Abstract
Recent molecular exploration of the Plasmodium species circulating in great apes in Africa has revealed the existence of a large and previously unknown diversity of Plasmodium. For instance, gorillas were found to be infected by parasites closely related to Plasmodium falciparum, suggesting that the human malignant malaria agent may have arisen after a transfer from gorillas. Although this scenario is likely in light of the data collected in great apes, it remained to be ascertained whether P. falciparum-related parasites may infect other nonhuman primates in Africa. Using molecular tools, we here explore the diversity of Plasmodium species infecting monkeys in Central Africa. In addition to previously described Hepatocystis and Plasmodium species (Plasmodium gonderi and Plasmodium sp DAJ-2004), we have found one African monkey to be infected by a P. falciparum-related parasite. Examination of the nuclear and mitochondrial genomes of this parasite reveals that it is specific of nonhuman primates, indicating that P. falciparum-related pathogens can naturally circulate in some monkey populations in Africa. We also show that at least two distinct genetic entities of P. falciparum infect nonhuman primates and humans, respectively. Our discoveries bring into question the proposed gorilla origin of human P. falciparum.
Keywords: human malaria, laverania clade, parasite host transfer, cytochrome b, biological evolution
Until very recently, only one species (Plasmodium reichenowi) was known to be a phylogenetic sister lineage of Plasmodium falciparum, the agent of malignant human malaria. These two species were the only known representatives of the subgenus Laverania (1). In 2009 and 2010, several studies have revealed the existence of a number of distinct phylogenetic species belonging to this subgenus and infecting chimpanzees, bonobos, and gorillas in Africa (2–6), drastically modifying our understanding of the evolution of the Laverania lineage, and P. falciparum in particular (7).
Liu et al. (4) have proposed that the human strains of P. falciparum arose after a single gorilla-to-human transmission (4, 7). This scenario is based on the discovery of P. falciparum-related strains naturally circulating in lowland gorillas (2, 4, 6) that display greater genetic diversity than human strains (4). In contrast, no P. falciparum strains were found in wild chimpanzees or bonobos despite similar sampling efforts (4). Strains of P. falciparum had been found only in chimpanzees and bonobos living either in captivity or sanctuaries, with the possibility of a cross-species infection from humans to them (2, 3).
It, however, remains possible that P. falciparum-related strains circulate in wild chimpanzees and bonobos at a lower prevalence than in gorillas (8) and/or in geographic areas not yet explored. Given the propensity of Plasmodium parasites to switch hosts (e.g., the transfer of P. knowlesi from macaques to humans (9) and P. vivax and P. malariae between humans and New World monkeys; ref. 10), one cannot exclude that P. falciparum-related strains also circulate in other African nonhuman primate species.
We report an analysis of the diversity of Plasmodium species infecting 10 monkey species in Africa and show that P. falciparum nonhuman primate-specific strains circulate in some of these primates. This finding challenges the gorilla origin of the human strains of P. falciparum.
Results and Discussion
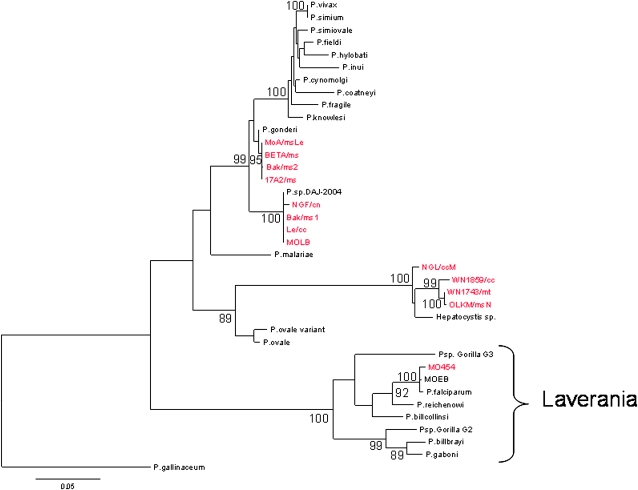
We analyzed blood samples from 338 monkeys from Gabon, including 10 different species of nonhuman primates, Cercopithecus nictitans, Cercopithecus cephus, Cercopithecus pogonias, Cercopithecus neglectus, Cercopithecus solatus, Cercocebus torquatus, Mandrillus sphinx, Miopithecus talapoin, Miopithecus ogouensis, and Lophocebus albigena. Approximately four percent (12 individuals) of the 338 monkeys were positive in a PCR array for Plasmodium cytochrome b (cyt b) (Table 1). As expected from previous studies (11–14), we detected infections by Hepatocystis spp. (12), Plasmodium gonderi (11, 12), and Plasmodium sp. DAJ-2004 (13, 14) (Fig. 1). Although Plasmodium sp. DAJ-2004 was originally isolated and molecularly characterized from a drill (Mandrillus leucophaeus) (14), we have now found that it also infects mandrills (M. sphinx) and some guenon species (C. nictitans and C. cephus).
