Abstract
PTPMT1 (PTP localized to the Mitochondrion 1) is a member of the protein tyrosine phosphatase superfamily that is localized exclusively to the mitochondrion. We recently reported that PTPMT1 dephosphorylates phosphatidylglycerol phosphate, an essential intermediate of cardiolipin biosynthesis. To gain further insights into the molecular basis of PTPMT1 function, we determined the crystal structures of the phosphatase domain of PTPMT1. PTPMT1 exhibits a canonical protein tyrosine phosphatase domain fold, resembling many dual-specificity phosphatases such as phosphatase and tensin homolog and vaccinia H1-related phosphatase. We also determined the structure of the catalytically inactive phosphatase in complex with a surrogate substrate, phosphatidylinositol 5-phosphate, which sheds light on the substrate recognition and specificity of PTPMT1. Comparison of the apo and substrate-bound structures of PTPMT1 suggests that it undergoes significant conformational change during catalysis, and we further demonstrated that an evolutionarily conserved EEYE loop is important for its activity.
The protein tyrosine phosphatase (PTP) superfamily features the highly conserved active site motif Cys-X5-Arg (CX5R). This family of catalysts contains more than 100 members in human genome, which play key roles in regulating many cellular signaling pathways (1, 2). Most PTPs dephosphorylate phospho-tyrosine, serine, or threonine residues in protein substrates. However, there are also PTPs known to have nonprotein substrates, including PTEN (phosphatase and tensin homolog) and myotubularins that dephosphorylate phosphoinositides (3, 4). We recently showed that laforin dephosphorylates a phosphorylated form of glycogen (5, 6). In addition, the RNGTT (RNA guanylyltransferase and 5′-phosphatase) phosphatase can remove a phosphate from the 5′-end of nascent mRNA (7, 8).
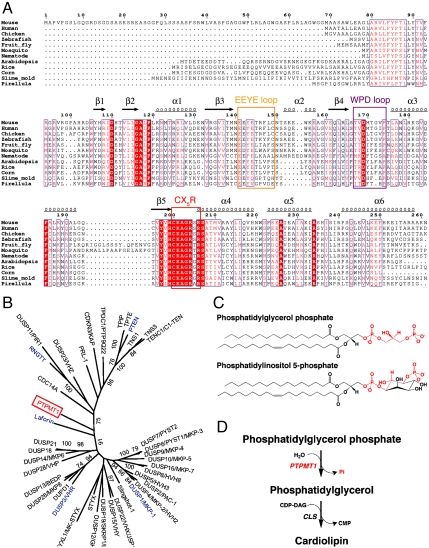
PTPMT1 (PTP localized to the Mitochondrion 1) was initially identified as a PTP that harbors a PTEN-like active site (9). It is highly conserved throughout most major eukaryotic lineages and even in a few bacteria such as Pirellula (Fig. 1A and Fig. S1). As suggested by its name, PTPMT1 is localized in the mitochondrion, and is anchored at the inner mitochondrial membrane with its phosphatase domain in the matrix (10). Disruption of PTPMT1 expression severely interferes with the function of mitochondria, leading to inhibition of mitochondrial respiration, reduction of electron transport chain complexes, and distortion of mitochondrial morphology (11). Knockout of PTPMT1 in mice results in early embryonic death. These observations suggest that PTPMT1 is indispensable for normal embryogenesis and cellular physiology. PTPMT1 belongs to the dual-specificity phosphatase (DSP) family of PTPs, which often have shallow catalytic pockets in their 3D structures (1, 12). In a phylogenetic tree generated using the phosphatase domains of all human DSPs, PTPMT1 is distant from members known to dephosphorylate proteins, such as vaccinia H1-related (VHR) and the MAP kinase phosphatases (MKPs) (Fig. 1B) (13). Indeed, PTPMT1 displays poor activity toward phosphoprotein substrates; instead, it can selectively work on phosphatidylinositol 5-phosphate [PI(5)P] in vitro (9). However, only trace amounts of PI(5)P are found in the mitochondria, and its level is not affected by the loss of PTPMT1, suggesting that PI(5)P is not its endogenous substrate (10). We recently demonstrated that the physiological target of PTPMT1 is phosphatidylglycerol phosphate (PGP) (11), which is structurally remarkably similar to PI(5)P (Fig. 1C).
Fig. 1.
PTPMT1 functions in the cardiolipin biosynthesis pathway. (A) Structure-based sequence alignment of PTPMT1. The EEYE loop, WPD loop, and CX5R residues are highlighted with orange, magenta, and red rectangles, respectively. Highly conserved residues are highlighted in red. Secondary structural elements are shown above the sequence block. Pirellula is a marine bacterium. (B) A phylogenetic tree of human DSPs built using maximum likelihood (ML) method in MEGA5.0. The tree was calculated based on the DSP domain regions only, using Poisson Correction model and partial deletion of gaps. Each branch was tested by 100 bootstrap replicates, and the branches with bootstrap values above 70% were shown. Proteins mentioned in the paper, including PTPMT1, Laforin, RNGTT, PTEN, VHR, and MKP-1 are highlighted in red and blue. (C) Chemical structures of PGP and PI(5)P. The head group of PGP and equivalent regions in PI(5)P are highlighted in red. (D) PTPMT1 dephosphorylates PGP in the cardiolipin biosynthesis pathway (shown in red).
