Abstract
Cancers frequently arise as a result of an acquired genomic instability and the subsequent clonal evolution of neoplastic cells with variable patterns of genetic aberrations. Thus, the presence and behaviors of distinct clonal populations in each patient's tumor may underlie multiple clinical phenotypes in cancers. We applied DNA content-based flow sorting to identify and isolate the nuclei of clonal populations from tumor biopsies, which was coupled with array CGH and targeted resequencing. The results produced high-definition genomic profiles of clonal populations from 40 pancreatic adenocarcinomas and a set of prostate adenocarcinomas, including serial biopsies from a patient who progressed to androgen-independent metastatic disease. The genomes of clonal populations were found to have patient-specific aberrations of clinical relevance. Furthermore, we identified genomic aberrations specific to therapeutically responsive and resistant clones arising during the evolution of androgen-independent metastatic prostate adenocarcinoma. We also distinguished divergent clonal populations within single biopsies and mapped aberrations in multiple aneuploid populations arising in primary and metastatic pancreatic adenocarcinoma. We propose that our high-definition analyses of the genomes of distinct clonal populations of cancer cells in patients in vivo can help guide diagnoses and tailor approaches to personalized treatment.
Keywords: clonal genomics, pancreatic cancer, prostate cancer
Large-scale investigations of cancer genomes are expected to lead to the discovery of common disease elements, including recurring genomic aberrations, that can guide the development of broadly applicable diagnostics and therapeutics (1–3). For example, studies of colorectal and breast carcinomas and glioblastoma multiforme (GBM) that surveyed somatic mutations in over 18,000 genes estimated an average of ∼80 gene-specific mutations in each cancer type (3). Strikingly few highly recurrent mutations were detected; the majority of mutations occur with a prevalence of <5% with little overlap between cancers. These and similar reports challenge the concept of collective cancer genomes (4–7). Therefore, there is a need for a unique approach (i.e., not reliant on prevalence-based studies) to identify and interpret sets of selected aberrations and define the clinical dependencies that arise in complex, highly variable carcinoma genomes in patients in vivo.
Cancer genome studies have relied on two main approaches for selecting and preparing patient samples for analyses. The first is to select samples exceeding a threshold for tumor cell content on the basis of histological methods such as evaluation of H&E-stained slides (2). However, many samples fail these criteria; notably, tumors arising in solid tissues exhibit high degrees of tissue heterogeneity, with varied admixtures of reactive stroma, inflammatory cells, and necrosis in immediate contact with tumor cells. It is well established that biopsies frequently contain multiple clonal populations of neoplastic cells that cannot be distinguished on the basis of morphology alone (8). Thus, the application of purification methods such as laser capture microdissection does not resolve the complexities of many samples. A second approach is to passage tumor biopsies in tissue culture or in xenografts (4, 9–11). These methods apply selective pressures on the complex mixtures of cells and clones present in a patient sample and are time-consuming and labor intensive and are not amenable to rapid deployment in most clinical settings. Consequently, the number of xenografts successfully grown varies from site to site, and the biological complexity and clinical context of the patient sample may not be reflected in the final processed sample.
Flow cytometry-based cell sorters can select, objectively measure, and sort individual particles such as cells or nuclei using desired features objectively defined by fluorescent and light-scattering parameters in a flow stream. Recent advances in this technology provide high-throughput flow rates and the detection of relatively rare events in dilute admixed samples, enabling the application of flow cytometry to in vivo high-definition analyses of human cancers (12). The combination of flow sorting and genomic analyses has been recently used for the enrichment of pancreas carcinoma cells and to study the clonal composition of primary breast tumors (13, 14). However, these studies relied on extensive bioinformatic analyses, platform-specific sample preparations, or relatively large amounts of input material to achieve an acceptable signal-to-noise ratio in their genome analyses. To overcome these limitations, we developed a methodology to adapt genomic DNA isolated from cytometrically purified nuclei for use with array comparative genomic hybridization (aCGH) and next-generation sequencing. Here we demonstrate the feasibility of this methodology for efficient high-resolution genomic analysis of clonal populations from even minute heavily admixed patient biopsies. We profiled patient samples of primary and metastatic pancreatic adenocarcinomas (PAs), a highly lethal tumor type that is difficult to molecularly characterize at the biopsy level due to complex genomes and heterogeneous cellularity, as cancer cells represent on average only 25% of the cells within the tumor (15). In contrast to recent studies of PA, we performed whole-genome discovery analyses with distinct clonal populations purified directly from patient biopsies (16, 17). Furthermore, we applied this approach to analyze distinct clonal tumor populations within clinically annotated prostate cancer specimens and were able to infer their clonal evolution and their mechanisms of therapy resistance. The results from this study demonstrate that our approach can study the clonal diversity and evolution of cancer genomes that arise in patients in vivo and reveal clinically relevant contexts at least in the cancer types profiled in this study.
