Abstract
Anaplastic lymphoma kinase (ALK), physiologically expressed only by certain neural cells, becomes highly oncogenic, when aberrantly expressed in nonneural tissues as a fusion protein with nucleophosphin (NPM) and other partners. The reason why NPM-ALK succeeds in transforming specifically CD4+ T lymphocytes remains unknown. The IL-2R common γ-chain (IL-2Rγ) is shared by receptors for several cytokines that play key roles in the maturation and growth of normal CD4+ T lymphocytes and other immune cells. We show that IL-2Rγ expression is inhibited in T-cell lymphoma cells expressing NPM-ALK kinase as a result of DNA methylation of the IL-2Rγ gene promoter. IL-2Rγ promoter methylation is induced in malignant T cells by NPM-ALK. NPM-ALK acts through STAT3, a transcription factor that binds to the IL-2Rγ gene promoter and enhances binding of DNA methyltransferases (DNMTs) to the promoter. In addition, STAT3 suppresses expression of miR-21, which selectively inhibits DNMT1 mRNA expression. Reconstitution of IL-2Rγ expression leads to loss of the NPM-ALK protein and, consequently, apoptotic cell death of the lymphoma cells. These results demonstrate that the oncogenic tyrosine kinase NPM-ALK induces epigenetic silencing of the IL-2Rγ gene and that IL-2Rγ acts as a tumor suppressor by reciprocally inhibiting expression of NPM-ALK.
Anaplastic large cell lymphoma tyrosine kinase (ALK) is physiologically expressed only in certain immature neuronal cells (1). However, aberrant expression of ALK has been identified in a number of histologically diverse malignancies including T- and B-cell lymphomas, inflammatory myofibroblastic tumors, nonsmall cell lung carcinomas, and familial and sporadic neuroblastomas (1–3). Among these various neoplasms, mechanisms of ALK-induced malignant cell transformation have been characterized in the greatest detail in T-cell lymphoma (ALK+ TCL). The expression of ALK results from chromosomal translocations involving the ALK gene and several different partners in the affected CD4+ T lymphocytes, by far the most frequent partner being the nucleophosmin (NPM) gene (4). The NPM-ALK chimeric protein is constitutively expressed and activated through autophosphorylation (5, 6). It is highly oncogenic (2, 7–9,). NPM-ALK acts by activating a number of signal transduction proteins, including STAT3 (1–3, 10). The continuous activity of these signal transmitters leads to the persistent modulation of expression of a number of genes, the protein products of which govern key cell functions such as cell proliferation, apoptosis, evasion of the immune response (11, 12), and preservation of NPM-ALK expression (3, 13). However, the reason why NPM-ALK specifically transforms CD4+ T lymphocytes remains unknown.
Several cytokines that are critical for normal maturation and function of CD4+ T lymphocytes (IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21) signal through receptors that share the common γ (IL-2Rγ)-chain (14, 15). In addition to the common γ-chain, the receptor for IL-2, the prototypical cytokine of the group, contains a second signal-transduction component, the β-chain, and, in the case of the high-affinity receptor, an IL-2–specific, nonsignaling α-chain. The IL-15 receptor is very similar to the IL-2 receptor by being composed of the same signal transducing γ- and β-chains combined with a third, IL-15–specific, nontransducing α-chain. The receptors for the remaining cytokines of this group contain the IL-2Rγ and one additional cytokine-specific, signal transducing chain. Expression of IL-2Rγ is essential to the proper function of the entire immune system, shown by the development of severe combined immunodeficiency in the IL-2Rγ absence in both humans and mice (16–18).
Transcriptional silencing through DNA methylation of gene promoter and enhancer regions is a common epigenetic phenomenon in malignant cells (19, 20). The silencing affects a diverse group of tumor suppressor genes, protein products of which regulate cell-cycle progression, signal transduction, DNA repair, oncogene expression, and other key features of neoplastic cells (3). It is directly mediated by three members of the DNA methyltransferase (DNMT) family, DNMT1, DNMT3a, and DNMT3b, which induce and maintain methylation of CpG dinucleotides. The silencing can be reversed by treatment with DNMT inhibitors such as 5-aza-2-deoxy-cytidine (ADC).
Here we report that the IL-2Rγ gene is silenced in ALK+ TCL due to methylation of its promoter. The silencing of the IL-2Rγ gene is induced by NPM-ALK at least in part via activation of STAT3, which binds to the IL-2Rγ promoter and supports binding of members of the DNMT family, leading to methylation of the promoter DNA. In addition, STAT3 enhances DNMT1 expression by suppressing expression of miR-21, which targets DNMT1 mRNA. Reconstitution of IL-2Rγ expression leads to inhibition of NPM-ALK expression by promoting proteosomal degradation of the NPM-ALK protein. Our results identify epigenetic gene silencing as a unique mechanism regulating IL-2Rγ expression. In addition, they show that IL-2Rγ is a unique tumor suppressor and, finally, provide insight into the mechanism by which the ectopically expressed NPM-ALK tyrosine kinase affects neoplastic transformation of CD4+ T lymphocytes.
Results
ALK+ TCL Cells Lack Expression of IL-2Rγ.