Phosphatidylglycerol (PG), the product of PTPMT1’s activity, is an essential component of pulmonary surfactant, and a precursor for cardiolipin biosynthesis (Fig. 1D and Fig. S2). Cardiolipin is a glycerophospholipid found predominantly in the mitochondrial inner membrane. It has a wide array of reported functions, including maintaining structure of membrane-embedded proteins and trapping protons during oxidative phosphorylation (14). Aberrant cardiolipin metabolism has been linked to a life-threatening inherited disease called Barth syndrome, as well as ischemia/reperfusion injury, heart failure, aging, neurodegeneration, and diabetes (14, 15). In eukaryotic cells, cardiolipin is synthesized in the mitochondria through an elaborate biochemical pathway (16). One step during this process involves the dephosphorylation of PGP to generate PG, before PG condenses with cytidinediphosphate-diacylglycerol (CDP-DAG) to form cardiolipin (Fig. 1D and Fig. S2). Although this event has been well documented by the pioneering work of Kennedy and his colleagues for about 50 years, the identity of the PGP phosphatase in eukaryotes had remained elusive (17). Recently, Osman et al. identified a haloacid dehydrogenase family protein, Gep4 as the PGP phosphatase in Saccharomyces cerevisiae (18); however, animals lack a Gep4 ortholog, yet still produce cardiolipin. We observed an accumulation of PGP in Ptpmt1-null MEF cells, along with reductions of PG and cardiolipin. Accordingly, recombinant PTPMT1 can also specifically dephosphorylate PGP in vitro (11). Therefore, we suggest that PTPMT1 is the long-sought animal PGP phosphatase.
The physiological importance of cardiolipin biosynthesis and the unique nature of PTPMT1’s substrate prompted us to further investigate the catalytic mechanism of PTPMT1. In this study, we determined crystal structures of the phosphatase domain of PTPMT1 in the apo state, and in complex with the surrogate substrate PI(5)P. The information gleaned from our results sheds light on the substrate recognition and activation mechanism of PTPMT1, and provides further knowledge in understanding cardiolipin metabolism.
Results and Discussion
Structure of apo PTPMT1.
We explored the catalytic mechanism of PTPMT1 by protein crystallography. Mouse PTPMT1 gene encodes a 261 amino acid protein [National Center for Biotechnology Information (NCBI) accession no. NP_079852]. However, residues 1–68 are absent in most PTPMT1 orthologs including the human protein (Fig. 1A) (9), and may represent a false prediction or mouse-specific extension. The rest of the amino acid sequence is highly conserved. Residues 69–99 contain a putative transmembrane helix shown to function as a mitochondrial targeting signal (10). We generated and crystallized a fragment encoding the phosphatase domain of PTPMT1 (residues 100–261). Despite extensive trials using available PTP structures as search models, we were not able to solve its structure by molecular replacement. This is likely due to the limited sequence similarity between PTPMT1 and other PTPs. To this end, we determined the structure of PTPMT1 by multiwavelength anomalous dispersion (MAD) using a selenomethionyl crystal (19). The structure was then refined against a native dataset to an R-factor of 22.7% and an Rfree of 24.3% at 1.93 Å resolution (Table S1). The rms deviations from ideal values in bond lengths and angles are 0.004 Å and 0.8°, respectively; and all of the residues are located in the favored and allowed regions of the Ramachandran plot.
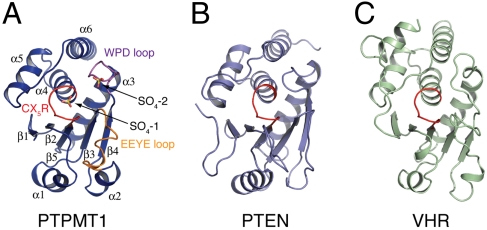
The structure of PTPMT1 reveals a canonical PTP domain fold, consisting of six α-helices (α1–α6) and five β-strands (β1–β5) (Figs. 1A and 2A). The five strands form a β-sheet in the order of β1-β2-β5-β3-β4, and the six helices are distributed on both sides of the sheet with α1–α2 on one side and α3–α6 on the other. Although PTPMT1 displays only limited protein sequence similarity to other PTPs, its overall structure is similar to many DSPs. For example, it can be superposed on PTEN [Protein Data Bank (PDB) ID code 1D5R (20); Fig. 2B and Fig. S3] with an rms deviation (rmsd) of 2.2 Å over 143 Cα atoms, or on VHR [PDB ID code 1VHR (21); Fig. 2C] with a rmsd of 2.3 Å over 146 Cα atoms; even though PTPMT1 shares only 17% and 19% sequence identity with them, respectively.