Results
We used DAPI-based DNA content measures to identify and sort distinct diploid and aneuploid tumor populations from each of 40 PAs and biopsies from patients with varying grades of prostate adenocarcinoma (PC), including a series from one patient who progressed to androgen-independent metastatic disease. Sorted tumor cell populations represented as few as 3% to over 91% of the cellular content in a biopsy. We then profiled each sorted population with aCGH to obtain high-definition clonal profiles of the copy-number aberrations in each tumor genome. Clonality was initially defined by flow cytometric DNA content (differences of ±0.2N) and by the genomic intervals in each sorted population that were defined by the aberration detection algorithm ADM2 (18). The use of purified flow-sorted populations enabled objective identification and ranking of copy-number aberrations, including homozygous deletions (log2 ratio ≤ −3.0) and high-level (log2 ratio ≥1.0) amplifications, regardless of tumor cell content in each biopsy (Figs. 1–5). High-scoring aberrations, which likely represent selected events in each sample, were discriminated from background events that typically arise in cancer genomes. The number of ADM2-defined intrachromosomal copy-number aberrations in sorted populations ranged from <5 in multiple genomes to >100 in a 3.8N PC genome (Fig. S1 A and B). Our ability to assign these aberrations to distinct ploidies within each patient sample provided high-resolution clinically relevant analyses of clonality on the basis of objective measures of distinct tumor populations in vivo.
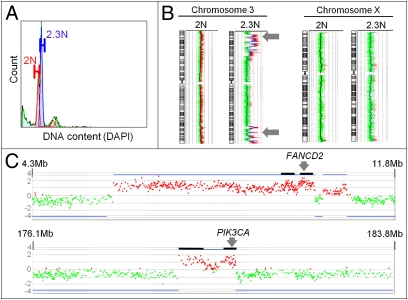
Fig. 1.
Clonal analyses of a PA genome (biopsy 4050489). (A) DAPI-based DNA content analysis detected a 2.3N clonal population. The diploid and aneuploid populations were sorted for subsequent aCGH studies. (B) Multiple focal amplicons were detected only on chromosome 3 in the 2.3N population. The diploid population had a nonaberrant genome. (C) Close-up view of FANCD2 and PIK3CA amplicons in the 2.3N clonal population. Black horizontal bars represent the cores of the amplicons. Genes included in the FANCD2 amplicon cores were the following: OGG1, CAMK1, TADA3L, ARPC4, TTLL3, RPUSD3, CIDEC, JAGN1, IL17RE, FANCD2, VHL, IRAK2, TATDN2, GHRL, SEC13. Genes included in the PIK3CA amplicon core: PIK3CA, KCNMB3.
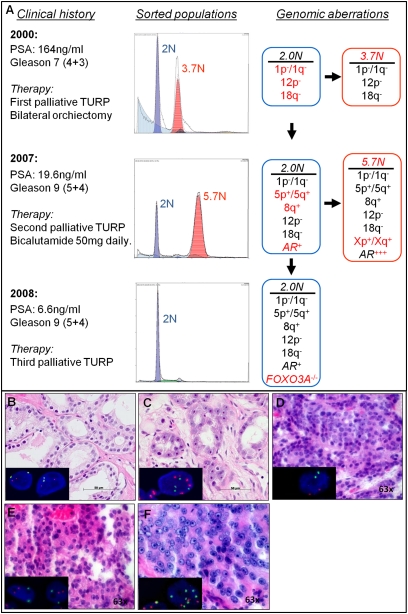
Fig. 5.
Clonal analyses of a multibiopsy series from a patient with PC. (A) Multiple populations were detected by DNA content flow cytometry over an 8-y time span during the clinical history of a prostate patient. Each population showed a series of shared (black) and unique (red) genomic aberrations. (B–F) Histological (HE) and FISH analysis (small box) (centromere 7, green; centromere 3, red) of the detected populations. (B) Diploid population from 2000. (C) Aneuploid population from 2000. (D) Diploid population from 2007. (E) Aneuploid population from 2007. (F) Unique (diploid) population from 2008.
Clonal Profiling of Patient Samples Unveils Masked Sets of Clinically Relevant Genomic Aberrations.
The high-resolution profiling of purified tumor cell populations unveiled genomic aberrations that would have been obscured by the presence of mixed tumor populations or normal cells in each biopsy of interest. This is highlighted by the detection of distinct homozygous deletions, including those targeting genes regulating different pathways in PA and PC genomes. These included NUMB, DDX10, PARK2, SMAD2, SMAD3, JARID2, PARD3, PARDG6, P2RY5, and MAP2K4 in PA and AIM1, FOXO3A, NEDD9, FEZF1, and PTEN in PC. In each case, the deletion was absent in the patient-matched diploid population, confirming their somatic nature. Significantly, the PA SMAD3 deletion, despite a high tumor cell content (>70% by flow cytometry and H&E) that is widely accepted for genome studies, was not detected in the matched unsorted tumor sample (Fig. S1 C–E) (2). In comparison, amplicons were detected in sorted and bulk sample preparations; however, the height and structural variation within amplicons further delineating regions of maximal selected copy number increase were compressed in unsorted samples (Fig. S2A). Thus, analyses of whole or histologically purified tumors cannot accurately distinguish the clonal patterns of aberrations present in a patient sample.
