additional plant LUT1 homologs were retrieved from The Institute of Genome Research Unique Gene Indices: TC76166 (Hordeum vulgare), TC163981 (G. max), and TC69886 (H. vulgare). The coding sequences of each were extracted, assembled, and corrected by the ESTSCAN program (http://tigrblast.tigr.org/tgi). Chlamydomonas CYP97A3 homolog (Scaffold1399) was obtained from the Department of Energy Joint Genome Institute (JGI) database (http://genome.jgi-psf.org/chlre1/chlre1.home.html). Truncated LUT1 homologs from Zea mays, lettuce, and cotton are also present in the databases but were not used for phylogenetic analysis because full-length assemblies were not possible.
The deduced amino acid sequences of LUT1 homologs were aligned by using the CLUSTALX algorithm (17). A neighbor-joining (18) tree was constructed based on the sequence alignment and tested further with 500 bootstrap resamplings by using the computer program MEGA2 2.1 (19). Poisson-correction distance was used with 340 amino acids after removing gaps.
Results
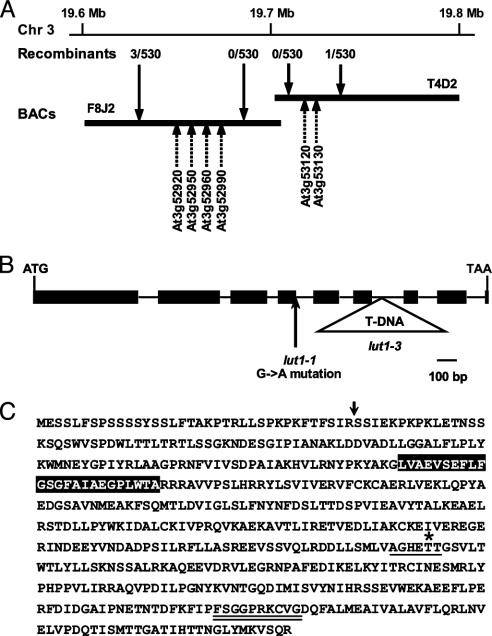
Fine-Mapping of the LUT1 Locus. The LUT1 locus has been mapped to the bottom arm of chromosome 3 at 67 ± 3 centimorgans (10). For fine-mapping of the locus, 530 plants homozygous for the lut1 mutation were identified from ≈2,000 plants in a segregating F2 mapping population. By using simple sequence length polymorphism markers, LUT1 was initially localized to an interval spanning two bacterial artificial chromosome clones (F8J2 and T4D2) and was delineated further to a 100-kb interval containing 30 predicted proteins (Fig. 2A). As with all other carotenoid biosynthetic enzymes, the LUT1 gene product is predicted to be chloroplast-targeted, and within the 100-kb interval containing LUT1, six proteins were predicted as being chloroplast-targeted by TARGETP prediction software (www.cbs.dtu.dk/services/TargetP). One of these chloroplast-targeted proteins, At3g53130, is a member of the cytochrome P450 monooxygenase family (CYP97C1). Cytochrome P450 monooxygenases are heme-binding proteins that insert a single oxygen atom into substrates (e.g., hydroxylation reactions), and therefore At3g53130 was considered to be a strong candidate for LUT1.
Fig. 2.
Positional cloning of the LUT1 locus. (A) Fine-mapping of the interval containing LUT1. Recombinants are indicated for specific simple sequence length polymorphism markers across the interval, and the position of chloroplast-targeted proteins are indicated by dashed arrows. BACs, bacterial artificial chromosomes. (B) Overview of the intron–exon organization of LUT1 and the locations of the lut1-1 and lut1-3 mutations. (C) Deduced amino acid sequence of LUT1. The cleavage site of the putative chloroplast-targeting sequence is indicated by an arrow, and the single predicted transmembrane domain is shaded in black. The conserved cytochrome P450 molecular oxygen-binding pocket and the cysteine motif are indicated by single and double underlines, respectively, and the conserved Thr is indicated by an asterisk.