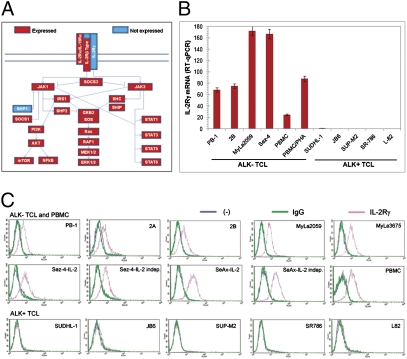
To identify potential suppressor genes that may be critical for NPM-ALK–mediated malignant cell transformation, we examined gene expression patterns in the ALK+ TCL-derived cell line SUDHL-1 using DNA oligonucleotide array-based genome-scale gene expression profiling. An important molecule not expressed by these cells was IL-2Rγ. Its absence was highly selective, because IL-2Rβ and IL-2Rα genes, and the receptors for all other IL-2-type cytokines except the IL-7 receptor-specific chain, were expressed (Fig. 1A). Furthermore, all proteins from the PI3K/AKT, MEK/ERK, and Jak/STAT signaling pathways known to be activated in normal CD4+ T cells by the IL-2Rγ–containing cytokine receptors were also expressed. The only exception was the SHP-1 phosphatase, the negative regulator of cell signaling previously shown to promote NPM-ALK degradation (21).
Fig. 1.
Lack of IL-2Rγ expression in NPM1/ALK-expressing (ALK+) T-cell lymphoma (TCL) cells. (A) The presence (red bar) or absence (blue bar) of expression of IL-2Rγ signaling pathway genes detected in the ALK+ TCL-derived SUDHL-1 cell line on the basis of results from genome-scale DNA oligonucleotide array analysis of total cellular RNA. (B) IL-2Rγ mRNA levels in ALK+ TCL cell lines and ALK− TCL cell lines detected by qRT-PCR. (C) IL-2Rγ protein expression in ALK+ TCL and ALK− TCL cell lines and peripheral blood mononuclear cells (PBMC), detected by immunostaining followed by flow cytometry.
To determine the frequency and specificity of loss of IL-2Rγ expression in ALK+ TCL, we used quantitative RT-PCR (RT-qPCR) to compare mRNA levels in five different ALK+ TCL-derived cell lines and four ALK− TCL-derived lines, with resting as well as mitogen-stimulated peripheral blood mononuclear cells (PBMC and PBMC/phytohaemagglutinin, respectively) serving as positive controls. Whereas all ALK− TCL cell lines strongly expressed IL-2Rγ, none of the ALK+ TCL lines showed appreciable levels of the mRNA (Fig. 1B). Furthermore, whereas IL-2Rγ protein was present in all nine ALK− TCL lines examined and the control PBMC, as determined by flow cytometry, it was not found in any of the ALK+ TCL cell lines (Fig. 1C).
Methylation of the IL-2Rγ Promoter in ALK+ TCL Cells.
To determine whether the IL-2Rγ gene is among the genes that are epigenetically silenced in ALK+ TCL (13, 22), we performed global-scale gene expression profiling of SUDHL-1 cells treated for 24, 48, and 72 h with the DNA methyltransferase inhibitor ADC or treated with the drug vehicle alone. As shown in SI Materials and Methods (Fig. S1), ADC treatment induced a marked, time-dependent increase in IL-2Rγ mRNA expression. The gradual ADC-induced increase in IL-2Rγ expression was confirmed at the mRNA level by RT-qPCR (Fig. 2A) and at the protein level by flow cytometry (Fig. 2B).
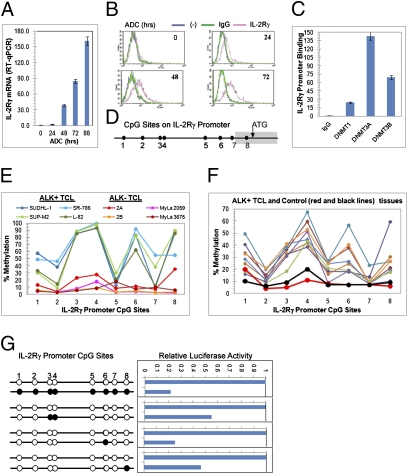
Fig. 2.
Lack of IL-2Rγ expression in ALK+ TCL cells results from methylation of the IL-2Rγ gene promoter. (A) Changes in IL-2Rγ mRNA expression in SUDHL-1 cells exposed to ADC for the depicted periods of time and detected by qRT-PCR. (B) Kinetic of the ADC-induced changes in IL-2Rγ protein expression in SUDHL-1 detected by flow cytometry. (C) Association of DNA methyltransferases (DNMT1, DNMT3a, and DNMT3b) with the IL-2Rγ gene promoter detected by ChIP assay. (D) Schematic map of the CpG sites in the IL-2Rγ gene promoter. (E) Methylation status of IL-2Rγ gene promoter in ALK+ TCL and control ALK− TCL cell lines determined by pyrosequence analysis of the bisulfate-converted DNA. (F) Methylation status of IL-2Rγ gene promoter in ALK+ TCL and control reactive lymphoid tissues determined by pyrosequence analysis of the bisulfate-converted DNA. (G) Impact of DNA methylation on activity of IL-2Rγ gene promoter. (Left) Patterns of DNA methylation introduced into the IL-2Rγ promoter (open circles depict the unmethylated and solid circles the methylated CpG sites). (Right) Impact of depicted CpG methylation status on the promoter's activity as detected by the luciferase gene reporter assay. Activity of the unmethylated IL-2Rγ promoter, examined in parallel, served as a reference.