Clonal genomic analyses of biopsies from PA patients allowed us to detect clinically relevant genomic aberrations. Examples are summarized in Fig. 1 and in Figs. S1–S3, and S4. In Fig. 1, the 2.3N aneuploid population from a male PA patient revealed focal genomic amplicons with loss of intervening sequences across chromosome 3. The amplicons found contain the FANCD2 and the PIK3CA genes and thus suggest an acquired resistance to DNA strand-breaking agents and a potential therapeutically relevant dependency on the AKT pathway. A further example (Fig. S2) of clonal genomic analyses identified a focal 6p21 amplicon in a 3.7N aneuploid tumor population that represented less than 8% of a PA biopsy. This amplicon included the VEGFA gene and the nucleoside transporter gene SLC29A1. These aberrations might point to a vulnerability to anti-angiogenesis agents and to responsiveness to gemcitabine because SLC29A1 can increase intracellular levels of gemcitabine (19, 20). Clonal analysis of a further PA biopsy (Fig. S3) revealed one single clonal tumor population (3.0N) that represented only 3% of the biopsy. Genomic analysis revealed high-level amplicons, including the insulin receptor (INSR) and the thymidylate synthetase (TYMS) genes. These findings might be of potential clinical relevance because INSR can be effectively targeted by different clinical agents and the genomic amplification of TYMS predicts resistance to existing chemotherapeutics like 5-fluorouracil, as recently suggested for colorectal cancer (21). However, amplified regions typically contain multiple genes and thus additional analyses would be necessary for testing biological hypotheses and providing clinical validation of these events.
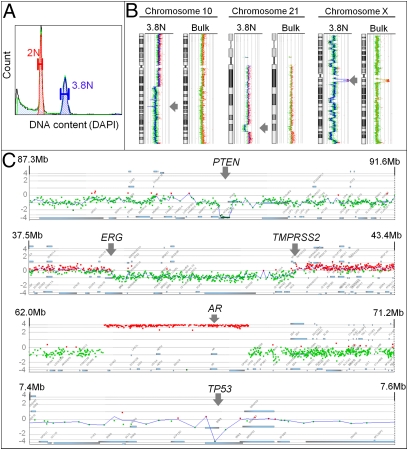
The precision of our clonal analyses can advance more accurate clinical diagnoses. For example, a 3.8N tumor population sorted from a patient originally diagnosed with an adrenal cortical carcinoma (ACC) contained a single-copy 21q22.2-q22.3 loss with boundaries at the 3′ end of ERG and the 5′ promoter region of TMPRSS2 genes (Fig. 2). These boundaries correspond to an interstitial deletion associated with the most common fusion gene in PC (22, 23). In addition, the same population had a high-level focal amplification of the AR gene (Fig. 2C). Given that this fusion gene responds to the androgen response elements in the TMPRSS2 promoter region, the concurrent presence of these independent genetic events in the same genome suggested a mechanistic interaction between the activation of ERG and the amplification of the AR gene in this clonal population. The genome of the 3.8N population also had homozygous deletions of PTEN (10q23.31), AIM1 (6q21), and, within exon 8, of TP53 (17p31.1) (Fig. 2 and Fig. S5). These lesions suggested a context of activated AKT and concomitant stimulation of AR-dependent transcriptional activation of ERG genes (Fig. S5C) (24). In addition, the homozygous deletion in AIM1 advocates a role for this melanoma-associated tumor suppressor gene (25). Significantly, none of the homozygous deletions nor the interstitial 21q loss were detected in the unsorted sample (Fig. S5D). Interestingly, both TMPRSS2/ERG rearrangement and AR gene amplification have been reported only in the context of PC (26, 27). A re-evaluation of diagnostic tumor blocks prompted by our clonal analyses showed that the tumor was positive for prostate-specific antigen and prostatic acid phosphatase. A review of the patient's history revealed a prior (>6 y) localized PC. As a result, the clinical diagnosis was changed from ACC to metastatic PC to the adrenal gland.
Fig. 2.
Clonal analysis of a PC metastasis initially diagnosed as adrenal cortical carcinoma. (A) DAPI-based DNA content analysis detected a 3.8N tumor population. (B) Examples of profiled chromosomes. The homozygous deletion on chromosome 10 and the interstitial deletion at 21q22 were detectable only in the flow-sorted population. (C) Locus-specific views of genomic aberrations in the 3.8N clonal population: concurrent homozygous deletions (PTEN, TP53), interstitial deletion targeting the ERG and the TMPRSS2 genes, and the amplification of the AR gene.