Fig. 2.
Apo structure of PTPMT1. (A) Ribbon representation of PTPMT1 structure. The CX5R motif, WPD loop, and EEYE loop are highlighted in red, magenta, and orange, respectively. The two sulfates present in the crystal structure are shown as sticks. The secondary structural elements of the molecule are indicated. (B) and (C) Structures of the phosphatase domain of PTEN (B) and VHR (C) are shown in the same orientation as PTPMT1. Their P-loops are highlighted in red.
PTPs contain a number of conserved structural elements that are critical for enzyme activity, including a P-loop formed by the CX5R motif, and a WPD loop featuring a highly conserved aspartate residue (22, 23). The cysteine in the P-loop covalently attacks the phosphate group during catalysis, whereas the WPD aspartate first acts as a general acid to assist the formation of the thiophosphoryl intermediate, and then as a general base to facilitate its hydrolysis. The P-loop is located between β5 and α4 (Fig. 1A, 2A). The active pocket formed by this loop has a depth of around 7 Å, similar to that of VHR and many other DSPs. A sulfate ion from the crystallization solution is bound at this region. The WPD loop contains a highly conserved Asp169. Mutating this residue to alanine completely abolished the activity of PTPMT1 (see below); therefore it likely functions as the general acid/base in PTPMT1. In the apo structure, the WPD loop adopts a wide-open conformation. This leads to the formation of a second anion-binding site that is occupied by another sulfate. A third important region in PTPMT1 is the β3–α2 loop, which we referred to as the “EEYE” loop due to the conserved residues of this region (Fig. 1A and Fig. S1). This loop is part of the “variable insert,” a variable regulatory region found between β3 and β4 in many PTP domains (21). In the apo structure, the EEYE loop has poor electron densities and high temperature factors, suggesting that it is very flexible.
PTPMT1-PI(5)P Complex Structure.
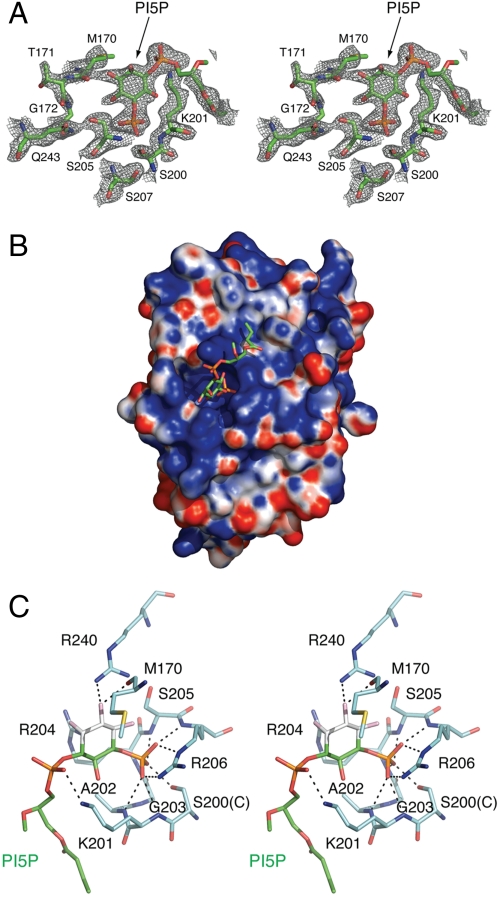
PGP could not readily be made in quantity and quality required for crystallization. Because PI(5)P can serve as a surrogate substrate of PTPMT1 and part of its structure mimics PGP (Fig. 1C), we reasoned that it likely interacts with PTPMT1 in a similar manner to PGP. To this end, we crystallized a catalytic-inactive mutant, Cys200Ser (C200S), of PTPMT1 in the presence of dioctanoyl PI(5)P. The structure was determined by molecular replacement using the apo structure as a search model. Clear density is present for the head group of PI(5)P (Fig. 3A). The diacylglycerol moiety of PI(5)P is less well defined, featured by a series of broken electron densities. As a result, only a portion of the diacylglycerol group is built into the final structural model. This probably reflects the flexibility of the acyl chains in the absence of a membrane context. The structure was further refined to an R-factor of 17.3% and an Rfree of 21.5% at 2.05 Å resolution (Table S1).
Fig. 3.