Table 1.
Plasmodium species determined by cytochrome b sequences in African monkeys
| Host species | Sample size | Number of cytochrome b sequences obtained | Plasmodium species (number of sequences) |
| Cercocebus torquatus | 14 | 0 | ― |
| Cercopithecus cephus | 67 | 3 | P.sp. DAJ-2004 (1) |
| Hepatocystis sp. (2) | |||
| Cercopithecus neglectus | 3 | 0 | ― |
| Cercopithecus nictitans | 29 | 2 | P.sp. DAJ-2004 (1) |
| P. falciparum (1) | |||
| Cercopithecus pogonias | 4 | 0 | ― |
| Cercopithecus solatus | 2 | 0 | ― |
| Lophocebus albigena | 4 | 0 | ― |
| Mandrillus sphinx | 212 | 6* | P.sp. DAJ-2004 (2) |
| P. gonderi (4) | |||
| Hepatocystis sp. (1) | |||
| Miopithecus ogouensis | 2 | 0 | ― |
| Miopithecus talapoin | 1 | 1 | Hepatocystis sp. (1) |
*One Mandrillus sphinx was mix-infected by Plasmodium sp. DAJ-2004 and P. gonderi.
Fig. 1.
Phylogenetic relationships among primate Plasmodium species based on a partial sequence (823 bp) of the cytochrome b gene. The tree was constructed by using maximum likelihood methods. The isolates genetically characterized in this study are highlighted in red. Bootstrap values >80% are indicated. P. gallinaceum, a parasite from birds, was used to root the tree. See Table S4 for the GenBank accession numbers of the sequences used in the tree. Note that P. sp. Gorilla G2 and P. sp. Gorilla G3 are the names given in ref. 4 to P.sp GorA and P.sp Gor B, respectively, as defined in ref. 6.
Surprisingly, we found one individual C. nictitans infected with a P. falciparum-related parasite (isolate MO454; Fig. 1). The presence of this P. falciparum-related parasite in C. nictitans was confirmed three times by using independent extraction products and PCRs. All PCR products were cloned before sequencing to avoid potential chimeric sequences. The individual carrying this parasite was a wild-born, adult C. nictitans kept as a pet in the Gabonese village of Oguewa (Moyen Ogooué province; Fig. S1). The host species was genetically confirmed by the analysis of its 12S mitochondrial sequence (Materials and Methods).
We tested the hypothesis that this monkey living in close contact with humans could have become infected by a human P. falciparum after its arrival at the village. We first tried to determine by qPCR the alleles present at two genes involved in chloroquine resistance: pfcrt and pfmdr1. In Gabon, the prevalence of the two point mutations conferring resistance (pfrct 76T and pfmdr1 86Y) in human P. falciparum populations is high (>85%) (15, 16). Hence, we hypothesized that if the MO454 isolate was of very recent human origin, there would be a high probability to observe at least one resistance allele at one or both genes. We were unable to determine the genotype at pfmdr1. For pfcrt, the MO454 isolate (note that Plasmodium blood stages are haploid) displayed the wild allele at codon 76 (pfcrt K76). This allele is absent in the human P. falciparum population in Lambaréné (Moyen Ogooué province), a city <30 km away from the village where MO454 was collected (16) (Fig. S1). In another village of Gabon (Bakoumba, Haut Ogooué province; Fig. S1), the wild allele at codon 76 is only 6% (15). Such low prevalence of the wild-type allele in the human P. falciparum population in Gabon suggests a nonhuman origin for the MO454 isolate. However, we considered this evidence by itself was not sufficient to rule out its human origin.