Genomic Profiles of Multiple Clonal Populations Within Single Tumors.
We detected more than one aneuploid population in a subset of individual biopsies in each tissue type. For example, two aneuploid populations in a single PA biopsy had distinct ploidies (2.3 and 3.8N) with common genomic aberrations, including two separate breakpoints in chromosome 8q and an amplicon on chromosome 19q13.2 (Fig. S4). The latter was the most significant aberrant interval in each genome and targets a region that contains a series of cancer-associated genes of interest (e.g., PAK4, AKT2, HIPK4, MAP3K10, RAB4B) to PA. These spatially admixed aneuploid populations represent 19.8% and 17.4% of the total cellular fraction in the sample. In contrast to common aCGH profiles in different ploidies, we also detected genomic aberration changes that occurred in the absence of changes in ploidy. For example, tumor populations in a primary and two metastatic lesions from the same patient each had a 2.8N ploidy (Fig. S6). However, the diaphragm metastasis had a focal AKT2 amplicon that was absent in the primary and in the liver metastasis.
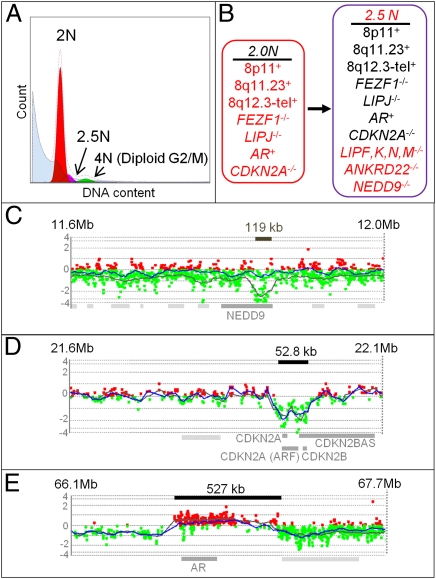
In comparison with the PA samples, the aCGH analysis of the 2N population sorted from a castration-resistant PC showed evidence of mixed tumor and genomically normal cells in the sample including compressed log2 ratios (e.g., > −2.0 for CDKN2A) compared with the aneuploid population (< −3.0 for CDKN2A). Therefore, we sorted the 4N fraction of the biopsy for analysis under the assumption that proliferating 2N cells accumulate in a 4N (G2/M) phase and are enriched for tumor cells (Fig. 3). The two tumor populations (2 and 2.5N) from this PC biopsy had common aberrations previously described in PC, including aberrations on chromosome 8 and a low-level amplicon on Xq12, which harbors only the AR gene (Fig. 3E and Fig. S7). In addition, these two populations shared homozygous deletions targeting the 9p21 CDKN2A and the 7q31 FEZF1 loci that further highlight their clonal relationship and confirmed the tumor purity of the analyzed G2/M fraction of the diploid population (Fig. 3D and Fig. S7). Interestingly, the 9p21 homozygous deletion could not be detected in either population with a commercially available FISH probe (Fig. S7). This discrepancy can be explained by the small size of the focal deletion (52.8 kb) and the 9p21/p16 FISH probe commonly used in diagnostics that encompasses a region of ∼200 kb (28). In addition to the shared aberrations, the aneuploid population (2.5N) is characterized by two additional genomic aberrations that were not present in the diploid tumor population: the extension of a homozygous deletion on 10q23 comprising several lipase genes and a deletion on 6p23 affecting only the NEDD9 gene (Fig. 3C and Fig. S7A). The presence of these unique deletions in a background of shared aberrations strongly suggested that the aneuploid clonal population originated from the 2N tumor population and that these additional homozygous deletions, targeting genes that have not been described in the context of PC, were selected in the evolution of the aneuploid tumor population (Fig. 3B). Interestingly, the down-regulation of the NEDD9 gene expression was recently described as a component of a lung metastatic signature of breast cancer and hypothesized to be responsible for enhancing migration in breast epithelial cells (29, 30).
Fig. 3.
Genomic profiling of distinct clonal populations within a castration-resistant PC. (A) DAPI-based DNA content analysis detected two distinct tumor populations: a diploid (2N) population and an aneuploid (2.5N) population. (B) Summary of the genomic aberrations present in the two clonal populations including homozygous deletions unique to the 2.5N population. Each population showed a series of shared (black) and unique (red) genomic aberrations. (C) The aneuploid population (dark gray line) harbors additional homozygous deletions, such as the deletion of the NEDD9 gene (6p25-24). (D) Both populations show the identical focal 9p21 deletion. (E) Both populations show the identical Xq12 amplification harboring only the AR gene.
Clonal Evolution and Metastases.