PI(5)P-bound PTPMT1 structure. (A) Stereo view of a simulated annealing omit map (contoured at 1.0σ) around PI(5)P in the active site of PTPMT1. PI(5)P is omitted to calculate the map. (B) PI(5)P is bound in a positively charged pocket of PTPMT1. PTPMT1 is shown as a surface representation, with saturating blue and red representing +10 and -10 kT/e, respectively. PI(5)P is shown as sticks. (C) A stereo view of PI(5)P interaction. Residues in PTPMT1 that interact with PI(5)P are indicated, and their carbon atoms are colored in cyan. S200 represents the catalytic Cys in the complex structure, which is indicated in parentheses. The carbon atoms in PI(5)P that reflect PGP are colored in green, whereas the carbon atoms that are not present in PGP are colored white. Hydrogen bond interactions are shown as dashed lines.
The head group of PI(5)P is bound in an extensive and conserved positively charged surface pocket of PTPMT1, with its D-5 phosphate group inserted deeply into the active site (Fig. 3B). A network of polar contacts is responsible for coordinating the interaction between PTPMT1 and PI(5)P, which involves three structural elements in PI(5)P: the D-5 phosphate, the D-1 phosphoryl group, and the D-3 hydroxyl group (Fig. 3C). Most of these interactions focus on the D-5 phosphate, the leaving group during dephosphorylation. In particular, the guanidinium group of the CX5R arginine (Arg206) makes parallel interactions with two nonbridge phosphoryl oxygens in the D-5 phosphate, using a hydrogen bond and a salt bridge, respectively. Ser200, which replaces the catalytic cysteine, forms a hydrogen bond with another oxygen in the D-5 phosphate. In addition, five other hydrogen bonds are formed between the backbone amide nitrogen atoms of the CX5R residues and the D-5 phosphate group. This type of bonding pattern between the CX5R motif and the phosphate group is characteristic of PTP superfamily. Besides the interactions seen at the D-5 position, Lys201 interacts with the D-1 phosphoryl group through a hydrogen bond, and both Met170 and Arg240 hydrogen bond with the D-3 hydroxyl group of PI(5)P.
Enlightened by the PTPMT1-PI(5)P complex structure, a picture of a putative PTPMT1-PGP complex can be easily deduced. It is very likely that the head group of PGP will bind to the active site of PTPMT1 in a similar fashion to the lower half of PI(5)P inositol ring shown in Fig. 3C. The phosphomonoester and phosphodiester groups of PGP likely occupy the positions of the D-5 and D-1 phosphates of PI(5)P, respectively; and their interactions with PTPMT1 are expected to echo the bonding pattern between the D-5/D-1 phosphates and PTPMT1. Although PGP lacks functional groups equivalent to the D2, D3, and D4 hydroxyl groups of PI(5)P, it is possible that water molecules may be present in similar positions to bridge hydrogen bond interactions.
A portion of the diacylglycerol moiety present in the structure packs onto a conserved hydrophobic surface patch of PTPMT1 (Fig. 3B), formed by residues including Trp105, Leu118, Leu120, and Leu148. In cells, the majority of the diacylglycerol moiety is expected to insert into the lipid bilayer, where the transmembrane region of PTPMT1 resides, which is not present in the current structure. Further interactions between PTPMT1 and PGP might occur within the mitochondrial membrane. By directly binding to or simply by colocalizing with the diacylglycerol chains, the N-terminal region of PTPMT1 may help to establish an additional level of substrate binding affinity/specificity.
Conformational Change Within PTPMT1.
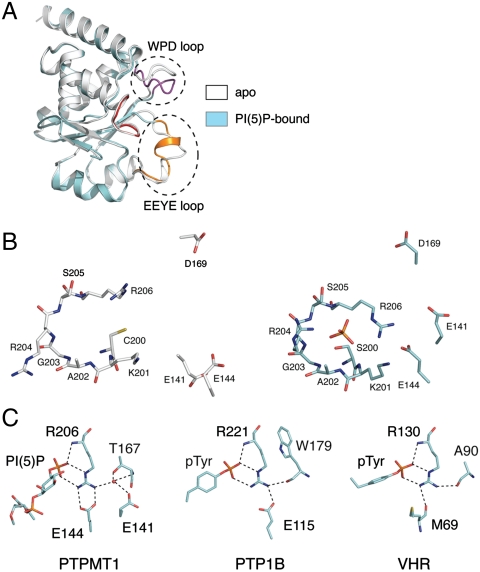
A comparison of the apo and PI(5)P-bound PTPMT1 structures reveals several substantial conformational changes within PTPMT1 upon substrate binding (Fig. 4A). In the PI(5)P-bound structure, the WPD loop swings approximately 15° from the apo conformation toward the active site. Curiously, further movement is still necessary for placing Asp169, the general acid/base residue, into the active pocket, suggesting that the WPD loop may move significantly during its enzymatic cycle (Fig. 4B). Residues in the CX5R motif undergo conformational changes as well. Ser200 adopts a g+ rotamer conformation, in contrast to the t-rotamer conformation seen for Cys200 in the apo structure. This subtle change is necessary to facilitate a hydrogen bond interaction with the phosphate group (Figs. 3C and 4B). Similarly, Arg206 changes its rotamer conformation and forms multiple interactions with the phosphate group (Figs. 3C and 4B). Other residues in the P-loop, including Arg204 and Ser205, also slightly move toward the center of the active site to promote hydrogen bonds formation between their main chain amide groups and the phosphate.
