Abstract
The ORFDB (http://orf.invitrogen.com/) represents an ongoing effort at Invitrogen Corporation to integrate relevant scientific data with an evolving collection of human and mouse Open Reading Frame (ORF) clones (Ultimate™ ORF Clones). The ORFDB serves as a central data warehouse enabling researchers to search the ORF collection through its web portal ORFBrowser, allowing researchers to find the Ultimate™ ORF clones by blast, keyword, GenBank accession, gene symbol, clone ID, Unigene ID, LocusLink ID or through functional relationships by browsing the collection via the Gene Ontology (GO) Browser. As of October 2003, the ORFDB contains 6200 human and 2870 mouse Ultimate™ ORF clones. All Ultimate™ ORF clones have been fully sequenced with high quality, and are matched to public reference protein sequences. In addition, the cloned ORFs have been extensively annotated across six categories: Gene, ORF, Clone Format, Protein, SNP and Genomic links, with the information assembled in a format termed the ORFCard. The ORFCard represents an information repository that documents the sequence quality, alignment with respect to public protein sequences, and the latest publicly available information associated with each human and mouse gene represented in the collection.
INTRODUCTION
Major challenges lie ahead as life science research focuses on efforts to convert genome sequence information resulting from numerous genome projects to functional information for each of the encoded genes. Key initiatives include the expression and characterization of all genome-encoded proteins, the determination of their 3D structures, the characterization of protein localization in vivo, and the elucidation of interactions and pathways that define the molecular architecture of the cell (1). An essential step in tackling these challenges is the generation of a set of high-quality clones representing the Open Reading Frames (ORFs) of each of the annotated and predicted gene transcripts. The preferred clone format enables the flexibility to express the ORF in multiple expression systems for use in subsequent protein expression and functional analyses (1–3).
Recombination-based cloning systems provide the required flexibility. In vivo homologous recombination-based cloning has been described in yeast. Over 6200 yeast ORFs have been cloned using in vivo homologuous recombination, enabling systematic analysis of expressed proteins on a genome-wide basis, the assignment and characterization of biochemical activities, the construction of protein arrays, the identification of interactions and the localization of proteins within cellular compartments (4–6). The Gateway™ recombination-based cloning technology is modeled after site-specific recombination reactions mediating bacteriophage λ lysogeny. DNA segments flanked by the appropriate recombination sequences can be transferred between vector systems using a simple in vitro recombination reaction (7). The utility of this system has been demonstrated through the construction of nearly 12 000 ORF clones representing >60% of the Caenorhabditis elegans genome (8–10). Using the Gateway® technology, transfer of the ORFs into a two-hybrid destination vector downstream of the sequence encoding the activation domain resulted in the AD-ORFeome library that allowed the large-scale mapping of protein–protein interaction using yeast two-hybrid technology (11). This same set of ORF sequences has been transferred into a number of different ORF-tagged expression systems for expression as fusions to maltose-binding protein, hexa-histidine and glutathione-S-transferase in bacterial or yeast systems for large-scale protein production (11), paving the way for large-scale biochemical analysis and protein chip experiments with C.elegans proteins (3).