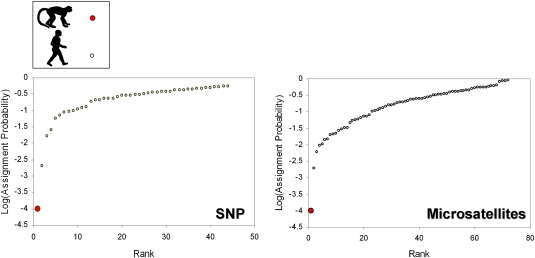
We then determined the genotype of the MO454 isolate at seven nuclear microsatellite markers and 96 nuclear single nucleotide polymorphisms (SNPs) distributed all over the genome, and compared the multilocus genotypes of the MO454 isolate to a set of human African isolates genotyped for the same markers (see Materials and Methods for additional details) and collected from the same region of Africa (Gabon, Cameroun, and Congo). We computed assignment probabilities and performed a principal component analysis (PCA) for the two sets of markers. An assignment probability corresponds to the probability that a multilocus genotype belongs to a population given the allelic frequencies observed in this population. If the MO454 isolate was a very recent transfer from humans, it should display a high assignment probability to the human P. falciparum population. As shown in Fig. 2, MO454 displays a very low probability of assignment to the human P. falciparum population, whether computed using the microsatellite (P = 0.0001) or the SNP markers (P = 0.0001). The assignment probabilities computed for the human isolates were always higher (Fig. 2).
Fig. 2.
Ranked logarithms of the assignment probabilities computed for human and nonhuman primate P. falciparum isolates obtained using SNP (Left) and microsatellite (Right) markers. The MO454’s probability is represented by the red solid circle at bottom left.
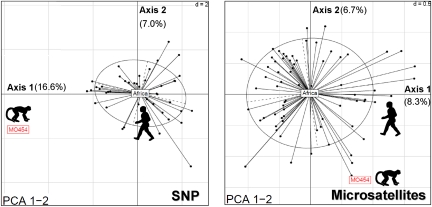
PCA results are congruent with the previous results. MO454 always displayed an outlier position on the first two axes compared with the human isolates, whether using the microsatellite or the SNP markers (Fig. 3). Thus, our analyses suggest that the MO454 isolate comes from a population of P. falciparum that is genetically differentiated and isolated from the current human P. falciparum population circulating in central Africa. These results, in turn, suggests that MO454 is not of very recent human origin and, therefore, that the monkey was most likely already infected before its arrival at the Gabonese village.
Fig. 3.
Projection of the principal component analysis (PCA) on axes 1 and 2. Dots represent individuals (haplotypes). Ellipses are a graphical summary of the dispersion of the human P. falciparum haplotypes. The position of the MO454 isolate is indicated by the red box. The barycentre of the human population (“Africa”) is located at the center of the ellipse. Left, SNPs; Right, microsatellites. The percent of the variance explained by each axis is given next to each axis.
Two scenarios can be considered regarding the nature of the population from which MO454 originates: (i) this population diverged from a population of human P. falciparum several generations ago, and had time to diverge from it because of adaptation to the host and/or genetic drift with limited exchanges of migrants. In this case, the population would have a human origin. (ii) The population has evolved for a long time in nonhuman primates. In this case, the population would have a simian origin.