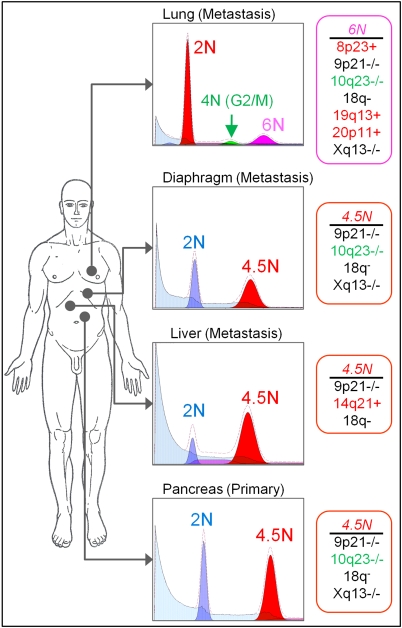
Recent studies have found that diverse patient-specific sets of genomic lesions arise in metastatic PA (16, 17). Therefore, we investigated the evolution of ploidy and genomic copy-number aberrations within clonal populations sorted from PA rapid autopsy samples from distinct anatomical sites. Our analyses revealed highly variable patterns of ploidies and genomic aberrations in each patient studied. For example, we detected common selected aberrations in each sorted tumor population from four anatomical sites, including homozygous deletions targeting CDKN2A and NRG3 (10q23) in a patient with advanced metastatic disease (Fig. 4 and Fig. S8). Populations from three of the sites had the same 4.5N ploidy whereas the lung metastases had a 6N ploidy that included a series of unique focal amplicons targeting kinases BLK (8p22-p23.1) and STK35 (20p13). However, the 4.5N population from the liver had a unique 14q21 amplicon and was the only population without the NRG3-specific homozygous deletion. These findings suggest that the 6N lung metastasis evolved from the 4.5N ploidy whereas the liver metastasis evolved at an earlier time point of carcinogenesis. In contrast, the 2N (all four sites) and the 4N (lung) populations from the same sorts were genomically normal by aCGH analyses.
Fig. 4.
Clonal analysis of a PA rapid autopsy. DAPI-based DNA content analyses of the four sites detected a 4.5N clonal population in the pancreas, the liver, and the diaphragm, but not in the lung. The lung metastasis consisted of a 6.0N clonal population. Genomic aberrations found in the distinct aneuploid populations are listed at the right. Aberrations in black are shared by all aneuploid populations, aberrations in red are specific for one organ site, and aberrations in green were found in all populations in addition to the liver.
Evolution of Clonal Populations in a Single Patient over Time.
The significance of clonal behavior is further illustrated in a multiple biopsy series from a patient diagnosed with PC in 2000 (Fig. 5 and Fig. S9). The patient received palliative transurethral resection of the prostate (TURP) and was treated with hormone deprivation by bilateral orchiectomy. Seven years later (2007), the patient relapsed. He underwent a second palliative TURP, and androgen blockade was completed by administration of bicalutamide (50 mg/day, Casodex). In 2008, the patient was subjected to a third palliative TURP. Five months later, he succumbed to the disease. The patient never received radiation or chemotherapy. DNA content-based flow sorting of the TURPs revealed populations with distinct ploidies in both 2000 (2 and 3.7N) and 2007 (2 and 5.7N), but only one population (2N) in 2008 (Fig. 5A). Histomorphological and FISH analysis of the biopsy specimens confirmed the presence of these distinct populations (Fig. 5 B–F).
The clonal populations shared aberrations, including copy-number transitions between ELK4 and SLC45A3 (1q32) and upstream of ETV6 (12p13) (Fig. 5 and Fig. S9 D, E, and L). These genes are fusion partners in epithelial tumors (31, 32). The co-occurrence of these aberrations in the original samples and their persistence in each population suggest that they arose as early selected events in the evolution of this PC (Fig. 5A). The 2007 populations were characterized by amplification of the AR gene, which can render advanced PCs more sensitive to remaining levels of androgen after castration. Strikingly, the diploid populations from 2007 and 2008 showed identical focal Xq12 amplicons, targeting only the AR gene, whereas the 5.7N population had multiple higher-level chromosome X amplicons, including a broad AR amplicon (Fig. S9 A–C). In each case, the clonally distinct AR amplicons arose after the chromosome 1q and 12p aberrations. This clonal heterogeneity and the presence of focal AR amplicons, barely visible in FISH assays, suggested that the prevalence of AR amplification in castration-resistant PC may be higher than the reported 20–30% (Fig. S9 H–K) (33, 34).