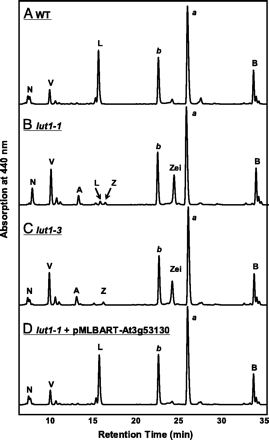
Mutant Complementation, Characterization, and the Identification of LUT1. The identity of At3g53130 as LUT1 was initially demonstrated by molecular complementation analysis. Homozygous lut1-1 mutants were transformed with a 4.2-kb genomic DNA fragment from wild-type Columbia (the background of lut1) containing the At3g53130 coding region, 1.0 kb upstream of the start codon and 0.7 kb downstream of the stop codon. Eight independent transformants were selected, and all showed a wild-type lutein level when analyzed by HPLC (Fig. 3D). These data indicate that At3g53130 genomic DNA can complement the lut1 mutation.
Fig. 3.
HPLC elution profiles of total leaf carotenoid extracts from wild type (A), lut1-1 (B), lut1-3 (C), and lut1-1 transformed with pMLBART-At3g53130 (D). Peaks correspond to neoxanthin (N), violaxanthin (V), antheraxanthin (A), lutein (L), zeaxanthin (Z), chlorophyll b (b), zeinoxanthin (Zei), chlorophyll a (a), and β-carotene (B).
To determine the molecular basis of the lut1 mutations, we sequenced both original EMS-derived lut1 alleles (11). The lut1-1 allele contains a G-to-A mutation at the highly conserved exon/intron splice junction (5′-AG/GT; the mutated G is in bold) that would cause an error in RNA splicing and lead to production of a mistranslated protein (Fig. 2B). The coding region of the lut1-2 allele was sequenced fully, but no mutations were identified. However, a rearrangement in the upstream region of the lut1-2 allele was identified by Southern blot analysis but was not characterized further (data not shown). A third lut1 allele, lut1-3, was identified by screening a T-DNA knockout population by using At3g53130-specific primers. Lut1-3 contains a T-DNA insertion in the sixth intron of the LUT1 gene (Fig. 2B).
To compare the impact of different lut1 alleles on carotenoid composition, total carotenoids were extracted from 4-week-old wild-type, lut1-1, lut1-2 (data not shown), and lut1-3 plants and separated by HPLC (Fig. 3 A–C). Lut1-1 and lut1-2 accumulated the monohydroxy biosynthetic intermediate zeinoxanthin and contained 8% of wild-type lutein, consistent with prior report (11). In contrast, although lut1-3 also accumulated zeinoxanthin, it lacked lutein (Fig. 3C), indicating that ε-ring hydroxylation function is eliminated by disruption of the At3g53130 gene. The lut1-3 phenotype also indicates that redundant ε-ring hydroxylation activities are not present in leaves and that the previously reported EMS-mutagenized lut1-1 and lut1-2 alleles are indeed leaky for ε-ring hydroxylation activity (11) (Fig. 3B). Taken together, the complementation of the lut1-1 mutation with a wild-type At3g53130 gene, the point mutation at a conserved splice site in the lut1-1 allele, and the phenotype of the At3g53130 T-DNA knockout mutant conclusively demonstrate that At3g53130 is the LUT1 locus.
LUT1 Encodes a Chloroplast-Targeted Cytochrome P450 with a Single Transmembrane Domain. The deduced amino acid sequence of LUT1 contains several features characteristic of cytochrome P450 enzymes (Fig. 2C). Cytochrome P450 monooxygenases contain a consensus sequence of (A/G)GX(D/E)T(T/S) that forms a binding pocket for molecular oxygen with the invariant Thr residue playing a critical role in oxygen binding in both prokaryotic and eukaryotic cytochrome P450s (20). In the deduced LUT1 protein sequence, this oxygen-binding pocket is highly conserved (Fig. 2C, single-underlined amino acids). The conserved sequence around the heme-binding cysteine residue for cytochrome P450-type enzymes is FXXGXXXCXG and is also present in LUT1 (Fig. 2C, double-underlined amino acids).