Fig. 4.
Conformational change of PTPMT1 upon substrate binding. (A) Overlay of apo (white) and PI(5)P-bound (cyan) PTPMT1. The CX5R motif, WPD loop, and EEYE loop in PI(5)P-bound PTPMT1 are highlighted in red, magenta, and orange. Dashed circles highlight the conformational changes in the WPD and EEYE loops. (B) Comparison of apo (Left) and PI(5)P-bound PTPMT1 (Right) in the active site region. The carbon atoms of apo and PI(5)P-bound PTPMT1 molecules are colored in white and cyan, respectively. The D-5 phosphate of PI(5)P is shown in the PI(5)P-bound conformation. (C) The CX5R arginine interactions in PTPMT1, PTP1B (PDB ID code 1EEO), and VHR (PDB ID code 1J4X) at their substrate-bound sates. All structures are shown with their CX5R arginines positioned in the same orientation. Hydrogen bond interactions are shown as dashed lines. The red sphere in PTPMT1 represents a water molecule.
The EEYE loop also undergoes significant conformational change. It is refolded in such a way that a helical turn in its C-terminal region unwinds and a new helical turn forms at the N-terminus (Fig. 4A). Consequently, Glu141 and Glu144 in this loop move dramatically closer to the active site (Fig. 4B). Both residues appear important to modulate the conformation of Arg206 (Fig. 4C): Glu144 directly interacts with Arg206 using a bidentate salt bridge, whereas Glu141 indirectly interacts with Arg206 through a water molecule. Arg206 assists catalysis by stabilizing the penta-coordinated transition state during the dephosphorylation reaction in all PTPs. It interacts with the leaving phosphate group by a signature bonding pattern: a salt bridge and a hydrogen bond using its guanidinium group, and another hydrogen bond using its main chain amide nitrogen (Fig. 4C). In many cases, this arginine also interacts with the WPD loop (usually with the residue in the “W” position) to modulate its movement. For example, in PTP1B, a prototypical phospho-tyrosine specific PTP, Arg221 forms a hydrogen bond with the main chain carbonyl group of Trp179 (Fig. 4C) (24, 25). Similarly, Arg130 in VHR forms a hydrogen bond with Ala90, the “W” residue in VHR (26). Although Arg206 in PTPMT1 does not interact with the “W” residue (T167) directly, it does so through a water molecule (Fig. 4C). Due to these critical functions, the conformation of this arginine is almost strictly superimposable among different PTP structures in their active states. Nevertheless, different PTP proteins appear to have evolved different strategies to stabilize the conformation of their CX5R arginines. In PTPMT1, two acidic residues, Glu141 and Glu144 in the EEYE loop indirectly or directly interact with Arg206 (above). PTP1B uses a single conserved glutamate, Glu115, to form a salt bridge with this arginine (Fig. 4C). Notably, Glu115 adopts almost the same position as Glu144 in PTPMT1, suggesting an evolutionary relationship between them. In VHR, Arg130 is anchored by the main chain carbonyl group of Met69 (Fig. 4C). Apparently, PTPMT1 is closer to PTP1B in this functional aspect, even though it belongs to the DSP family of PTPs and more resembles VHR in 3D structure. In a broader context, all these structural elements that function to stabilize this arginine belong to the variable insert region, which differ significantly in length and conformation in different PTP proteins. Although the variable insert regions have undergone substantial transformation during evolution, the function to secure the correct conformation of the active site arginine is “invariable,” reflecting a strong selective pressure exerted by this functional requirement.
Glu141 and Glu144 Are Important for the Activity of PTPMT1.
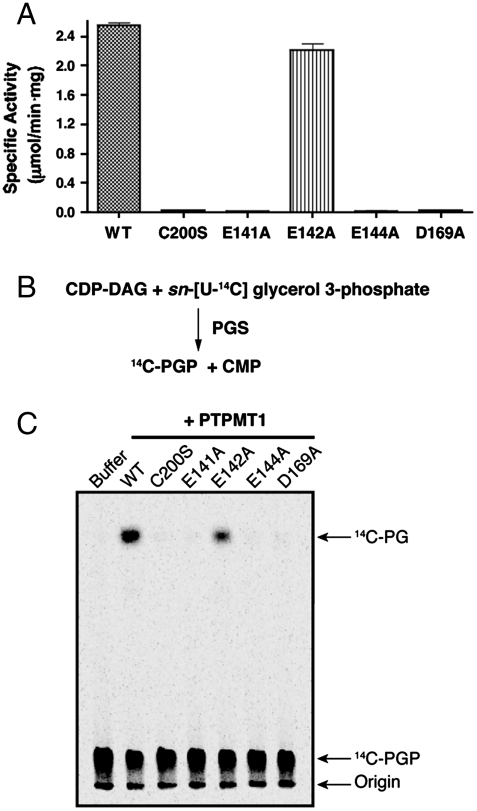
To assess the functional importance of Glu141 and Glu144, we generated single point mutants and tested their phosphatase activities. Both Glu141Ala (E141A) and Glu144Ala (E144A) mutants display a dramatic decrease in activity against the artificial substrate p-nitrophenyl phosphate (pNPP) (Fig. 5A). In fact, their activities are indistinguishable from the activities of the catalytic Cys mutant (C200S) and the general acid/base mutant (D169A). In contrast, mutation of Glu142, which does not make contact with Arg206, has no significant effect on activity.