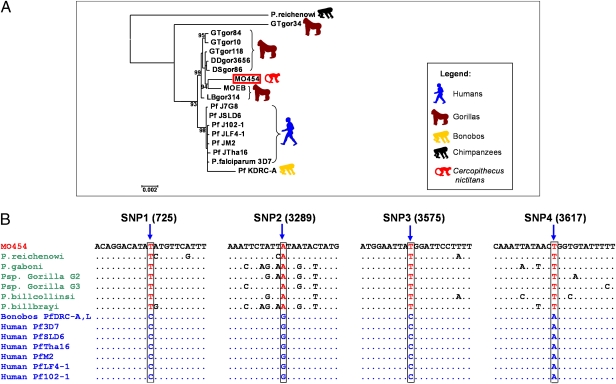
To test these alternatives, we sequenced the entire mitochondrial genome of the MO454 isolate. Liu et al. (4) have shown that four SNPs of the Laverania mitochondrial genome make it possible to distinguish the Plasmodium infecting nonhuman primates from the Plasmodium infecting humans. We found that, for these four SNPs, the MO454 isolate displayed the same alleles as the ones found in nonhuman primate Plasmodium (Fig. 4), thus indicating that the MO454 strain did not come from a population of human origin. This hypothesis is further sustained by the high bootstrap value of the cluster of P. falciparum isolates of simian origin (which includes the MO454 isolate), which is clearly genetically differentiated from the cluster formed by the human isolates of worldwide origin (Fig. 4 and Fig. S2). In contrast, all strains isolated from captive bonobos by Krief et al. (3) fall within the Plasmodium genetic diversity found in humans and all display the four SNPs specific of the human P. falciparum (Fig. 4 and Fig. S2). This result, added to the presence of double or triple mutations associated with resistance to pyrimethamine in the dhfr gene (3), indicates that the bonobo strains arose from a direct transfer from humans.
Fig. 4.
(A) Phylogenetic relationships among P. falciparum isolates obtained with mitochondrial genomes (≈5,800 bp). The tree was constructed by using maximum likelihood methods. Bootstrap values (in %) are indicated at each node. P. reichenowi was used to root the tree. See Table S4 for GenBank accession numbers of the different sequences used in the tree. (B) The four diagnostic mitochondrial SNPs observed in MO454 compared with those present in Plasmodium isolates of nonhuman (i.e., chimpanzees, gorillas, and bonobos) and human origin. Plasmodium isolated from bonobos by Krief et al. (3), PfDRC-A, and PfDRC-L (as well as those from PfDRC-E and PfDRC-C, not shown in the figure) display the same SNP as human isolates. Note that P. sp. Gorilla G2 and P. sp. Gorilla G3 are the names given by ref. 4 to P.sp GorA and P.sp Gor B, respectively, as defined by ref. 6.
Our results show that African monkeys can be infected by P. falciparum nonhuman primate-specific strains, in addition to the previously described Hepatocystis and Plasmodium species. Whether the P. falciparum infection we report in this study was due to a recent transmission from apes (e.g., gorillas) to monkeys or whether independent transmission foci exist in monkeys remains to be determined. In any case, our findings indicate that systematic surveys of the diversity of Plasmodium species circulating in primates and other mammals are needed to resolve the evolution of the Laverania subgenus. Our finding of a P. falciparum-related strain in a monkey suggests that P. falciparum may not be strictly specific to gorillas and humans, and opens up the possibility that other nonhuman primate species might also be naturally infected by P. falciparum parasites.
Our discovery brings into question Liu et al.’s (4) proposal that human P. falciparum arose after a single gorilla-to-human transmission event. Indeed, if P. falciparum appeared in humans after a single cross-species transmission from a nonhuman primate species, then a transfer from a monkey is as likely as a transfer from an ape. An alternative possibility is that P. falciparum diverged from P. reichenowi after the divergence of the human and chimpanzee lineages and then experienced a recent radiation in primates in Africa, including great apes and monkeys (6, 17). The fact that the genetic diversity observed in human isolates is lower than the genetic diversity observed in nonhuman primates might be due to recent demographic events (see e.g., ref. 18). There is a need for additional investigations to decide among these alternative hypotheses. Determining the mosquito species involved in the transmission of the nonhuman primate P. falciparum (which are not known) and their feeding behavior and host preferences might allow the assessment of the likelihood of an ape-to-human transmission. Similarly, estimating with accuracy the dates of divergence between the different P. falciparum lineages and between P. reichenowi and P. falciparum would help to clarify the evolutionary history of P. falciparum.