Because members of the DNMT family are known to be directly responsible for DNA methylation, we investigated whether these proteins associate with the IL-2Rγ gene promoter region. Using a combination of chromatin immunoprecipitation (ChIP) and qPCR, we determined that DNMT1, DNMT3a, and DNMT3b all bind within this region (Fig. 2C). We demonstrated IL-2Rγ gene promoter DNA methylation directly by pyrosequencing of the gene to locate bisulfate-modified CpG sites. The IL-2Rγ gene promoter contains eight CpG sites (Fig. 2D) and most of them—in particular the sites 3, 4, 6, and 8—were found to be highly methylated in the SUDH-L1 and three other ALK+ TCL cell lines (Fig. 2E). In contrast, little or no CpG methylation was observed within the IL-2Rγ promoter in four ALK− TCL lines. DNA from eight ALK+ TCL tissue samples evaluated by pyrosequencing showed essentially the same CpG methylation pattern that we observed in ALK+ TCL cell lines (Fig. 2F) in all samples. There was some quantitative variation, likely due to variation in degree of sample contamination with nonmalignant cells. In contrast, two samples from reactive lymphoid tissue showed no or limited methylation. Given the ability of ADC to induce expression of IL-2Rγ mRNA (Fig. 2A) and protein (Fig. 2B), we evaluated methylation of the IL-2Rγ promoter following treatment of ALK+ TCL cells with ADC. Not surprisingly, we observed a clear, time-dependent decrease in IL-2Rγ promoter methylation at all sites in ADC-treated cells (Fig. S2).
To obtain direct evidence that IL-2Rγ promoter methylation impairs transcriptional activity of the gene, and to evaluate the impact of methylation at selected CpG sites within the promoter, we performed luciferase reporter assays by transient transfection of constructs made with variably methylated IL-2Rγ gene promoter segments driving transcription of a firefly luciferase gene (Fig. 2G, Left). Concomitant methylation of all eight CpG sites within the promoter reduced transcriptional activity by fivefold compared with the unmethylated promoter (Fig. 2G, Right). Targeted methylation of selected promoter CpG sites that are preferentially methylated in ALK+ TCL (3 and 4, 6, or 8) also markedly reduced IL-2Rγ gene promoter activity, indicating that methylation of even a single CpG site within its promoter contributes significantly to IL-2Rγ gene silencing.
NPM-ALK Induces Epigenetic Silencing of the IL-2Rγ Gene.
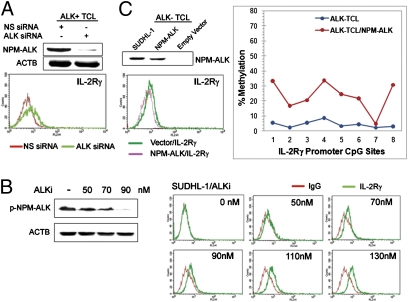
Because silencing of the IL-2Rγ gene is unique to ALK+ TCL (Fig. 1), we evaluated the role of NPM-ALK in silencing of the gene. As shown in Fig. 3A, siRNA-mediated NPM-ALK depletion consistently led to increased IL-2Rγ expression in the SUDH-L1 cells. In addition, treatment of the cells with a highly selective ALK inhibitor, designated compound 13 (23), not only suppressed NPM/ALK autophosphorylation in a dose-dependent fashion, but also proportionately induced expression of the IL-2Rγ protein (Fig. 3B, Left and Right, respectively). To provide yet another piece of evidence that NPM-ALK plays a key role in IL-2Rγ gene silencing, we transfected an NPM-ALK gene into cells of the ALK− TCL cell line 2B. Expression of NPM-ALK in these cells led to both depressed levels of NPM-ALK and IL-2Rγ protein (Fig. 3C, Left) and increased level of methylation of the IL-2Rγ gene promoter (Fig. 3C, Right).
Fig. 3.
NPM-ALK promotes epigenetic silencing of the IL-2Rγ gene. (A) Effect of the siRNA-mediated NPM-ALK depletion on expression of IL-2Rγ. (B) Dose-dependent effect of small-molecule ALK inhibitor on IL-2Rγ expression. (C) Effect of the enforced NPM-ALK expression on expression of IL-2Rγ protein and methylation status of the IL-2Rγ gene promoter.
NPM-ALK Induces IL-2Rγ Gene Silencing Through STAT3.
Given accumulating evidence that STAT3 is the main effector of NPM-ALK–mediated oncogenesis (3) capable of promoting epigenetic gene silencing in malignant T cells (24, 25), we next looked for possible involvement of that transcription factor in IL-2Rγ gene silencing. Sequence analysis of the IL-2Rγ promoter region identified two potential STAT3 binding sites containing the canonical TT and AA dinucleotides (Fig. 4A). Using electrophoretic mobility shift assays (EMSA), we observed binding of proteins in cell extracts from all five ALK+ TCL cell lines to the biotin-labeled 23-mer DNA oligonucleotides corresponding to the STAT3 binding sites (Fig. 4B). The binding was specific, as demonstrated by its abrogation by either addition of unlabeled probe or selective mutation of the TT and AA dimers within the probe sequences (Fig. S3). Identity of STAT3 as the protein bound was firmly established using a “supershift” EMSA assay with a STAT3-specific antibody (Fig. 4B).
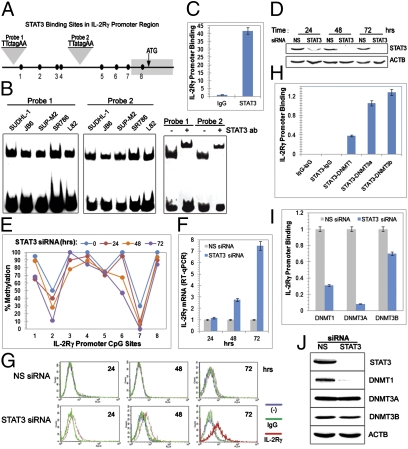
Fig. 4.