The chloroplast transit peptide prediction software CHLOROP 1.1 (www.cbs.dtu.dk/services/ChloroP) predicts an N-terminal transit peptide in LUT1 that is cleaved between Arg-36 and Ser-37 (Fig. 2C). The predicted chloroplast localization for LUT1 is consistent with the subcellular localization of carotenoid biosynthesis in higher plants (6) but is uncommon for a plant cytochrome P450. Of the 272 predicted cytochrome P450s in the Arabidopsis genome, only nine, including LUT1, are predicted to be chloroplast-targeted (21). LUT1 also contains a single predicted transmembrane domain (Fig. 2C, shaded box), which contrasts with the four transmembrane domains predicted for the nonheme diiron β-hydroxylases (6). Initial attempts to express and assay LUT1 protein in yeast were unsuccessful.
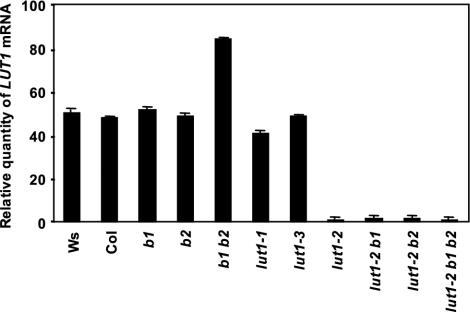
LUT1 Gene Expression and in Vivo Activity in the β-Hydroxylase-Deficient Backgrounds. Characterization of previously isolated T-DNA knockouts in the two Arabidopsis β-hydroxylase genes suggested that β- and ε-hydroxylases have overlapping functions in vivo (12). To investigate whether ε-hydroxylase expression is affected in the various carotenoid hydroxylase mutant backgrounds, steady-state LUT1 mRNA levels were quantified by real-time PCR (Fig. 4). The LUT1 TaqMan probe hybridizes 336 bp downstream from the start codon. LUT1 mRNA levels are not significantly different from wild type in the β-hydroxylase single mutants (b1 and b2) but are increased significantly in the β-hydroxylase double mutant b1 b2 (Fig. 4). LUT1 mRNA levels in lut1-2 alone and in combination with various β-hydroxylase mutant loci (i.e., lut1-2 b1, lut1-2 b2, and lut1-2 b1 b2) are similar and reduced to 2% of wild-type levels, consistent with the rearrangement of the upstream region in lut1-2 negatively impacting LUT1 transcription. The steady-state levels of modified LUT1 transcript in lut1-1 and lut1-3 are similar to wild-type transcript levels suggesting that, although LUT1 activity is negatively impacted in each mutant, LUT1 transcription is not.
Fig. 4.
The relative wild-type or mutant LUT1 transcript level detected in each genotype by real-time PCR (refer to Materials and Methods). The relative quantity of the LUT1 mRNA has been corrected with elongation factor 1α. Data shown are means with SD (n = 6). Ws, Wassilewskija; Col, Columbia.
The phenotype of the previously isolated lut1-2 b1 b2 mutant was not conclusive because of the leaky nature of the EMS-derived lut1-2 allele. Cloning of LUT1 and isolation of the LUT1 knockout mutant, lut1-3, allow for the complete elimination of LUT1 activity in vivo. Lut1-3 was crossed to b1 b2, and homozygous lut1-3 b1 b2 mutants were isolated. There was no lutein production in the lut1-3 b1 b2 triple mutant (data not shown), consistent with the lut1-3 single mutant phenotype (Fig. 3C). The total moles of β-carotene-derived xanthophylls produced are not significantly different between lut1-2 b1 b2 and lut1-3 b1 b2 (Table 1). However, when one considers the total moles of hydroxylated β-rings produced in each mutant (which includes hydroxylated β-ring in zeinoxanthin), total hydroxylated β-rings are reduced significantly in lut1-2 b1 b2 and lut1-3 b1 b2 compared to b1 b2, suggesting that LUT1 also has β-ring hydroxylation activity in vivo (Table 1). In addition, the presence of β-carotene-derived xanthophylls in the triple-knockout mutant lut1-3 b1 b2 indicates that a third β-hydroxylase must exist in vivo (Table 1).
Table 1. β-Xanthophyll production and β-ring hydroxylation in leaf tissue of wild-type and carotenoid hydroxylase mutants.
| Genotype | β-Xanthophylls* | Hydroxylated β-rings† |
|---|---|---|
| Ws | 54.0 ± 2.7a‡ | 48.5 ± 1.0a |
| Col | 60.7 ± 7.6a | 48.7 ± 0.9a |
| b1 b2 | 20.5 ± 4.8b | 40.2 ± 1.4b |
| lut1—2 b1 b2 | 26.5 ± 3.4b | 33.6 ± 2.4c |
| lut1—3 b1 b2 | 28.3 ± 4.6b | 31.1 ± 1.2c |
Total carotenoids were extracted from 5-week-old plants and quantified by HPLC as described (12).