The remaining androgen-independent diploid population (2008) retained the aberrations present in the previous 2N populations and acquired an intragenic homozygous deletion targeting the FOXO3A (6q21) gene (Fig. 5A and Fig. S9 F and G). Although the FOXO3A transcription factor has been implicated in regulation of proapoptotic genes in human PC and its interference has been shown to favor androgen-independent growth of PC cells in model systems, in vivo homozygous deletion of FOXO3A has never been demonstrated (35). To further characterize the diploid 2008 population, we sequenced each of the 8 exons of the AR gene and deep-sequenced a validated set of ∼4,000 exons from ∼500 genes directly or indirectly involved in cancer in the sorted populations from 2007 and 2008. The sequences for each of the genes surveyed, including wild-type AR, were identical in each of the three clonal populations. Thus, our clonal genomic data suggested that loss of FOXO3A rather than mutation of the AR or other cancer-related genes was selected as a result of complete androgen blockage therapy and was permissive for the evolution of advanced cancer in this patient. This report represents a unique in vivo analysis of the clonal evolution of androgen-independent metastatic disease in a PC patient.
Discussion
A fundamental hypothesis in cancer biology is that cancers frequently arise as a result of acquired genomic instability and the evolution of neoplastic populations with variable patterns of random and selected aberrations (8, 36). Consequently, each patient's cancer may evolve and become dependent on distinct sets of selected aberrations during its clinical history. The genes and cellular pathways deregulated by these selected events represent enriched candidates for developing diagnostic markers and therapeutic targets. However, the cellular heterogeneity of clinical samples and the genetic diversity of cancer genomes limit the translation of genomics for improved patient care. A common approach for identifying clinically relevant cancer genome aberrations is to characterize lesions, including any loss or any gain for each chromosome occurring at rates that are statistically significant in samples of interest (2, 37–41). Single-copy losses and gains occur as part of the random events associated with genomic instability present in tumor genomes. Thus, many cancer genome studies determine a statistical threshold based on the background rates of losses and gains for detecting selected copy-number changes. This requires relatively large numbers of patient samples to account for the genomic instability and the patient-specific variations typically present in cancer genomes.
In contrast to single-copy gains and losses, events such as homozygous deletions and focal high-level amplifications typically require multiple independent genomic events and thus can represent biologically selected events in cancer genomes that target known and putative tumor suppressor genes and activated oncogenes. Recent reports have identified somatic mutations of NUMB in breast carcinoma and of PARK2 in GBM, colon, and lung cancers (42, 43). Thus, our detection of homozygous deletions provides evidence that their tumor suppressor function extends to PA and identifies a series of genes that are selectively disrupted in PA (e.g., SMAD2, SMAD3, JARID2) and PC (e.g., AIM1, NEDD9, FOXO3A). The detection of the complete loss of genomic sequence is highly sensitive to as little as 5% admixtures of nontumor cells in clinical samples (44). This is consistent with recurring observations that fewer aberrations can be detected in patient samples in vivo than in passaged model systems (4, 9, 45). However, a limitation in many cancer genome studies is the inability to objectively discriminate single-copy losses from homozygous deletions and to accurately map and determine high-level amplification in cancer genomes arising in individual patient samples. Thus, copy-number data are typically reduced to reporting frequencies of any loss or any gain for each chromosome in relatively large cohorts of samples. This further limits the translational potential of the genomic study of cancers, including those with variable complex genomes, rare cancers, and biopsies with high admixtures of genomically normal cells and insufficient tumor cell content (38). Furthermore, histology-based methods cannot readily distinguish whether aberrations in a tumor are present in a single cancer genome or if they are distributed in multiple clonal populations. Consequently, current approaches for the analyses of cancer genomes are limited in their ability to determine the clinical context of each patient's tumor.
The detection and sorting of more than one tumor population in a biopsy and the availability of multiple biopsies from individual patients extends the study of clinical phenotypes and the behaviors of clonal populations in vivo. For example, the aneuploid populations with adverse histological features and multiple selected genomic aberrations, including high-level AR amplification that arose during the clinical history of a patient who developed advanced PC, were uniquely sensitive to therapeutic regimen and were erased after hormone withdrawal (Fig. 5). The patterns of acquired clonal aberrations suggest that the aneuploid populations arose from diploid progenitors during the evolution of disease. Strikingly, the co-occurring diploid cells acquired a low-level focal AR amplicon after bilateral orchiectomy leading to increased sensitivity to remaining levels of adrenal testosterone, followed by homozygous deletion of FOXO3A in response to androgen blockage, resulting in the evolution of androgen-independent metastatic disease. The distinct AR amplicons present in the diploid and the aneuploid populations that arose during the evolution of androgen-independent metastatic PC would have been obscured in a conventional histologically prepared sample.