STAT3-mediated inhibition of IL-2Rγ gene expression. (A) Schematic map of the STAT3 binding sites in the IL-2Rγ gene promoter. (B) Binding of STAT3 to the IL-2Rγ gene promoter in vitro detected by EMSA. (C) Binding of STAT3 to the IL-2Rγ gene promoter in vivo detected by ChIP. (D) Efficiency of siRNA-mediated STAT3 depletion. (E) STAT3 depletion-induced changes in methylation of the IL-2Rγ gene promoter. (F) Kinetics of the STAT3 siRNA-induced expression of IL-2Rγ mRNA detected by quantitative RT-PCR. (G) Kinetics of STAT3 siRNA-induced IL-2Rγ protein expression detected by flow cytometry. (H) Association of STAT3 with members of the DNMT family at the IL-2Rγ gene promoter as detected by two-step ChIP assay. (I) Effect of STAT3 depletion on DNMT binding to the IL-2Rγ gene promoter detected by ChIP assay. (J) Effect of STAT3 depletion on DNMT protein expression detected by Western blotting.
To demonstrate that STAT3 binds to the IL-2Rγ promoter in vivo as well, we performed ChIP assays using an antibody to STAT3 and an antibody to murine IgG as a control and PCR primers that amplify a promoter segment containing both STAT3 binding sites. The presence of STAT3 in promoter chromatin was demonstrated by precipitation of >40-fold more promoter DNA using the STAT3-specific antibody than with the nonspecific control antibody (Fig. 4C). In addition, depletion of STAT3 protein by transfection of cells with siRNA (Fig. 4D) led to time-dependent demethylation of the IL-2Rγ promoter (Fig. 4E) with concomitant gradual induction of expression of IL-2Rγ mRNA demonstrated by standard (Fig. S4) and quantitative RT-PCR (Fig. 4F) and IL-2Rγ protein detected by flow cytometry (Fig. 4G). To determine whether the STAT3 effect on IL-2Rγ expression was cell replication dependent, we treated ALK+ TCL cells with an mTORC1 inhibitor rapamycin. Rapamycin decreased the percentage of cells in the S and G2 phases by ∼50% (Fig. S5A). Simultaneous treatment of the cells with STAT3 siRNA and rapamycin not only depleted STAT3 and inhibited phosphorylation of mTORC1 target p70SK (Fig. S5B), but also diminished IL-2Rγ expression on both mRNA (Fig. S5C) and protein (Fig. S5D) levels proportionately to the degree of cell-cycle inhibition compared with the cells treated with STAT3 siRNA alone. Because activity of the DNA methylation enzyme DNMT1 is also largely replication dependent, we examined the effect of its depletion on IL-2Rγ expression. Not surprisingly, cell treatment with DNMT1 siRNA led to induction of IL-2Rγ expression at both mRNA (Fig. S6A) and protein (Fig. S6B) levels.
Because not only DNMT1 but also other members of the DNMT family are directly responsible for DNA methylation (19, 20) and associate with IL-2Rγ promoter (Fig. 2C), we explored next the exact relationship between STAT3’s repressive effects and DNMT expression and binding at the IL-2Rγ gene promoter. Two-step ChIP (“re-ChIP”) assays indicated a physical association of STAT3 protein with all three key members of the DNMT family, DNMT1, DNMT3a, and DNMT3b, at the promoter (Fig. 4H). Binding of all three DNMTs, especially DNMT3a, to the IL-2Rγ promoter was greatly diminished following transfection of cells with STAT3 siRNA (Fig. 4I), indicating that STAT3 is required for effective DNMT binding to the promoter. Diminished binding by DNMT1 is due at least in part to the dependence of DNMT1 gene expression on STAT3 as reported by us previously (25) and shown here at both mRNA (Fig. S7) and protein (Fig. 4J) levels. However, STAT3 depletion did not affect expression of DNMT3a and DNMT3b proteins, indicating that STAT3 exerts its effects in the case of these DNMTs solely by enhancing their binding to the IL-2Rγ gene promoter.
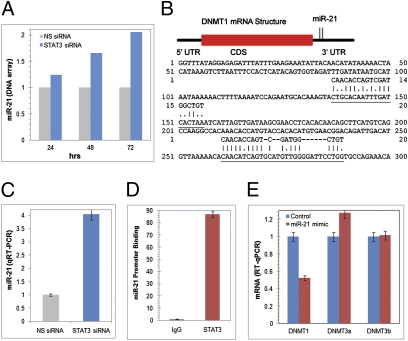
STAT3 Enhances DNMT1 Expression by Inhibiting Expression of miR-21.
While examining the time-dependent effect of STAT3 depletion on global-scale gene expression in the ALK+ TCL SUDH-L1 cells, we observed that the gene for miR-21 is suppressed by STAT3 (Fig. 5A). Sequence analysis of the DNMT1 mRNA identified two putative binding sites for miR-21 in its 3′-UTR (Fig. 5B). These two pieces of information suggest that STAT3 could inhibit expression of the miR-21 gene and that miR-21 could modulate DNMT1 expression. To evaluate this possibility, we first performed miR-21–specific RT-qPCR on RNA isolated from SUDHL-1 cells transfected with either STAT3 siRNA or control nonspecific siRNA. Depletion of STAT3 led to markedly increased miR-21 expression (Fig. 5C). Furthermore, STAT3 associated with miR-21 promoter (Fig. 5D), indicating that STAT3 inhibits miR-21 transcription. Importantly, transduction of SUDHL-1 cells with a miR-21 mimicker led to depletion of DNMT1 mRNA, but not DNMT3a or DNMT3b mRNA (Fig. 5D), indicating that miR-21 selectively targets DNMT1 expression.