β-Xanthophylls are the sum of zeaxanthin, antheraxanthin, violaxanthin, and neoxanthin as mmol of pigment/mol of chlorophyll a + b
Data are given as percentage of total ring hydroxylation
All values are means ± SD (n = 6). Values marked with the same letters are not significantly different from each other within a column (Student's t test, P > 0.05)
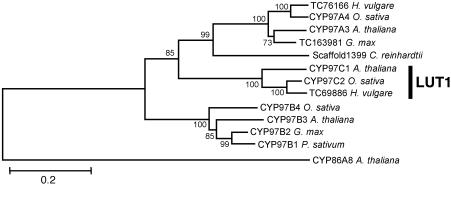
CYP97 Homologs in Other Species. Arabidopsis LUT1 was designated previously as CYP97C1 according to the standardized cytochrome P450 nomenclature (www.biobase.dk/P450). The Arabidopsis genome also contains two other CYP97 family members, CYP97A3 and CYP97B3, which are 49% and 42% identical to the LUT1 protein, respectively. Interestingly, CYP97A3 (At1g31800) is also one of the nine cytochrome P450s in Arabidopsis predicted to be chloroplast-targeted, whereas CYP97B3 (At4g15110) is predicted to be targeted to the mitochondria (21). Additional CYP97 family proteins were identified in the EST and genomic databases from a wide variety of monocots and dicots including Arabidopsis, barley, rice, soybean, and pea (Fig. 5).
Fig. 5.
Phylogenetic analysis of LUT1. A rooted neighbor-joining tree was constructed by using the fatty acid ω-hydroxylase (CYP86A8) from A. thaliana as an outgroup. Bootstrap values are indicated adjacent to the branches. Accession numbers for the sequences used are listed in Materials and Methods.
Discussion
Several independent lines of evidence confirm that the Arabidopsis ε-hydroxylase/LUT1 locus is a cytochrome P450 monooxygenase, encoded by At3g53130. The cytochrome P450 ε-hydroxylase carries out a type of hydroxylation reaction that is mechanistically distinct from the nonheme diiron β-hydroxylases and has evolved independently of the β-hydroxylases. The absence of lutein in the LUT1-null knockout allele, lut1-3, demonstrates that LUT1 is the only ε-hydroxylase activity in photosynthetic tissues. Thus, although β-hydroxylases have been shown to use ε-ring substrates in vitro with low efficiency (9, 10), they do not contribute to ε-ring hydroxylation activity in vivo and cannot compensate for the lack of ε-ring hydroxylation activity in lut1-3.
Isolation of a Hydroxylase Triple-Knockout Mutant Indicates the Existence of a Third, Previously Uncharacterized β-Hydroxylase in Vivo. Previous work with mutant genotypes defective in one or more of the carotenoid hydroxylases (β-hydroxylases 1 and 2 and LUT1) suggested that LUT1 may be active toward β-rings (12). The b1 b2 double mutant lacks both known β-hydroxylases, but the level of hydroxylated β-rings is only reduced 25% relative to wild type, indicating that other β-ring hydroxylation activity must exist. The introduction of the lut1-2 allele (now known to be a leaky mutation) into the b1 b2 background led to an additional 15% reduction of hydroxylated β-rings relative to wild type, consistent with LUT1 contributing to β-ring hydroxylation in vivo. Induction of LUT1 expression in the b1 b2 double mutant is also consistent with the hypothesis that the β-ring hydroxylation deficiency in b1 b2 is partially compensated by up-regulating expression of the LUT1 gene (Fig. 4).