Recent reports have shown that clinical phenotypes such as acquired therapeutic resistance and metastases may be mediated by the biological behaviors of distinct cellular populations in individual tumors (46, 47). However, rather than inferring the presence of preexisting clones, we isolated distinct populations arising during the clinical history of the disease and then profiled their genomes at high definition. The cataloguing of mutations in patient samples provides insights into the genomic basis of disease and the identification of therapeutic targets for personalized medicine. Our clonal analyses identified selected aberrations that would otherwise be obscured by the complexity of clinical samples containing variant clonal populations in each biopsy of interest. Our data suggest that the distribution of somatic lesions, including mutations, in tumors that contain more than one clonal tumor population may have a profound effect on the clinical behavior of human cancers. This information cannot be obtained from conventional genomic profiling studies and is fundamental for the identification of selected aberrations that are essential for the evolution and clinical behaviors of a cancer.
Our current work has focused on DNA content-based sorting strategies. A major advantage of current flow cytometry technology is the ability to apply multiparameter assays that can identify and purify nuclei from both diploid and aneuploid tumor populations on the basis of features such as proliferation, cell specificity, and therapeutic targets (12, 48, 49). Furthermore, we have validated the use of DAPI-based flow assays with formalin fixed, paraffin embedded (FFPE) samples, providing a rich resource of clinical samples for our clonal analyses of clinical phenotypes.
In summary, we have shown that flow cytometric-based separation followed by precise genomic characterization of sorted tumor (sub)populations provides a deep clonal analysis of the genomic heterogeneity present in human biopsies. We propose that this comprehensive clonal analysis of clinical samples provides insights into the evolution of each patient's tumor and their responses to therapeutic treatment and, in the era of personalized medicine, might also be considered for advancing effective therapeutic decisions.
Materials and Methods
Clinical Samples.
PA samples were obtained under a Western Institutional Review Board protocol (20040832) for a National Institutes of Health-funded biospecimens repository (National Cancer Institute P01 Grant CA109552). Participating centers are listed in SI Materials and Methods. PC samples and additional PA samples were obtained with approved consent of the Ethics Committee of Basel (252/08, 302/09). All samples were collected in liquid nitrogen and stored at –80 °C. All tumor samples were histopathologically evaluated before analysis.
Flow Cytometry.
Biopsies were minced in the presence of NST buffer [146 mM NaCl buffer containing 10 mM Tris-HCl (pH 7.5), 0.2% Nonidet P40] and DAPI according to published protocols (8, 49). Nuclei were disaggregated and then filtered through a 40-μm mesh before flow sorting with a Cytopeia Influx cytometer (Becton-Dickinson) with UV excitation and DAPI emission collected at >450 nm. DNA content and cell cycle were analyzed using the software program MultiCycle (Phoenix Flow Systems).
aCGH.
DNAs were extracted using Qiagen micro kits. For each hybridization, 100 ng of genomic DNA from each sample and of pooled commercial 46, XX reference (Promega) were amplified using the GenomiPhi amplification kit (GE Healthcare). Subsequently, 1 μg of amplified sample and 1 μg of amplified reference template were digested with DNaseI and then labeled with Cy-5 dUTP and Cy-3 dUTP, respectively, using a BioPrime labeling kit (Invitrogen). All labeling reactions were assessed using a Nanodrop assay before mixing and hybridization to either 244,000 or 400,000 feature CGH arrays (Agilent Technologies).
Supplementary Material
Acknowledgments
This project was supported by Grant CA 109552 from the National Cancer Institute. C.R. was supported by the Swiss National Science Foundation (SNF für angehende Forschende) and by the Oncosuisse Foundation (OCS-02285-08-2008).
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray files reported in this paper have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus (accession no. GSE21660).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104009108/-/DCSupplemental.
References
- 1.Thomas RK, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 2.Weir BA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 4.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shah SP, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 6.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 7.Kan Z, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 8.Maley CC, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38:468–473. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 9.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2009;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pleasance ED, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2009;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ibrahim SF, van den Engh G. Flow cytometry and cell sorting. Adv Biochem Eng Biotechnol. 2007;106:19–39. doi: 10.1007/10_2007_073. [DOI] [PubMed] [Google Scholar]
- 13.Navin N, et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 2010;20:68–80. doi: 10.1101/gr.099622.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyd ZS, Raja R, Johnson S, Eberhard DA, Lackner MR. A tumor sorting protocol that enables enrichment of pancreatic adenocarcinoma cells and facilitation of genetic analyses. J Mol Diagn. 2009;11:290–297. doi: 10.2353/jmoldx.2009.080124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seymour AB, et al. Allelotype of pancreatic adenocarcinoma. Cancer Res. 1994;54:2761–2764. [PubMed] [Google Scholar]