Fig. 5.
STAT3 inhibits miR-21 expression and miR-21 selectively inhibits DNMT1 RNA expression. (A) Kinetics of the STAT3 siRNA-induced miR-21 expression detected by the DNA oligonucleotide array. The data are depicted as fold increase in the miR-21 mRNA expression in the STAT3 siRNA-treated compared with the control siRNA-treated cells. (B) Schematic representation of the miR-21 binding sites in 3′-UTR of DNMT1 mRNA and the sequence complementarity of miR-21 and the DNMT1 3′-UTR target sequence. (C) STAT3 siRNA-induced miR-21 expression detected by qRT-PCR. (D) Binding of STAT3 to the miR-21 promoter in vivo detected by ChIP. (E) Effect of miR-21 on expression of members of the DNMT family detected by RT-qPCR.
Expression of IL-2Rγ Results in Inhibition of NPM-ALK Expression.
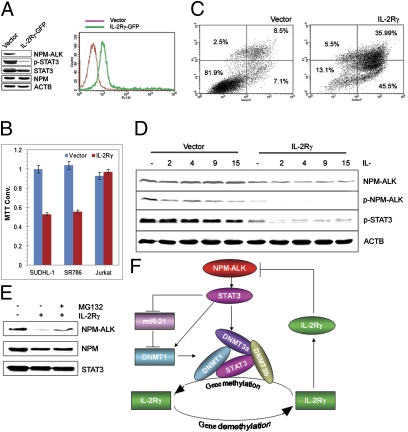
To determine the effect of IL-2Rγ expression on the ALK+ TCL cells, we transfected SUDHL-1 cells with the vector containing the IL-2Rγ–GFP construct (Fig. 6A). Strikingly, IL-2Rγ expressed at the concentration matching that seen in ALK− TCL cells (Fig. 6A, Right and Fig. 1C, respectively) led to the loss of the NPM-ALK expression and dephosphorylation of the crucial target of the kinase, STAT3 (Fig. 6A, Left). Expression of total STAT3, wild-type NPM, and actin was not affected, indicating the selective nature of inhibition of the NPM-ALK expression. Not surprisingly given the oncogenic role of NPM-ALK, IL-2Rγ expression led as well to inhibition of cell growth of the two ALK+ TCL cell lines but not the control ALK− TCL Jurkat cells as demonstrated in the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide enzymatic conversion assay (Fig. 6B). This failure to thrive of the IL-2Rγ–expressing cells was primarily due to apoptotic cell death as shown in the Annexin V cell-surface expression detection assay (Fig. 6C).
Fig. 6.
IL-2Rγ inhibits NPM-ALK expression and induces apoptosis of ALK+ TCL cells. (A) Transfection efficiency (Right) and impact (Left) of the transfected IL-2Rγ on the expression of NPM-ALK and the other depicted proteins detected by flow cytometry and Western blotting, respectively. (B) Effect of transfected IL-2Rγ on growth of the ALK+ TCL (SUDH-L1 and SR786) and control ALK− TCL (Jurkat) cells. (C) Effect of transfected IL-2Rγ on the apoptotic rate of the ALK+ TCL (SUDHL-1) cells. (D) Effect of IL-2Rγ–signaling cytokines on NPM-ALK expression in the vector or IL-2Rγ–transfected SUDHL-1 cells. (E) Effect of proteosome inhibitor MG132 on NPM-ALK expression in the IL-2Rγ–transfected SUDHL-1 cells. (F) Schematic diagram of NPM-ALK and IL-2Rγ expression regulation loop. STAT3 activated by the oncogenic NPM-ALK tyrosine kinase induces DNMT1 expression by activating DNMT1 gene transcription (25) and inhibition of miR-21 expression. STAT3, DNMT1, DNMT3a, and DNMT3b bind to the IL-2Rγ gene promoter and foster the promoter's silencing. Expression of IL-2Rγ protein induced by demethylation of the IL-2Rγ gene inhibits NPM-ALK expression.
To determine whether the IL-2Rγ–signaling cytokines affect the IL-2Rγ–mediated inhibition of NPM-ALK expression, SUDH-L1 cells transfected with either IL-2Rγ or vector were stimulated with four different IL-2Rγ–signaling cytokines or medium (Fig. 6D). Whereas expression of IL-2Rγ alone inhibited NPM-ALK expression and, consequently, signaling, possibly due to some degree of endogeneous secretion of the IL-2Rγ–signaling cytokines by the ALK+ TCL cells, all four exogenous IL-2Rγ–signaling cytokines further increased the inhibition of NPM-ALK expression and signaling. To determine whether IL-2Rγ affects NPM-ALK expression at the mRNA or protein level, we first examined by RT-qPCR the NPM-ALK mRNA concentration in SUDH-L1 cells transfected with IL-2Rγ or vector. However, no major difference was observed (Fig. S8). In contrast, cotreatment of the IL-2Rγ–transfected cells with a proteosome inhibitor MG132 largely rescued the NPM-ALK protein expression, indicating that IL-2Rγ inhibits expression of the NPM-ALK protein by promoting its degradation.