To test whether LUT1 is the only additional β-ring hydroxylation activity in the b1 b2 background, the LUT1-null allele, lut1-3, was introduced into the b1 b2 double-mutant background. Were LUT1 the only additional β-ring hydroxylation activity in vivo, the lut1-3 b1 b2 triple mutant would lack all xanthophylls and likely be lethal. However, hydroxylated β-ring groups are still produced in lut1-3 b1 b2 at levels similar to lut1-2 b1 b2 (Table 1), which clearly indicates that a fourth unknown carotenoid hydroxylation activity exists that is specific for β-rings in vivo. This activity is not a nonheme diiron β-hydroxylase, because no additional nonheme diiron β-hydroxylase homologs are present in the Arabidopsis genome (10). Phylogenetic analysis of LUT1 homologs reveals that the closest homolog in Arabidopsis, CYP97A3 (Fig. 5), is also chloroplast-targeted. CYP97A3 does not have ε-ring hydroxylation activity based on the lut1-3 phenotype (Fig. 2C) but is a likely candidate for the additional β-hydroxylase in the lut1-3 b1 b2 triple mutant.
β- and ε-Ring Hydroxylases Have Different Hydroxylation Mechanisms. Carotenoid β- and ε-hydroxylases add hydroxyl groups to the β- and ε-rings of carotenes, respectively. β- and ε-rings are quite similar in structure and differ only in the placement of a double bond on the ring structure (Figs. 1 and 6). In a β-ring this double bond is conjugated with the polyene chain, whereas in an ε-ring it is not, and hence the relative conformations of the two rings to the polyene chain differ. Given the degree of similarity of the two substrates, why would two different types of monooxygenases be required for carotene hydroxylation reactions in Arabidopsis?
Fig. 6.
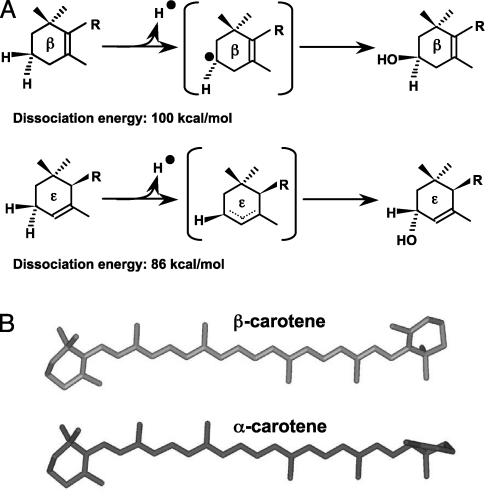
The substrates and proposed mechanisms of carotenoid hydroxylation reactions. (A) The hydroxylation reactions of β- and ε-rings. R, polyene chain. (B) Three-dimensional structures of α- and β-carotene hydroxylation substrates. The left rings of both molecules are β-rings, and the right rings are β- and ε-rings, respectively, for β- and α-carotene.
Hydroxylation of both β- and ε-rings requires the abstraction of a hydrogen atom from the C-3 position of each ring (Fig. 6A). However, it is energetically more favorable to withdraw a C-3 hydrogen atom from an ε-ring than from a β-ring (dissociation energy 86 kcal/mol vs. 100 kcal/mol, respectively) (22). This is because the ε-ring C-3 is an allylic carbon and produces a resonance-stabilized allylic radical after hydrogen abstraction, whereas the nonallylic β-ring C-3 cannot (Fig. 6A). This suggests that β-rings may require a stronger oxidant for hydrogen abstraction, raising the possibility that the nonheme diiron β-hydroxylase is a stronger oxidant than the cytochrome P450 ε-hydroxylase.
Both cytochrome P450-type and nonheme diiron-type monooxygenases hydroxylate substrates by a hydrogen atom-abstraction/oxygen-rebound mechanism with a short-lived ironoxo intermediate. The key cytochrome P450 intermediate is an FeIV=O porphyrin π-radical cation (23), whereas the key nonheme diiron monooxygenase intermediate is a di-FeIV unit with bridging oxos (24). Experimental evidence has shown that both types of enzymes are able to add oxygen to allylic and nonallylic C—H bonds (25, 26), and hence both β- and ε-hydroxylases can produce oxidants sufficient to extract a hydrogen atom from both types of rings. Therefore, the requirement of two fundamentally different types of hydroxylases for β- and ε-rings cannot simply be explained by their oxidation capacities relative to their different substrates.