Fig. 5.
Glu141 and Glu144 are important for the activity of PTPMT1. (A) pNPP assay. Activity was calculated from the change in absorbance at 410 nm. Data are represented as the mean ± SD of triplicate measurements. (B) A diagram of PGP synthesis. (C) Activity of PTPMT1 mutants measured using PGP as a substrate. PGP synthesized above is incubated with the PTPMT1 proteins as indicated. The reaction products are separated by TLC and analyzed by autoradiography.
We further tested the activities of these mutants against PGP, the physiological substrate of PTPMT1. For the PGP phosphatase assay, we first enzymatically synthesized radiolabeled PGP as previously described, by incubating CDP-DAG with sn-[U-14C] glycerol 3-phosphate in the presence of affinity purified recombinant Escherichia coli PGP synthase (Fig. 5B) (27). The resulting 14C-PGP was then separated from the reaction mixture and incubated with wild type or mutant PTPMT1. Production of 14C-PG was examined by using thin layer chromatography (TLC) and autoradiography. Similar to their behavior in the pNPP assay, C200S, D169A, E141A, and E144A mutants failed to dephosphorylate PGP in vitro, whereas D142A retained normal activity (Fig. 5C). These data corroborate our structure-based analysis, and demonstrate that both Glu141 and Glu144 in the EEYE loop are crucial for the catalytic activity of PTPMT1.
In summary, we have determined the crystal structures of the PGP phosphatase PTPMT1, by itself and in complex with a surrogate substrate. The apo structure adopts a catalytically inactive conformation, and we further identified an evolutionally conserved EEYE loop that is essential for the enzyme activation. To fully understand the functional mechanism of this enzyme, it will ultimately be required to characterize its structure and function in the context of a membrane environment. Nonetheless, the present study advances our understanding of the mitochondrial PGP phosphatase PTPMT1 at the structural level, and provides a platform under which the function of PTPMT1 could be studied by mutagenesis and other methods. The intimate link of PTPMT1 and cardiolipin to many disease states has prompted initial efforts in developing PTPMT1-specific compounds (28). In this regard, understanding the structure of this unique enzyme also has significant practical relevance.
Methods
Protein Expression and Purification.
Murine PTPMT169–261 was expressed using a modified pET41a vector (pSJ6) as a GST fusion protein. PTPMT1100–261 was expressed using a modified pET28a vector as a His6-SUMO fusion protein. Plasmids encoding PTPMT1 proteins were expressed in E. coli strain BL21-CodonPlus(DE3)-RIPL (Stratagene). Cultures were grown at 37 °C in LB medium to an OD600 of 0.7 before induction with 0.4 mM IPTG at room temperature for 16 h. Cells were harvested by centrifugation and frozen at -80° C.
GST-PTPMT169–261 was purified by GST affinity chromatography. His6-SUMO-PTPMT1100–261 was purified by Ni-NTA affinity chromatography and digested with the small ubiquitin-like modifier (SUMO)-specific protease ULP1. His6-SUMO and the ULP1 protease were removed by a second Ni-NTA affinity chromatography step. Untagged PTPMT1100–261 was further purified by cation exchange chromatography. More details of protein purification are presented in SI Text.
Crystallization and Structure Determination.
PTPMT1100–261 crystals were grown at 4 °C via sitting-drop vapor diffusion method, using a 1∶1 ratio of protein: reservoir solution containing 21–26% PEG 5000 MME, 100 mM Bis-Tris (pH 5.5), and 100 mM (NH4)2SO4. Crystals grew to full size in several days and were transferred to 28% PEG 5000 MME, 100 mM Bis-Tris (pH 5.5), 200 mM (NH4)2SO4 and 10% ethylene glycol before flash-frozen under liquid nitrogen. The Se-Met derivative crystals used for phase determination were obtained similarly.