Discussion
Results from the experiments described here demonstrate that ALK+ TCL cells universally lack expression of IL-2Rγ mRNA and protein due to epigenetic silencing of the IL-2Rγ gene. As schematically depicted in Fig. 6F, IL-2Rγ gene silencing is induced by NPM-ALK through activation of STAT3. STAT3 affects IL-2Rγ expression at several levels. It acts as a transcriptional inhibitor by enhancing binding of key members of the DNMT family to the IL-2Rγ gene promoter. It also up-regulates expression of the DNMT1 gene both directly, by enhancing transcription of the gene, as we showed previously (25), and indirectly, by inhibiting expression of miR-21, which suppresses expression of the DNMT1 mRNA. Finally, IL-2Rγ acts as a tumor suppressor, selectively reducing levels of the NPM-ALK oncoprotein, leading to impaired growth and viability of ALK+ TCL cells. Our results provide insight into the mechanism by which ALK transforms CD4+ T lymphocytes and provide a rationale for therapeutic targeting of NPM-ALK, STAT3, and DNMT for ALK+ TCL treatment. Finally, they may contribute to a better understanding of the regulation of expression of IL-2Rγ and other key immune molecules in the normal immune response and, possibly, some immunodeficiencies.
It is now well established that ALK+ TCL originates from normal CD4+ T lymphocytes (1–3). However, why and how the oncogenic ALK enzyme, which is expressed in its native, nononcogenic form exclusively in immature neural cells, is able to neoplastically transform CD4+ T cells remain unknown. Physiologically, CD4+ T lymphocytes act as helper cells for B lymphocytes, CD8+ effector T cells, and other immune cells, and as such are critical for normal functioning of the entire adaptive immune system. They act at least in part by releasing and responding to various cytokines, in particular the cytokines that signal through receptors containing the common IL-2Rγ chain (14, 15). Consequently, expression of IL-2Rγ is critical to immune system function, and its loss through mutation of the IL-2Rγ gene locus leads to severe combined immunodeficiency in affected patients and in genetically engineered mice. It is therefore surprising that CD4+ T-cell–derived ALK+ TCL cells not only do not require IL-2Rγ expression but also, on the contrary, survive only when the IL-2Rγ gene is silenced. Several studies have demonstrated that constitutively expressed and self-activated NPM-ALK induces chronic activation of key intracellular signal transduction pathways in CD4+ T lymphocytes (3, 26). Of note, the main NPM-ALK–activated cell signaling pathways are the same as the ones activated by IL-2–type cytokines, including the PI3K/AKT, MEK/ERK, STAT3, and STAT5B pathways. This finding suggests that NPM/-ALK transforms CD4+ T cells by rendering them independent of external stimuli provided by the IL-2Rγ–signaling cytokines through epigenetic silencing of the IL-2Rγ gene and, at the same time, by constitutively activating intracellular signaling pathways normally regulated by these cytokines. Recent reports indicate that NPM-ALK also inhibits signaling from another key activator of normal CD4+ T cells, the T-cell antigen receptor, by epigenetically silencing the gene of at least one critical member of the TCR signaling pathway, ZAP70 (27). However, the exact mechanism of the silencing as well as the consequences of ZAP70 reexpression remain to be examined.
It is also remarkable that, in addition to making ALK-transformed CD4+ T lymphocytes “deaf” to cytokine signaling, IL-2Rγ gene silencing leads to uninterrupted expression of NPM-ALK. This finding identifies IL-2Rγ as a bona fide tumor suppressor by placing it within a “double-negative” feedback loop in which NPM-ALK succeeds in being persistently expressed by inhibiting expression of its inhibitor, IL-2Rγ (Fig. 6F). This unique regulatory mechanism complements and markedly expands the interactions between NPM-ALK and STAT5a (13) and SHP-1 (24, 21), revealing a highly complex mechanism underlying cell transformation by NPM-ALK, aimed at securing the protein's own unperturbed expression (Fig. 6F). Future studies should address whether similar mechanisms for self-preservation apply to other oncogenic forms of ALK and, perhaps more importantly, to oncogenic kinases in general. In principle, there is no reason to suspect that NPM-ALK would be unique in this regard and, therefore, in all likelihood other oncogenic kinases are also involved in the epigenetic silencing of tumor suppressor genes, possibly also by activating STAT3.
Our observation that STAT3 promotes DNMT1 expression after being activated by NPM-ALK illuminates another facet of NPM-ALK–mediated cell transformation, uncovering another link between a kinase and epigenetic gene silencing. We reported previously (25) that STAT3 directly activates the DNMT1 gene. In this study we show that STAT3 also enhances DNMT1 expression indirectly in ALK+ TCL cells, by inhibiting expression of miR-21, a selective suppressor of DNMT1 mRNA (Fig. 5E). Recent studies have found that two other miRs affect DNMT expression. Members of the miR-29, in particular miR-29b, inhibit expression of DNMT3a and DNMT3b in lung cancer (28) and leukemic cells (29) by binding to the 3′-UTR of their mRNAs. miR-29b also depresses DNMT1 expression indirectly by interfering with expression of Sp-1, a transcription factor involved in DNMT1 gene activation in mice (30). In contrast, in the mouse model, the miR-290 cluster plays an opposite role, up-regulating the expression of all three DNMTs by inhibiting expression of the retinoblastoma-like 2 (RBL2) protein, an inhibitor of DNMT gene expression (31). Our observation that expression of miR-21 under the influence of STAT3 negatively regulates DNMT1 mRNA expression not only identifies miR-21 as a direct and selective posttranscriptional DNMT1 inhibitor but also establishes a unique functional link between a kinase and a kinase-activated transcription factor in regard to regulating miR expression. It also defines a unique interaction between members of the miR and DNMT families.