What can be readily concluded from previous studies and from this one is that the diversity of P. falciparum is large, and so is the range of host species that it can naturally infect. It now becomes important to ascertain the origin of this diversity and to determine which genomic characteristics have allowed this lineage to become a generalist parasite. At least two distinct genetic entities of P. falciparum are present, one infecting apes and other nonhuman primates and another one infecting humans (Fig. 4). It will be worthwhile to ascertain the specific genomic adaptations of P. falciparum to the human host (19, 20).
The capacity of P. falciparum to switch hosts represents a risk that new strains may be transferred from nonhuman primates to humans, and vice versa. Although there is still no definitive evidence that nonhuman primates can serve as reservoirs of P. falciparum for humans, the contrary is not true. It is now apparent that the bonobos described by Krief et al. (3) were infected by human strains because they lived in close contact with humans. Should Plasmodium infections also prove seriously deleterious for nonhuman primate health, this additional factor could have major implications for the conservation of these already threatened species (International Union for Conservation of Nature, http://www.iucn.org/), although the evidence available indicates that P. falciparum is much less malignant for apes than it is for humans (19, 20). Given the steadily increasing contacts between humans and nonhuman primate populations due to human population growth and the increasing development of anthropic activities (e.g., logging, mining, commercial hunting) in tropical regions, the risk of emergence of these pathogens may not only have an impact on public health but also seriously compromise the preservation and persistence of nonhuman primate populations in the long term. There is a need to evaluate this risk. The identification as well as the description of the ecology and trophic behaviors of the vectors of the nonhuman primate Plasmodium species seems now all important.
Materials and Methods
Nonhuman Primate Sampling.
All procedures in this study involving animals were approved by the Government of the Republic of Gabon and the Game and Wildlife Department of Libreville, Gabon (no. CITES 00956). The procedures complied with the relevant national and international guidelines. The samples were collected from wild-born nonhuman primates kept as pets by villagers in Gabon, as well as from 142 mandrills kept in a semifree ranging enclosure at the Centre International de Recherches Médicales de Franceville (CIRMF, Gabon). Blood samples were collected in 7-mL EDTA vacutainers from sedated animals (Ketamine; 10 mg/kg i.m.). Clots and plasma were obtained by centrifugation and stored at −20 °C until transported to CIRMF and stored at −80 °C until further analyses.
DNA Extraction, PCR Conditions, and Sequencing.
Total DNA (Plasmodium and host) was isolated and purified from blood by using the DNeasy blood kit (Qiagen) according to the manufacturer's instructions. The Plasmodium cytochrome b (cyt b) gene was amplified and sequenced by using the protocols described in Prugnolle et al. (6) for diagnostic purposes. In addition to the cyt b gene, a set of other loci were amplified, genotyped, or sequenced in the isolate related to P. falciparum (see below).