To obtain the PTPMT1-PI(5)P complex crystals, diC8 PI(5)P (Avanti Polar Lipids) was added to the PTPMT1100–261 C200S protein to a final concentration of 2 mM. The mixture was allowed to incubate on ice for 2 h before subjected to crystal screen. The crystal was obtained at 16 °C via sitting-drop vapor diffusion using a 1∶1 ratio of protein: reservoir solution containing 18–20% PEG 3350, and 100 mM MES (pH 6.5). Crystals grew to full size in several weeks. For cryo protection, the crystals were transferred to a solution containing 26% PEG 3350, 10% ethylene glycol, 100 mM MES (pH 6.5), and 2 mM diC8 PI(5)P, left in the same solution for 30 min, and then flash-frozen under liquid nitrogen.
The structure of apo PTPMT1 was determined by the multiple-wavelength anomalous diffraction (MAD) method using the data collected from a Se-Met crystal. The structure of the PTPMT1–PI(5)P complex was determined by molecular replacement. More details for structure determination are presented in SI Text.
Phosphatase Assays.
Phosphatase assays using para-nitrophenylphosphate (pNPP) or radiolabeled PGP were carried in assay buffer containing 50 mM sodium acetate, 25 mM bis-Tris, 25 mM Tris, and 2 mM dithiothreitol at pH 5.5. For pNPP assays, reactions were carried out at 37 °C for 10 min as previously described (29). For PGP assays, 14C-labeled PGP was synthesized as previously described (27). Briefly, the reaction was carried out in the presence of 0.1 M Tris (pH 8.0), 0.1% Triton X-100, 0.2 mM CDP-DAG (Avanti Polar Lipids), 0.5 mM glycerol 3-phosphate (Sigma) and 13 μM sn-[U-14C] glycerol 3-phosphate (2 ∼ 4 μCi/μmol, American Radiolabeled Chemicals). The reaction is initiated by the addition affinity purified of recombinant E. coli PGP synthase and 0.1 M MgCl2. After incubation at 37 °C for 2 h, 0.5 mL methanol containing 0.1 N HCl, 2 mL chloroform, and 3 mL of 1 M MgCl2 were added to stop the reaction and separate the radiolabeled PGP. The organic phase was then dried under vacuum and resuspended in assay buffer by water bath sonication, treated with PTPMT1 for 10 min at 37 °C. Reactions were stopped by the addition of 0.5 mL methanol (0.1 N in HCl), 2 mL chloroform, and 3 mL of 1 M MgCl2, followed by lipid extraction and thin layer chromatography (TLC) separation. Details for TLC are presented in SI Text.
Phylogenetic Analysis.
The phylogenetic tree of human DSPs was built by using the region of phosphatase domains only. Protein sequences were aligned by PROMALS3D (30) and adjusted manually. Phylogenetic trees were inferred by using maximum likelihood (ML) and neighbor joining (NJ) methods in MEGA 5.0 (31). Different models of Poisson Correction, Dayhoff matrix, Jones–Taylor–Thornton (JTT) matrix, and JTT+ frequency (F) were used, and they yield similar trees. Support for each interior branch was tested with 100 and 1,000 bootstrap replicates for ML and NJ analyses, respectively.
Supplementary Material
Acknowledgments.
We thank Dr. Carolyn Worby and Dr. Ping Zhang for critically reading the manuscript, and members of the Dixon laboratory for helpful discussions. We thank the Advanced Light Source (beam line 8.2.1) staff of Lawrence Berkeley National Laboratory for beam access and help with data collection. This work was supported by the National Institutes of Health Grants DK18024 and DK18849 (J.E.D.) and HG004164 (G.M.) and a Larry L. Hillblom Foundation postdoctoral fellowship (J. Z.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 3RGO and 3RGQ for apo and PI(5)P-bound PTPMT1, respectively].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109290108/-/DCSupplemental.