Another key factor that determines enzyme catalysis is the access to and binding of substrates. Neither the β- nor the ε-hydroxylase has been crystallized; therefore the precise conformations of their substrate-binding sites are not known. However, small differences in the carotenoid ring structures may contribute to the different substrate specificity of the enzymes. β-Rings have a double bond in conjugation with the polyene chain and hence are restrained to the same plane as the polyene chain, whereas the double bond of ε-rings is not in conjugation and has relatively free rotation around the C6′–C7′ carbon (Fig. 6B). One possible explanation for the differing hydroxylase substrate specificity is that the substrate-binding pocket of β-hydroxylases can only accommodate a straight-chain hydro-carbon (e.g., β-rings) and not a ring that is tilted from the polyene chain (e.g., ε-rings). The opposite could be true of substrate binding by the ε-hydroxylase. Another possible explanation is that although both hydroxylases may bind β- and ε-ring substrates, only one of the ring structures is in the correct orientation for efficient C-3 hydrogen atom abstraction and subsequent oxidation by the respective enzyme. Therefore, it is likely that the stereochemistry of substrates leads to the specificity of each hydroxylase enzyme.
Evolution of the ε-Hydroxylase. The identification of the LUT1 locus as a cytochrome P450 enzyme makes it clear that the ε-hydroxylases evolved independently of β-hydroxylases, which have invariably been shown to be nonheme diiron enzymes (10). Putative ε-hydroxylase homologs have been identified in monocot (e.g., rice) and dicot (e.g., soybean and pea) databases, suggesting that the enzyme is widespread in the plant kingdom. The obvious question becomes: What is the evolutionary origin of the ε-hydroxylase in plants?
Although the exact driving force for the evolution of the ε-hydroxylase is unknown, it may have evolved in parallel with the evolution of its substrate, α-carotene (β,ε-carotene). The ε-hydroxylase product, lutein, has only been identified in land plants and the green algae that gave rise to land plants (27). Although cyanobacteria do not contain lutein, the biosynthetic precursor of lutein, α-carotene, is present in two cyanobacteria: Prochlorococcus and Acaryochloris (28, 29). However, no CYP97 family homologs could be identified in any cyanobacterial databases, including Prochlorococcus and Acaryochloris (data not shown), which suggests that the acquisition of ε-hydroxylase activity must have occurred sometime between the evolution of α-carotene-containing cyanobacteria and the evolution of lutein-containing green algae.
ε-Hydroxylase is a member of the cytochrome P450 family. Plant cytochrome P450s are generally divided into the A type, which is highly conserved and specific to plants, and the non-A type, which is more divergent and similar to nonplant cytochrome P450s (20). Previous phylogenetic analysis has shown that the CYP97 family (which includes the ε-hydroxylase) belongs to the non-A-type clade and is related most closely to the CYP86 and CYP94 families, both of which use fatty acids as substrates (30). Carotenoids and fatty acids have similar structures and solubilities, and it is conceivable that the ε-hydroxylase evolved from cytochrome P450-type fatty acid hydroxylases. This would be analogous to the nonheme diiron carotenoid β-hydroxylases that have consensus iron-binding histidine motifs shared with the membrane fatty acid desaturases (24).
In conclusion, we have identified LUT1 as a member of the cytochrome P450 monooxygenase family that utilizes a hydroxylation mechanism distinct from that of nonheme diiron β-hydroxylases. Cloning of LUT1 is fundamental for our understanding of lutein biosynthesis, the overlapping functions of different carotenoid hydroxylases, and the regulation of ring hydroxylations in vivo. In addition, isolation of a carotenoid hydroxylase triple-knockout mutant has defined the existence of an additional β-hydroxylase in vivo.
Acknowledgments
The University of Wisconsin Arabidopsis T-DNA knockout facility provided mutant screening services. This work was supported by National Science Foundation Grant IBN-0131253.
Abbreviations: EMS, ethyl methane sulfonate; T-DNA, portion of the tumor-inducing plasmid that is transferred to plant cells.
Data deposition: The cDNA sequence reported in this paper has been deposited in the GenBank database (accession no. AY424805).