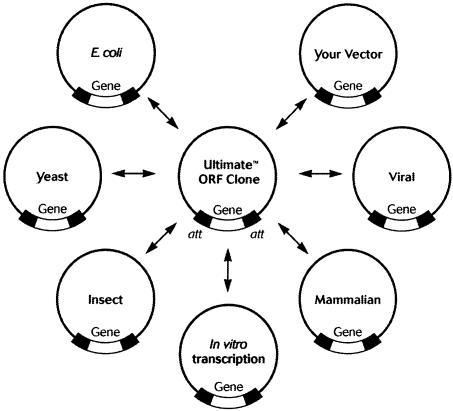
The complete sequencing of the human genome has provided the first complete catalog of annotated and predicted gene transcripts (12) (http://www.ncbi.nlm.nih.gov/genome/seq/HsHome.shtml; http://www.ensembl.org; http://genome.ucsc.edu/cgi-bin/hgGateway). In addition, the Mammalian Gene Collection (MGC) program has generated nearly 26 000 full-length human and mouse cDNA sequences (13) (http://mgc.nci.nih.gov/). The vast number of candidate proteins and cDNA resources generated from the various genome projects, combined with the ongoing MGC effort has created enormous opportunities in basic biological research. However, the resultant cDNA clones are not inherently amenable for direct use in protein expression, production or biochemical characterization studies because they contain variable length 5′ and 3′ untranslated regions and natural stop codons that may interfere with the expression of fusion proteins. Clones comprising only the ORF sequences are better suited for such studies but to date only a limited number of such ORF clones have been made available to address the needs of functional proteomics research (14,15). In response to the need for these essential clones, Invitrogen is engaged in a program to create a high-quality ORF collection (Ultimate™ ORFs) for all known genes and genes with predicted ORFs from human and mouse. The Ultimate™ ORF collection is constructed in the form of Gateway® Entry clones, making them compatible with the recombination-based technology and thus enabling the transfer of each ORF into any expression background. The availability of this critical resource will facilitate analysis of protein activity on a genome scale, protein interaction analysis using a genome-wide two-hybrid approach, systematic protein localization and bioproduction of hundreds of proteins in high-throughput formats (Fig. 1).
Figure 1.
The flexibility of Gateway® technology.
ORF CONSTRUCTION AND PIPELINE
Taking advantage of in-house, full-length clone collections, including full-length verified cDNA clones from the MGC program and full-length cDNA libraries, the collection is being built using cDNA clones as template for PCR amplification of the ORF sequences. Full-length template clones are identified based on the coding region (CDS) defined in the RefSeq database. Candidate clones are arranged in a 96-well format and pairs of ORF-specific primers are designed and synthesized. Each ORF is PCR-amplified from a sequence-verified cDNA template and the PCR products are recombined in vitro into the Gateway® Entry vector using a high-throughput process. The resultant ORF clones are then subjected to full-insert sequence verification.
ORF CLONE FORMAT
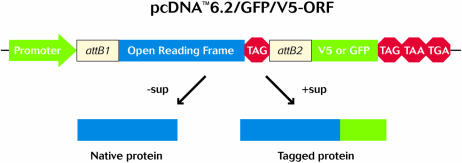
Each ORF sequence is contained within the pENTR221 Gateway® Entry vector (http://www.invitrogen.com/content/sfs/vectors/pentr221_map.pdf). The Entry clone carries the ORF flanked by the appropriate att recombination sites. The 5′ attL1 recombination sequence is followed by a consensus Kozak sequence (CACC) immediately upstream of the ATG start site. The Kozak consensus sequence enables optimal expression of the ORF after recombination with any eukaryotic Gateway™ destination vector (16). At the 3′ end, each ORF is designed to contain an amber stop codon (TAG), followed by the attL2 recombination sequence. The amber stop is compatible with the Invitrogen Tag-on-Demand™ tRNA suppression technology that allows the expression of native or C-terminal-tagged protein from a single clone (17) (Fig. 2).
Figure 2.
The versatility of Tag-On-Demand™ technology.
ORF QUALITY CONTROL
Each ORF Entry clone is qualified based on two criteria: (i) generation of a high-quality, full-insert consensus sequence and (ii) exact match between the consensus sequence and the corresponding public sequence. Each clone is fully sequenced with a high quality at each consensus base. For the >8000 currently available ORF clones, the average Phred score is 84. ORF clones passing the first criteria are then examined for precise nucleotide sequence match at attL recombination sites, Kozak site, amber stop codon and perfect match to public protein sequence. The ORFCard provides a link to view quality scores for each nucleotide in the ORF, and alignment of each ORF protein sequence to public protein databases.