References
- 1.Alonso A, et al. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 2.Tonks NK. Protein tyrosine phosphatases: From genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 3.Wishart MJ, et al. PTEN and myotubularin phosphoinositide phosphatases: Bringing bioinformatics to the lab bench. Curr Opin Cell Biol. 2001;13:172–181. doi: 10.1016/s0955-0674(00)00195-2. [DOI] [PubMed] [Google Scholar]
- 4.Maehama T, Taylor GS, Dixon JE. PTEN and myotubularin: Novel phosphoinositide phosphatases. Annu Rev Biochem. 2001;70:247–279. doi: 10.1146/annurev.biochem.70.1.247. [DOI] [PubMed] [Google Scholar]
- 5.Worby CA, Gentry MS, Dixon JE. Laforin, a dual specificity phosphatase that dephosphorylates complex carbohydrates. J Biol Chem. 2006;281:30412–30418. doi: 10.1074/jbc.M606117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tagliabracci VS, et al. Laforin is a glycogen phosphatase, deficiency of which leads to elevated phosphorylation of glycogen in vivo. Proc Natl Acad Sci USA. 2007;104:19262–19266. doi: 10.1073/pnas.0707952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wen Y, Yue Z, Shatkin AJ. Mammalian capping enzyme binds RNA and uses protein tyrosine phosphatase mechanism. Proc Natl Acad Sci USA. 1998;95:12226–12231. doi: 10.1073/pnas.95.21.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shuman S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog Nucleic Acid Res Mol Biol. 2001;66:1–40. doi: 10.1016/s0079-6603(00)66025-7. [DOI] [PubMed] [Google Scholar]
- 9.Pagliarini DJ, Worby CA, Dixon JE. A PTEN-like phosphatase with a novel substrate specificity. J Biol Chem. 2004;279:38590–38596. doi: 10.1074/jbc.M404959200. [DOI] [PubMed] [Google Scholar]
- 10.Pagliarini DJ, et al. Involvement of a mitochondrial phosphatase in the regulation of ATP production and insulin secretion in pancreatic beta Cells. Mol Cell. 2005;19:197–207. doi: 10.1016/j.molcel.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, et al. Mitochondrial phosphatase PTPMT1 is essential for cardiolipin biosynthesis. Cell Metab. 2011;13:690–700. doi: 10.1016/j.cmet.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patterson KI, et al. Dual-specificity phosphatases: Critical regulators with diverse cellular targets. Biochem J. 2009;418:475–489. doi: 10.1042/bj20082234. [DOI] [PubMed] [Google Scholar]
- 13.Gentry MS, Dixon JE, Worby CA. Lafora disease: Insights into neurodegeneration from plant metabolism. Trends Biochem Sci. 2009;34:628–639. doi: 10.1016/j.tibs.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houtkooper RH, Vaz FM. Cardiolipin, the heart of mitochondrial metabolism. Cell Mol Life Sci. 2008;65:2493–2506. doi: 10.1007/s00018-008-8030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chicco AJ, Sparagna GC. Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am J Physiol Cell Physiol. 2007;292:33–44. doi: 10.1152/ajpcell.00243.2006. [DOI] [PubMed] [Google Scholar]
- 16.Voet D, Voet JG. Biochemistry. 3rd Ed. Vol. 1. Hoboken, NJ: Wiley; 2004. [Google Scholar]
- 17.Kiyasu JY, et al. The biosynthesis of phosphatidylglycerol. J Biol Chem. 1963;238:2293–2298. [PubMed] [Google Scholar]
- 18.Osman C, et al. A mitochondrial phosphatase required for cardiolipin biosynthesis: The PGP phosphatase Gep4. EMBO J. 2010;29:1976–1987. doi: 10.1038/emboj.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendrickson WA, Horton JR, LeMaster DM. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): A vehicle for direct determination of three-dimensional structure. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JO, et al. Crystal structure of the PTEN tumor suppressor: Implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 21.Yuvaniyama J, et al. Crystal structure of the dual specificity protein phosphatase VHR. Science. 1996;272:1328–1331. doi: 10.1126/science.272.5266.1328. [DOI] [PubMed] [Google Scholar]
- 22.Zhang ZY. Protein tyrosine phosphatases: Structure and function, substrate specificity, and inhibitor development. Annu Rev Pharmacol Toxicol. 2002;42:209–234. doi: 10.1146/annurev.pharmtox.42.083001.144616. [DOI] [PubMed] [Google Scholar]
- 23.Barford D, Das AK, Egloff MP. The structure and mechanism of protein phosphatases: Insights into catalysis and regulation. Annu Rev Biophys Biomol Struct. 1998;27:133–164. doi: 10.1146/annurev.biophys.27.1.133. [DOI] [PubMed] [Google Scholar]
- 24.Sarmiento M, et al. Structural basis of plasticity in protein tyrosine phosphatase 1B substrate recognition. Biochemistry. 2000;39:8171–8179. doi: 10.1021/bi000319w. [DOI] [PubMed] [Google Scholar]
- 25.Jia Z, et al. Structural basis for phosphotyrosine peptide recognition by protein tyrosine phosphatase 1B. Science. 1995;268:1754–1758. doi: 10.1126/science.7540771. [DOI] [PubMed] [Google Scholar]
- 26.Schumacher MA, et al. Structural basis for the recognition of a bisphosphorylated MAP kinase peptide by human VHR protein phosphatase. Biochemistry. 2002;41:3009–3017. doi: 10.1021/bi015799l. [DOI] [PubMed] [Google Scholar]
- 27.Dowhan W. Phosphatidylglycerophosphate synthase from Escherichia coli. Methods Enzymol. 1992;209:313–321. doi: 10.1016/0076-6879(92)09039-6. [DOI] [PubMed] [Google Scholar]
- 28.Doughty-Shenton D, et al. Pharmacological targeting of the mitochondrial phosphatase PTPMT1. J Pharmacol Exp Ther. 2010;333:584–592. doi: 10.1124/jpet.109.163329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor GS, Maehama T, Dixon JE. Inaugural article: Myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc Natl Acad Sci USA. 2000;97:8910–8915. doi: 10.1073/pnas.160255697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pei J, Kim BH, Grishin NV. PROMALS3D: A tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar S, et al. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.