Results of the studies reported here have potential therapeutic implications for NPM-ALK–expressing lymphomas and, possibly, other kinase-driven malignancies. Whereas current efforts concentrate on inhibition of ALK enzymatic activity (32–34), as in the breakpoint cluster region/V-abl Abelson murine leukemia viral oncogene homolog 1 model (35), this approach by itself is not curative and is often complicated by development of drug resistance. Suppression of expression of oncogenic kinases in combination with inhibition of their enzymatic activities may provide more therapeutic benefit, possibly effecting elimination of the chimeric kinase and, as a result, eradication of the malignant cells. DNMT inhibitors such as ADC, which have already proven effective in treating certain myeloid malignancies (36), might offer a complementary therapeutic approach by directly inducing expression of the epigenetically silenced tumor suppressor(s) that might target the oncogenic kinase genes. This notion is supported by the ability of NPM-ALK–transformed T cells to express IL-2Rγ following treatment with ADC, which leads to abrogation of NPM-ALK expression and, as a consequence, inhibition of cell growth and induction of apoptotic cell death.
Finally, our model describing a kinase-activated transcription factor capable of fostering epigenetic gene silencing (Fig. 6F) may contribute to a better understanding of T-cell physiology. Whereas many key immune molecules including CD4, CD8, IL-2, and perforin are silenced epigenetically at various stages during the maturation and functional differentiation of normal T lymphocytes (37, 38), the exact mechanisms regulating the underlying gene promoter methylation are still poorly defined. Whether activated STAT3 or a similar transcriptional inhibitor fosters silencing of these genes by facilitating binding of members of the DNMT family to their promoters remains to be evaluated. Moreover, evaluation of IL-2Rγ gene promoter methylation patterns during development of normal immune cells and analysis of methylation patterns associated with immunodeficiency would both be very interesting extensions of the past studies characterizing mutations of the gene (16–18).
Materials and Methods
Detailed technical information is provided in SI Materials and Methods. Most cell lines used in this study have been described previously (13). ALK+ TCL and reactive tissues were from biopsies obtained for diagnostic purposes.
Supplementary Material
Acknowledgments
This work was supported in part by National Cancer Institute Grants R01-CA89194 and R01-CA96856 and the Leukemia and Lymphoma Society.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100319108/-/DCSupplemental.
References
- 1.Li R, Morris SW. Development of anaplastic lymphoma kinase (ALK) small-molecule inhibitors for cancer therapy. Med Res Rev. 2008;28:372–412. doi: 10.1002/med.20109. [DOI] [PubMed] [Google Scholar]
- 2.Chiarle R, Voena C, Ambrogio C, Piva R, Inghirami G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat Rev Cancer. 2008;8:11–23. doi: 10.1038/nrc2291. [DOI] [PubMed] [Google Scholar]
- 3.Wasik MA, et al. Anaplastic lymphoma kinase (ALK)-induced malignancies: Novel mechanisms of cell transformation and potential therapeutic approaches. Semin Oncol. 2009;36(2) Suppl 1:S27–S35. doi: 10.1053/j.seminoncol.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Morris SW, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 5.Shiota M, et al. Hyperphosphorylation of a novel 80 kDa protein-tyrosine kinase similar to Ltk in a human Ki-1 lymphoma cell line, AMS3. Oncogene. 1994;9:1567–1574. [PubMed] [Google Scholar]
- 6.Morris SW, et al. ALK, the chromosome 2 gene locus altered by the t(2;5) in non-Hodgkin's lymphoma, encodes a novel neural receptor tyrosine kinase that is highly related to leukocyte tyrosine kinase (LTK) Oncogene. 1997;14:2175–2188. doi: 10.1038/sj.onc.1201062. [DOI] [PubMed] [Google Scholar]
- 7.Fujimoto J, et al. Characterization of the transforming activity of p80, a hyperphosphorylated protein in a Ki-1 lymphoma cell line with chromosomal translocation t(2;5) Proc Natl Acad Sci USA. 1996;93:4181–4186. doi: 10.1073/pnas.93.9.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bischof D, Pulford K, Mason DY, Morris SW. Role of the nucleophosmin (NPM) portion of the non-Hodgkin's lymphoma-associated NPM-anaplastic lymphoma kinase fusion protein in oncogenesis. Mol Cell Biol. 1997;17:2312–2325. doi: 10.1128/mcb.17.4.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuefer MU, et al. Retrovirus-mediated gene transfer of NPM-ALK causes lymphoid malignancy in mice. Blood. 1997;90:2901–2910. [PubMed] [Google Scholar]
- 10.Zhang Q, et al. Multilevel dysregulation of STAT3 activation in anaplastic lymphoma kinase-positive T/null-cell lymphoma. J Immunol. 2002;168:466–474. doi: 10.4049/jimmunol.168.1.466. [DOI] [PubMed] [Google Scholar]
- 11.Kasprzycka M, Marzec M, Liu X, Zhang Q, Wasik MA. Nucleophosmin/anaplastic lymphoma kinase (NPM/ALK) oncoprotein induces the T regulatory cell phenotype by activating STAT3. Proc Natl Acad Sci USA. 2006;103:9964–9969. doi: 10.1073/pnas.0603507103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marzec M, et al. Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD-L1, B7-H1) Proc Natl Acad Sci USA. 2008;105:20852–20857. doi: 10.1073/pnas.0810958105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Q, Wang HY, Liu X, Wasik MA. STAT5A is epigenetically silenced by the tyrosine kinase NPM1-ALK and acts as a tumor suppressor by reciprocally inhibiting NPM1-ALK expression. Nat Med. 2007;13:1341–1348. doi: 10.1038/nm1659. [DOI] [PubMed] [Google Scholar]