The ORF DATABASE AND SEARCH INTERFACE
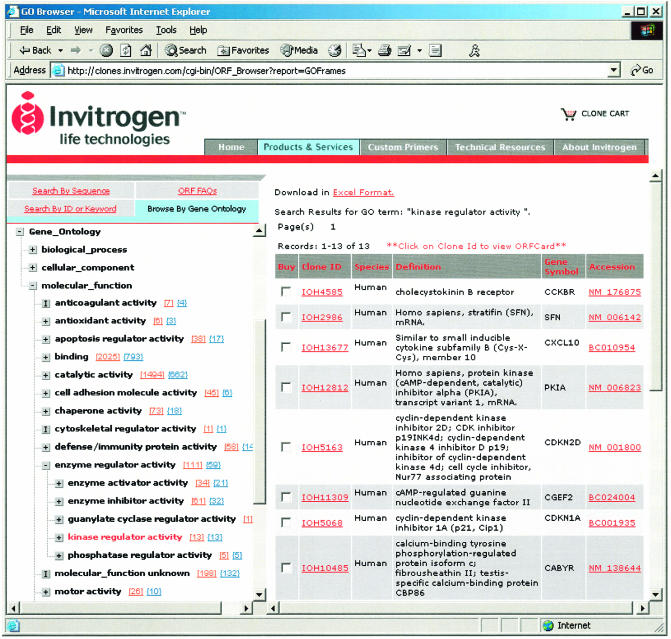
The ORFDB is implemented with a distributed multi-tiered J2EE application consisting of various application components such as an enterprise information system (EIS) tier, business logic tier, web tier and client tier. In order to enable researchers to find ORFs in our database, we have developed a cgi-bin-based ORFBrowser. This ORFBrowser allows researchers to find the ORF clone of interest, blasting with sequence, searching by keyword, accession number, clone ID, Unigene ID, LocusLink ID or browsing through Gene Ontology (GO) (18). Using the GO Browser, researchers can easily browse through the ORF collection based on biological processes, cellular component and molecular function (Fig. 3).
Figure 3.
ORF Gene Ontology Browser.
ORFCARD
Each ORF has been annotated extensively and the annotation is captured in the ORFCard. The ORFCards are dynamically linked to public databases and capture all current information related to each ORF clone. Each ORF clone has an associated clone ID linking it to an ORFCard containing continuously updated information on Gene, ORF, Clone, Protein, single nucleotide polymorphism (SNP) and Genomic links (Fig. 4).
Figure 4.
An example of Ultimate™ ORFCard IOH4878.
(i) Gene information contains the gene definition, function annotation, related accessions, gene symbol, GO classification, links for CGAP gene expression profile and PubMed references.
(ii) ORF information contains the ORF size, nucleotide and protein sequences as well as Phred quality values for each consensus base.
(iii) Clone information has the vector type and the source of the clone collection.
(iv) Protein annotation includes the basic features of the protein, function annotation, related accessions, protease digestion profile, secondary structure prediction as well as the links to domain mapping sites like PFAM, Prosite and SMART.
(v) SNP information contains the links to the NCBI SNP database.
(vi) Genomic links include links to Unigene, LocusLink, Ensembl, as well as the links to map the gene sequence to human and mouse genomic backbones.
Access to this information is absolutely free of charge with no obligation to buy the clones.
FUTURE DIRECTIONS
The current build schedule for the collection results in the release of ∼2000 sequence-verified Ultimate™ ORF clones per calendar quarter. The availability of additional Gateway®-enabled expression options continues to be a focus of ongoing product development effort at Invitrogen. In addition, a simple conversion method allows any vector system to be modified to a Gateway®-compatible format. The high quality associated with the Ultimate™ ORF collection provides a critical resource for life science researchers engaged in the challenge of understanding protein function at the cellular level. We will continue to incorporate the annotations from the research community into ORFCard and enhance the searching capability of ORFDB.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Tim Hensley, Siamak Barhaloo, Aruna Myneni, Josh Lopez, Peter Rifkin, Christine Wong and Christian Wip for integrating the ORFDB web portal into Invitrogen’s external website, and the Gene Index Group at The Institute for Genomic Research (TIGR) for making the cdbfasta/cdbyank package, which was used in creating the ORF retrieving system, freely available to the community. This article was accepted for publication following peer review. However, because the ORFDB is commercial in nature, and not freely available, publication of the article was funded through payment of commercial rates by Invitrogen Corporation.