First, we characterized the mutations present at codon 76 of the gene pfcrt and codon 86 of the gene pfmdr1 by using real-time PCR assays. Detection of pfcrt T76 mutations was done by using a real-time PCR method described (21). Detection of pfmdr-1 Y86 mutations was performed with the same quantitative real-time PCR assay by using fluorescence resonance energy transfer (FRET) hybridization probes. The method included amplification of a fragment of the pfmdr1 gene of P. falciparum, coupled with simultaneous detection of the product by probe hybridization and analysis of the melting curve by using real-time PCR. The pfmdr1 gene was amplified by using primers F186 (5′-GTATTATCAGGAGGAACATTACC-3′) and R186 (5′-CCACAAACATAAATTAACGCA-3′). The amplified product was detected with two probes: the sensor 86 probe, 3′-labeled with fluorescein (5′-TTAATATCATCACCTAAATACATg–FL-3′), which hybridized with the region containing the mutation sites, and the anchor 86 probe, 5′-labeled with LC-Red 640 and 3′-phosphorylated (5′-LC640-TCTTTAATATTACACCAAACACAgATAT–PH-3′). Primers and probes were provided by Tib MolBiol (Berlin). The PCR and hybridization reactions were carried out on a LightCyler carousel-based system (Roche Diagnostics), in glass capillaries in a volume of 10 μL containing 2.5 μL of template DNA, 1 μL of MgCl2 (25 mM), 1 μL of forward and reverse primers (5 μM each), 0.1 μL of sensor and anchor probes (20 μM each), and 1 μL of FastStart DNA Master Hybridization Probes (Roche Diagnostics). Briefly, qPCR was performed under standard conditions (1 cycle of 95 °C for 9 min; 50 cycles of 95 °C for 15 s, 50 °C for 25 s, and 68 °C for 7 s; 1 cycle of 95 °C for 15 s, 40 °C for 2 min, and a slow rise in the temperature to 65 °C at a rate of 0.1 °C/s with continuous acquisition of fluorescence decline).
Each run of pfcrt and pfmdr1 PCR contained two DNA control samples of P. falciparum, F32-Tanzania (a chloroquine-sensitive, wild-type pfcrt K76 and pfmdr1 N86 strain) and FcM29-Cameroon (a chloroquine-resistant strain mutated for codon 76 of pfcrt and codon 86 of pfmdr1).
Second, we genotyped seven microsatellite markers. These microsatellites were originally discovered and designed on human P. falciparum strains but worked well on the primate P. falciparum (see Table S1 for a description of the microsatellites). PCR amplifications followed the protocols described in ref. 22. Fluorescence-labeled PCR products were sized on ABI Prism3110xl genetic analyzer (Applied Biosystems), with a Genescan500LIZ internal size standard. For comparison purposes, the same microsatellites were genotyped in human P. falciparum samples coming from Gabon, Cameroon, and Congo (see Table S2 for a detail of the populations and sample sizes).
Third, we investigated the polymorphism at 96 SNPs distributed all over the genome and ascertained on human P. falciparum. This set of SNPs included SNPs located in coding (synonymous and nonsynonymous; introns and exons) as well as in noncoding regions (see Table S3 for a list of the SNPs). The SNP primers were defined according to the 3D7 sequence and genotyped by using the Golden Gate Illumina technology (Integragen Company). For comparison, the same SNPs were genotyped in a subset of the human samples genotyped with the microsatellites (Table S3).
Finally, we sequenced the entire mitochondrial genome of the P. falciparum isolate. The amplification was performed by following the protocol detailed in ref. 3. Approximately 5,800 bp (of 6,000) were amplified by using the oligos Forward 5′-GAGGATTCTCTCCACACTTCAATTCGTACTTC and Reverse 5′-CAGGAAAATWATAGACCGAACCTTGGACTC with Takara LA Taq Polymerase (TaKaRa Mirus Bio). PCR products were cloned in the PGem-T vector (Promega) and sequenced by Eurofins. Note that in addition to the MO454 and for comparison purposes, we used the same method to sequence the entire mitochondrial genome of the P. falciparum isolate MOEB (isolated from a gorilla in Gabon) and we completed the mitochondrial genome of P. gaboni (GenBank accession no. FJ895307) described in Prugnolle et al. (6).
The host species Cercopithecus nictitans of the MO454 isolate was confirmed by the amplification and the sequencing of the host partial 12S rRNA gene using the protocol described in ref. 23.
Statistical Analyses.
Phylogenetic tree.
For phylogenetic analyses, we constructed trees by using maximum likelihood (ML) methods as described in ref. 6. The best-fitting ML model under the Akaike Information Criterion was GTR (General Time Reversible) + Γ (gamma distribution) for nucleotides as identified by ModelTest (24). The most likely DNA tree and corresponding bootstrap support values were obtained by PhyML (freely available at the ATGC bioinformatics platform; http://www.atgc-montpellier.fr) using nearest neighbor interchange branch swapping and 100 bootstrap replicates (25). Parasite isolates with their GenBank accession numbers are given in Table S4.