- 16.Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipson D, Aumann Y, Ben-Dor A, Linial N, Yakhini Z. Efficient calculation of interval scores for DNA copy number data analysis. J Comput Biol. 2006;13:215–228. doi: 10.1089/cmb.2006.13.215. [DOI] [PubMed] [Google Scholar]
- 19.Farrell JJ, et al. Human equilibrative nucleoside transporter 1 levels predict response to gemcitabine in patients with pancreatic cancer. Gastroenterology. 2009;136:187–195. doi: 10.1053/j.gastro.2008.09.067. [DOI] [PubMed] [Google Scholar]
- 20.Giovannetti E, et al. Transcription analysis of human equilibrative nucleoside transporter-1 predicts survival in pancreas cancer patients treated with gemcitabine. Cancer Res. 2006;66:3928–3935. doi: 10.1158/0008-5472.CAN-05-4203. [DOI] [PubMed] [Google Scholar]
- 21.Watson RG, et al. Amplification of thymidylate synthetase in metastatic colorectal cancer patients pretreated with 5-fluorouracil-based chemotherapy. Eur J Cancer. 2010;46:3358–3364. doi: 10.1016/j.ejca.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomlins SA, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 23.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 24.Carver BS, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ray ME, Wistow G, Su YA, Meltzer PS, Trent JM. AIM1, a novel non-lens member of the betagamma-crystallin superfamily, is associated with the control of tumorigenicity in human malignant melanoma. Proc Natl Acad Sci USA. 1997;94:3229–3234. doi: 10.1073/pnas.94.7.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esgueva R, et al. Prevalence of TMPRSS2-ERG and SLC45A3-ERG gene fusions in a large prostatectomy cohort. Mod Pathol. 2010;23:539–546. doi: 10.1038/modpathol.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheble VJ, et al. ERG rearrangement is specific to prostate cancer and does not occur in any other common tumor. Mod Pathol. 2010;23:1061–1067. doi: 10.1038/modpathol.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korshunov A, Sycheva R, Golanov A. The prognostic relevance of molecular alterations in glioblastomas for patients age < 50 years. Cancer. 2005;104:825–832. doi: 10.1002/cncr.21221. [DOI] [PubMed] [Google Scholar]
- 29.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simpson KJ, et al. Identification of genes that regulate epithelial cell migration using an siRNA screening approach. Nat Cell Biol. 2008;10:1027–1038. doi: 10.1038/ncb1762. [DOI] [PubMed] [Google Scholar]
- 31.Rickman DS, et al. SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer. Cancer Res. 2009;69:2734–2738. doi: 10.1158/0008-5472.CAN-08-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tognon C, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2:367–376. doi: 10.1016/s1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 33.Bubendorf L, et al. Survey of gene amplifications during prostate cancer progression by high-throughout fluorescence in situ hybridization on tissue microarrays. Cancer Res. 1999;59:803–806. [PubMed] [Google Scholar]
- 34.Linja MJ, et al. Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res. 2001;61:3550–3555. [PubMed] [Google Scholar]
- 35.Lynch RL, et al. The progression of LNCaP human prostate cancer cells to androgen independence involves decreased FOXO3a expression and reduced p27KIP1 promoter transactivation. Mol Cancer Res. 2005;3:163–169. doi: 10.1158/1541-7786.MCR-04-0163. [DOI] [PubMed] [Google Scholar]
- 36.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 37.Bredel M, et al. A network model of a cooperative genetic landscape in brain tumors. JAMA. 2009;302:261–275. doi: 10.1001/jama.2009.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aguirre AJ, et al. High-resolution characterization of the pancreatic adenocarcinoma genome. Proc Natl Acad Sci USA. 2004;101:9067–9072. doi: 10.1073/pnas.0402932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen S, et al. Copy number alterations in pancreatic cancer identify recurrent PAK4 amplification. Cancer Biol Ther. 2008;7:1793–1802. doi: 10.4161/cbt.7.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimmelman AC, et al. Genomic alterations link Rho family of GTPases to the highly invasive phenotype of pancreas cancer. Proc Natl Acad Sci USA. 2008;105:19372–19377. doi: 10.1073/pnas.0809966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu W, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15:559–565. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colaluca IN, et al. NUMB controls p53 tumour suppressor activity. Nature. 2008;451:76–80. doi: 10.1038/nature06412. [DOI] [PubMed] [Google Scholar]
- 43.Veeriah S, et al. Somatic mutations of the Parkinson's disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat Genet. 2010;42:77–82. doi: 10.1038/ng.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao X, et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 2004;64:3060–3071. doi: 10.1158/0008-5472.can-03-3308. [DOI] [PubMed] [Google Scholar]
- 45.Leary RJ, et al. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci USA. 2008;105:16224–16229. doi: 10.1073/pnas.0808041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mullighan CG, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loo LW, et al. Array comparative genomic hybridization analysis of genomic alterations in breast cancer subtypes. Cancer Res. 2004;64:8541–8549. doi: 10.1158/0008-5472.CAN-04-1992. [DOI] [PubMed] [Google Scholar]
- 49.Rabinovitch PS, Longton G, Blount PL, Levine DS, Reid BJ. Predictors of progression in Barrett's esophagus III: Baseline flow cytometric variables. Am J Gastroenterol. 2001;96:3071–3083. doi: 10.1111/j.1572-0241.2001.05261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.