REFERENCES
- 1.Phizicky E., Bastiaens,P.I.H., Zhu,H., Snyder,M. and Fields,S. (2003) Protein analysis on a proteomic scale. Nature, 422, 208–215. [DOI] [PubMed] [Google Scholar]
- 2.Tyers M. and Mann,M. (2003) From genomics to proteimics. Nature, 422, 193–195. [DOI] [PubMed] [Google Scholar]
- 3.Boone C. and Andrews,B. (2003) ORFeomics: correcting the wiggle in worm genes. Nature Genet., 34, 8–9. [DOI] [PubMed] [Google Scholar]
- 4.Zhu H., Bilgin,M., Bangham,R., Hall,D., Casamayor,A., Bertone,P., Lan,N., Jansen,R., Bidlingmaier,S., Houfek,T. et al. (2001) Global analysis of protein activities using proteome chips. Science, 293, 2101–2105. [DOI] [PubMed] [Google Scholar]
- 5.MacBeath G. and Schreiber,S.L. (2000) Printing proteins as microarrays for high-throughput function determination. Science, 289, 1760–1763. [DOI] [PubMed] [Google Scholar]
- 6.Zhu H. and Snyder,M. (2002) ‘Omic’ approaches for unraveling signaling networks. Curr. Opin. Cell Biol., 14, 173–179. [DOI] [PubMed] [Google Scholar]
- 7.Hartley J.L., Temple,G.F. and Brasch,M.A. (2000) DNA cloning using in vitro site-specific recombination. Genome Res., 10, 1788–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walhout A.J., Temple,G.F., Brasch,M.A., Hartley,J.L., Lorson,M.A., van den Heuvel,S. and Vidal,M. (2000) GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol., 328, 575–592. [DOI] [PubMed] [Google Scholar]
- 9.Vaglio P., Lamesch,P., Reboul,J., Rual,J.F., Martinez,M., Hill,D. and Vidal,M. (2003) WorfDB: the C.elegans ORFeome Database. Nucleic Acids Res., 31, 237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reboul J., Vaglio,P., Tzellas,N., Thierry-Mieg,N., Moore,T., Jackson,C., Shin-i,T., Kohara,Y., Thierry-Mieg,D., Thierry-Mieg,J. et al. (2001) Open-reading-frame sequence tags (OSTs) support the existence of at least 17 300 genes in C. elegans. Nature Genet., 27, 332–336. [DOI] [PubMed] [Google Scholar]
- 11.Reboul J., Vaglio,P., Rual,J.F., Lamesch,P., Martinez,M., Armstrong,C.M., Li,S., Jacotot,L., Bertin,N., Janky,R. et al. (2003) C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nature Genet., 34, 35–41. [DOI] [PubMed] [Google Scholar]
- 12.Baxevanis A.D. (2003) The molecular biology database collection: 2003 update. Nucleic Acids Res., 31, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strausberg R.L., Feingold,E.A., Grouse,L.H., Derge,J.G., Klausner,R.D., Collins,F.S., Wagner,L., Shenmen,C.M., Schuler,G.D., Altschul,S.F. et al. Mammalian Gene Collection Program Team (2002) Generation and initial analysis of more than 15 000 full-length human and mouse cDNA sequences. Proc. Natl Acad. Sci. USA, 99, 16899–16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braun P., Hu,Y., Shen,B., Halleck,A., Koundinya,M., Harlow,E. and LaBaer,J. (2002) Proteome-scale purification of human proteins from bacteria. Proc. Natl Acad. Sci. USA, 99, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammarstrom M., Hellgren,N., van Den Berg,S., Berglund,H. and Hard,T. (2002) Rapid screening for improved solubility of small human proteins produced as fusion proteins in Escherichia coli. Protein Sci., 11, 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozak M. (1987) An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res., 15, 8125–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drabkin H.J., Park,H.J. and RajBhandary,H.L. (1996) Amber suppression in mammalian cells dependent upon expression of an Escherichia coli aminoacyl-tRNA synthetase gene. Mol. Cell. Biol., 16, 907–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T. et al. The Gene Ontology Consortium (2000) Gene Ontology: tool for the unification of biology. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]