Assignment probability.
Assignment probabilities were calculated for each individual according to the reference human P. falciparum population composed by the isolates coming from Gabon, Congo, and Cameroon.
To compute the assignment probability, we used the following procedure: first, genotype likelihood was computed by using the criterion of ref. 26. This criterion simply corresponds to the multinomial probability of the multilocus genotype given the allelic frequencies at each locus in the reference population, under an assumption of independence between loci. Note that each individual was removed from the reference population before calculating the observed allele frequencies (the leave-one-out procedure; ref. 27).
The assignment probability was then computed for each individual by using a MonteCarlo resampling method developed by ref. 28. This probability was computed by using the software GeneClass-2 (29) and 10,000 randomizations for both microsatellite and SNP markers.
Principal component analysis.
Principal component analysis (PCA) was used to gain complementary insights into the relative position of the MO454 P. falciparum haplotype relatively to the diversity found in the human isolates from Central Africa. The PCA was performed on the matrix of binary allele profiles by using the R-packages Ade4 (30) and Adegenet (31). Missing values were replaced by the average frequencies of the corresponding alleles (31).
Supplementary Material
Acknowledgments
We thank two anonymous referees for very helpful comments and suggestions, the staff of the CIRMF Primate Centre for providing the mandrill samples, and Jérôme Clain and Karen Day for helpful discussions and suggestions. This work was jointly supported by Centre National de la Recherche Scientifique, Institut de Recherche pour le Développement (IRD), and Centre International de Recherches Médicales de Franceville, as well as the Agence Nationale de la Recherche Malaria Genetic Adaptation to New Environments Programme Santé-Environnement et Santé-Travail 07 2012 and the Programme Maladie Infectieuses Emergentes 2009. B.O. was financed by an Institut de Recherche pour le Développement fellowship for 6 mo.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109368108/-/DCSupplemental.
References
- 1.Bray RS. Studies on malaria in chimpanzees. VI. Laverania falciparum. Am J Trop Med Hyg. 1958;7:20–24. doi: 10.4269/ajtmh.1958.7.20. [DOI] [PubMed] [Google Scholar]
- 2.Duval L, et al. African apes as reservoirs of Plasmodium falciparum and the origin and diversification of the Laverania subgenus. Proc Natl Acad Sci USA. 2010;107:10561–10566. doi: 10.1073/pnas.1005435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krief S, et al. On the diversity of malaria parasites in African apes and the origin of Plasmodium falciparum from Bonobos. PLoS Pathog. 2010;6:e1000765. doi: 10.1371/journal.ppat.1000765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu W, et al. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature. 2010;467:420–425. doi: 10.1038/nature09442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ollomo B, et al. A new malaria agent in African hominids. PLoS Pathog. 2009;5:e1000446. doi: 10.1371/journal.ppat.1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prugnolle F, et al. African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. Proc Natl Acad Sci USA. 2010;107:1458–1463. doi: 10.1073/pnas.0914440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prugnolle F, et al. A fresh look at the origin of Plasmodium falciparum, the most malignant malaria agent. PLoS Pathog. 2011;7:e1001283. doi: 10.1371/journal.ppat.1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holmes EC. Malaria: The gorilla connection. Nature. 2010;467:404–405. doi: 10.1038/467404a. [DOI] [PubMed] [Google Scholar]