References
- 1.Niyogi, K. K. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 333-359. [DOI] [PubMed] [Google Scholar]
- 2.Hirschberg, J. (2001) Curr. Opin. Plant Biol. 4, 210-218. [DOI] [PubMed] [Google Scholar]
- 3.Walton, T. J., Britton, G. & Goodwin, T. W. (1969) Biochem. J. 112, 383-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milborrow, B. V., Swift, I. E. & Netting, A. G. (1982) Phytochemistry 21, 2853-2857. [Google Scholar]
- 5.Britton, G. (1998) in Carotenoids: Biosynthesis and Metabolism, eds. Britton, G., Liaaen-Jensen, S. & Pfander, H. (Birkhäuser, Basel), Vol. 3, pp. 13-147. [Google Scholar]
- 6.Cunningham, F. X., Jr., & Gantt, E. (1998) Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 557-583. [DOI] [PubMed] [Google Scholar]
- 7.Shanklin, J., Whittle, E. & Fox, B. G. (1994) Biochemistry 33, 12787-12794. [DOI] [PubMed] [Google Scholar]
- 8.Bouvier, F., Keller, Y., d'Harlingue, A. & Camara, B. (1998) Biochim. Biophys. Acta 1391, 320-328. [DOI] [PubMed] [Google Scholar]
- 9.Sun, Z., Gantt, E. & Cunningham, F. X. (1996) J. Biol. Chem. 271, 24349-24352. [DOI] [PubMed] [Google Scholar]
- 10.Tian, L. & DellaPenna, D. (2001) Plant Mol. Biol. 47, 379-388. [DOI] [PubMed] [Google Scholar]
- 11.Pogson, B., McDonald, K. A., Truong, M., Britton, G. & DellaPenna, D. (1996) Plant Cell 8, 1627-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian, L., Magallanes-Lundback, M., Musetti, V. & DellaPenna, D. (2003) Plant Cell 15, 1320-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer, K., Benning, G. & Grill, E. (1996) in Genome Mapping in Plants, ed. Peterson, A. G. (Landes Bioscience, Austin, TX), pp. 137-154.
- 14.Gleave, A. P. (1992) Plant Mol. Biol. 20, 1203-1207. [DOI] [PubMed] [Google Scholar]
- 15.Clough, S. J. & Bent, A. F. (1998) Plant J. 16, 735-743. [DOI] [PubMed] [Google Scholar]
- 16.Livak, K. J. (1997) User Bulletin No. 2: ABI PRISM 7700 Sequence Detection System (PE Applied Biosystems, Foster City, CA), pp. 11-15.
- 17.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 24, 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saitou, N. & Nei, M. (1987) Mol. Biol. Evol. 4, 406-425. [DOI] [PubMed] [Google Scholar]
- 19.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244-1245. [DOI] [PubMed] [Google Scholar]
- 20.Chapple, C. (1998) Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 311-343. [DOI] [PubMed] [Google Scholar]
- 21.Schuler, M. A. & Werck-Reichhart, D. (2003) Annu. Rev. Plant Biol. 54, 629-667. [DOI] [PubMed] [Google Scholar]
- 22.Berkowitz, J., Ellison, G. B. & Gutman, D. (1994) J. Phys. Chem. 98, 2744-2765. [Google Scholar]
- 23.Ortiz de Montellano, P. R. (1995) Drug Metab. Dispos. 23, 1181-1187. [PubMed] [Google Scholar]
- 24.Shanklin, J. & Cahoon, E. B. (1998) Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 611-641. [DOI] [PubMed] [Google Scholar]
- 25.Sono, M., Roach, M. P., Coulter, E. D. & Dawson, J. H. (1996) Chem. Rev. (Washington, D.C.) 96, 2841-2887. [DOI] [PubMed] [Google Scholar]
- 26.Yoshizawa, K. (2000) J. Inorg. Chem. 78, 23-34. [DOI] [PubMed] [Google Scholar]
- 27.Johnson, E. A. & Schroeder, W. A. (1995) Adv. Biochem. Eng. Biotechnol. 53, 119-178. [DOI] [PubMed] [Google Scholar]
- 28.Hess, W. R., Rocap, G., Ting, C. S., Larimer, F., Stilwagen, S., Lamerdin, J. & Chisholm, S. W. (2001) Photosynth. Res. 70, 53-71. [DOI] [PubMed] [Google Scholar]
- 29.Miyashita, H., Adachi, K., Kurano, N., Ikemoto, H., Chihara, M. & Miyachi, S. (1997) Plant Cell Physiol. 38, 274-281. [Google Scholar]
- 30.Paquette, S. M., Bak, S. & Feyereisen, R. (2000) DNA Cell Biol. 19, 307-317. [DOI] [PubMed] [Google Scholar]