- 14.Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol. 2009;9:480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Overwijk WW, Schluns KS. Functions of gammaC cytokines in immune homeostasis: Current and potential clinical applications. Clin Immunol. 2009;132:153–165. doi: 10.1016/j.clim.2009.03.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noguchi M, et al. Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell. 1993;73:147–157. doi: 10.1016/0092-8674(93)90167-o. [DOI] [PubMed] [Google Scholar]
- 17.DiSanto JP, Müller W, Guy-Grand D, Fischer A, Rajewsky K. Lymphoid development in mice with a targeted deletion of the interleukin 2 receptor gamma chain. Proc Natl Acad Sci USA. 1995;92:377–381. doi: 10.1073/pnas.92.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao X, et al. Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity. 1995;2:223–238. doi: 10.1016/1074-7613(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 19.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Best JD, Carey N. Epigenetic opportunities and challenges in cancer. Drug Discov Today. 2010;15:65–70. doi: 10.1016/j.drudis.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 21.Honorat JF, Ragab A, Lamant L, Delsol G, Ragab-Thomas J. SHP1 tyrosine phosphatase negatively regulates NPM-ALK tyrosine kinase signaling. Blood. 2006;107:4130–4138. doi: 10.1182/blood-2005-06-2421. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Q, et al. Lack of TNFalpha expression protects anaplastic lymphoma kinase-positive T-cell lymphoma (ALK+ TCL) cells from apoptosis. Proc Natl Acad Sci USA. 2009;106:15843–15848. doi: 10.1073/pnas.0907070106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu L, et al. ALK mutants in the kinase domain exhibit altered kinase activity and differential sensitivity to small molecule ALK inhibitors. Biochemistry. 2009;48:3600–3609. doi: 10.1021/bi8020923. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q, et al. STAT3- and DNA methyltransferase 1-mediated epigenetic silencing of SHP-1 tyrosine phosphatase tumor suppressor gene in malignant T lymphocytes. Proc Natl Acad Sci USA. 2005;102:6948–6953. doi: 10.1073/pnas.0501959102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Q, et al. STAT3 induces transcription of the DNA methyltransferase 1 gene (DNMT1) in malignant T lymphocytes. Blood. 2006;108:1058–1064. doi: 10.1182/blood-2005-08-007377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu L, et al. Autocrine release of interleukin-9 promotes Jak3-dependent survival of ALK+ anaplastic large-cell lymphoma cells. Blood. 2006;108:2407–2415. doi: 10.1182/blood-2006-04-020305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ambrogio C, et al. NPM-ALK oncogenic tyrosine kinase controls T-cell identity by transcriptional regulation and epigenetic silencing in lymphoma cells. Cancer Res. 2009;69:8611–8619. doi: 10.1158/0008-5472.CAN-09-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fabbri M, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garzon R, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kishikawa S, Murata T, Kimura H, Shiota K, Yokoyama KK. Regulation of transcription of the Dnmt1 gene by Sp1 and Sp3 zinc finger proteins. Eur J Biochem. 2002;269:2961–2970. doi: 10.1046/j.1432-1033.2002.02972.x. [DOI] [PubMed] [Google Scholar]
- 31.Benetti R, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:268–279. doi: 10.1038/nsmb.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marzec M, et al. Inhibition of ALK enzymatic activity in T-cell lymphoma cells induces apoptosis and suppresses proliferation and STAT3 phosphorylation independently of Jak3. Lab Invest. 2005;85:1544–1554. doi: 10.1038/labinvest.3700348. [DOI] [PubMed] [Google Scholar]
- 33.Wan W, et al. Anaplastic lymphoma kinase activity is essential for the proliferation and survival of anaplastic large-cell lymphoma cells. Blood. 2006;107:1617–1623. doi: 10.1182/blood-2005-08-3254. [DOI] [PubMed] [Google Scholar]
- 34.Galkin AV, et al. Identification of NVP-TAE684, a potent, selective, and efficacious inhibitor of NPM-ALK. Proc Natl Acad Sci USA. 2007;104:270–275. doi: 10.1073/pnas.0609412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hehlmann R, Berger U, Hochhaus A. Chronic myeloid leukemia: A model for oncology. Ann Hematol. 2005;84:487–497. doi: 10.1007/s00277-005-1039-z. [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Manero G. Demethylating agents in myeloid malignancies. Curr Opin Oncol. 2008;20:705–710. doi: 10.1097/CCO.0b013e328313699c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson CB, Makar KW, Shnyreva M, Fitzpatrick DR. DNA methylation and the expanding epigenetics of T cell lineage commitment. Semin Immunol. 2005;17:105–119. doi: 10.1016/j.smim.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 38.Narasimhan S, Falkenberg VR, Khin MM, Rajeevan MS. Determination of quantitative and site-specific DNA methylation of perforin by pyrosequencing. BMC Res Notes. 2009;2:104. doi: 10.1186/1756-0500-2-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.