- 9.Singh B, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–1024. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- 10.Tazi L, Ayala FJ. Unresolved direction of host transfer of Plasmodium vivax v. P. simium and P. malariae v. P. brasilianum. Infect Genet Evol. 2011;11:209–221. doi: 10.1016/j.meegid.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Coatney GR, Collins WE, Warren M, Contacos PG. The Primate Malarias. Washington, DC: US Government Printing Office; 1971. [Google Scholar]
- 12.Garnham PCC. Malaria Parasites and Other Haemosporidia. Oxford: Blackwell Scientific Publications; 1966. [Google Scholar]
- 13.Mu J, et al. Host switch leads to emergence of Plasmodium vivax malaria in humans. Mol Biol Evol. 2005;22:1686–1693. doi: 10.1093/molbev/msi160. [DOI] [PubMed] [Google Scholar]
- 14.Escalante AA, Freeland DE, Collins WE, Lal AA. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc Natl Acad Sci USA. 1998;95:8124–8129. doi: 10.1073/pnas.95.14.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maïga-Ascofaré O, et al. Adaptive differentiation of Plasmodium falciparum populations inferred from single-nucleotide polymorphisms (SNPs) conferring drug resistance and from neutral SNPs. J Infect Dis. 2010;202:1095–1103. doi: 10.1086/656142. [DOI] [PubMed] [Google Scholar]
- 16.Mayengue PI, Kalmbach Y, Issifou S, Kremsner PG, Ntoumi F. No variation in the prevalence of point mutations in the Pfcrt and Pfmdr1 genes in isolates from Gabonese patients with uncomplicated or severe Plasmodium falciparum malaria. Parasitol Res. 2007;100:487–493. doi: 10.1007/s00436-006-0287-8. [DOI] [PubMed] [Google Scholar]
- 17.Silva JC, et al. Genome sequences reveal divergence times of malaria parasite lineages. Parasitology. 2010 doi: 10.1017/S0031182010001575. 10.1017/S0031182010001575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanabe K, et al. Plasmodium falciparum accompanied the human expansion out of Africa. Curr Biol. 2010;20:1283–1289. doi: 10.1016/j.cub.2010.05.053. [DOI] [PubMed] [Google Scholar]
- 19.Martin MJ, Rayner JC, Gagneux P, Barnwell JW, Varki A. Evolution of human-chimpanzee differences in malaria susceptibility: Relationship to human genetic loss of N-glycolylneuraminic acid. Proc Natl Acad Sci USA. 2005;102:12819–12824. doi: 10.1073/pnas.0503819102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varki A, Gagneux P. Human-specific evolution of sialic acid targets: Explaining the malignant malaria mystery? Proc Natl Acad Sci USA. 2009;106:14739–14740. doi: 10.1073/pnas.0908196106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vessière A, Berry A, Fabre R, Benoit-Vical F, Magnaval JF. Detection by real-time PCR of the Pfcrt T76 mutation, a molecular marker of chloroquine-resistant Plasmodium falciparum strains. Parasitol Res. 2004;93:5–7. doi: 10.1007/s00436-004-1096-6. [DOI] [PubMed] [Google Scholar]
- 22.Anderson TJ, et al. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- 23.Kocher TD, et al. Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proc Natl Acad Sci USA. 1989;86:6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 25.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 26.Paetkau D, Calvert W, Stirling I, Strobeck C. Microsatellite analysis of population structure in Canadian polar bears. Mol Ecol. 1995;4:347–354. doi: 10.1111/j.1365-294x.1995.tb00227.x. [DOI] [PubMed] [Google Scholar]
- 27.Paetkau D, Shields GF, Strobeck C. Gene flow between insular, coastal and interior populations of brown bears in Alaska. Mol Ecol. 1998;7:1283–1292. doi: 10.1046/j.1365-294x.1998.00440.x. [DOI] [PubMed] [Google Scholar]
- 28.Paetkau D, Slade R, Burden M, Estoup A. Genetic assignment methods for the direct, real-time estimation of migration rate: A simulation-based exploration of accuracy and power. Mol Ecol. 2004;13:55–65. doi: 10.1046/j.1365-294x.2004.02008.x. [DOI] [PubMed] [Google Scholar]
- 29.Piry S, et al. GENECLASS2: A software for genetic assignment and first-generation migrant detection. J Hered. 2004;95:536–539. doi: 10.1093/jhered/esh074. [DOI] [PubMed] [Google Scholar]
- 30.Dray S, Dufour AB. The ade4 package: Implementing the duality diagram for ecologists. J Stat Softw. 2007;22:1–20. [Google Scholar]
- 31.Jombart T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.