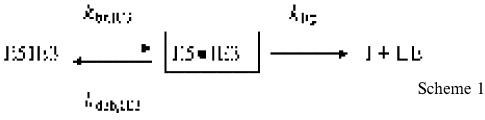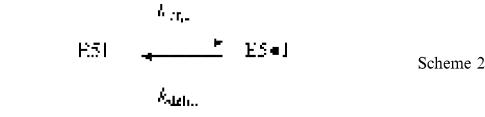Abstract
We have used chemical footprinting, kinetic dissection of reactions and comparative sequence analysis to show that in self-splicing introns belonging to subgroup IIB, the sites that bind the 5′ and 3′ exons are connected to one another by tertiary interactions. This unanticipated arrangement, which contrasts with the direct covalent linkage that prevails in the other major subdivision of group II (subgroup IIA), results in a unique three-dimensional architecture for the complex between the exons, their binding sites and intron domain V. A key feature of the modeled complex is the presence of several close contacts between domain V and one of the intron–exon pairings. These contacts, whose existence is supported by hydroxyl radical footprinting, provide a structural framework for the known role of domain V in catalysis and its recently demonstrated involvement in binding of the 5′ exon.
Keywords: group II intron/hydroxyl radical probing/intron–exon interactions/ribozyme/RNA structure modeling
Introduction
Progress in understanding group II self-splicing has been hindered by our ignorance of how key components of the large group II ribozyme are brought together. For instance, it has long been known that group II introns recognize their 5′ exon by means of two distinct exon-binding sequences, EBS1 and EBS2, that form (typically) six base pairs each with two sequence stretches called IBS1 and IBS2 at the 3′ end of the exon (Jacquier and Michel, 1987). Unfortu nately, subsequent studies have mostly failed to provide insight into how the EBS elements, which are located far away in secondary structure models from those sections of the intron that are conserved in sequence, may be connected to the active center of the ribozyme. Only recently could it be demonstrated (Costa and Michel, 1999) that tight binding of the 5′ exon requires not only intron domain I, of which the EBS sequences are part, but also the distal part of domain V, a small yet major component that is believed to be involved in catalysis (Chanfreau and Jacquier, 1994; Peebles et al., 1995; Abramovitz et al., 1996; Konforti et al., 1998a). Even so, it has remained unclear whether domain V contacts the exon, one of the EBS segments or yet another intron component that is itself involved in exon binding.
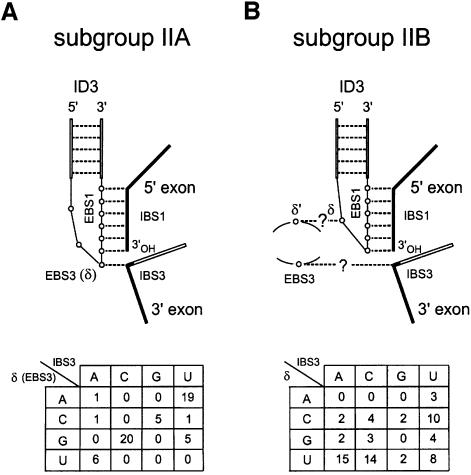
Another source of uncertainty has been the identity of EBS3, the intron-contained binding site for the 3′ exon. It is often regarded as well established that like group I introns, the group II ribozymes guide the ligation of their exons by getting them to bind next to one another to a continuous ‘internal guide sequence’ (IGS). The fact is that the nucleotide immediately 5′ of EBS1 (δ or EBS3 in Figure 1A) can more often than not form a canonical pair with the first nucleotide (IBS3) of the 3′ exon (Michel and Jacquier, 1987) and experiments have shown that under some conditions cleavage at the 3′ splice site can be redirected by base substitutions at the δ position (Jacquier and Jacquesson-Breuleux, 1991). Moreover, reverse splicing of some group II introns into their DNA target site, which involves pairing of EBS1 and EBS2 with DNA counterparts of IBS1 and IBS2, is significantly more efficient when the δ base can pair with the nucleotide 3′ of IBS1 (Guo et al., 1997; Mohr et al., 2000). However, a potential complication to this picture stems from the existence of two major subdivisions of group II introns (Michel et al., 1989). Among features that differentiate the two subgroups is the relative location of the EBS1 sequence within the ID3 terminal loop (Figure 1). Contrary to subgroup IIA introns, which have at least three unpaired nucleotides 5′ of EBS1, most members of subgroup IIB have only one base available for pairing with the 3′ exon and would seem unlikely to stack it on top of EBS1 without unwinding part of the ID3 helix [compare Figure 1A and B; the helical continuity of the IGS is an integral part of group I and group II guide models and is supported by experiments in which the hydrolytic cleavage reaction catalyzed by a group II intron was shown to strongly prefer extended double-stranded structures with canonical base pairs on both sides of the reactive bond (Jacquier and Jacquesson-Breuleux, 1991; see also Michel and Ferat, 1995; Jacquier, 1996)]. As already noted (Michel et al., 1989), this difference between the two subclasses appears to be reflected in the distribution of bases at the δ and IBS3 sites. Whereas only three out of 58 subgroup IIA introns possess non-matching δ:IBS3 combinations (Figure 1A), thorough examination of subgroup IIB sequences fails to reveal any statistical evidence of base pairing between the δ base and the first residue of the 3′ exon (Figure 1B).
Fig. 1. Distinct modes of guiding exon ligation in subgroup IIA and IIB introns. (A) Subgroup IIA. Hollow lines and circles indicate stretches of intron sequence and individual intron residues; (---), canonical base pairs; EBS1 and EBS3, exon-binding sites; IBS1 and IBS3, intron-binding sites; ID3, a helix that is part of domain I (see Figure 3); δ is the nucleotide immediately 5′ of EBS1. The table at the bottom shows the distribution of bases at the δ (EBS3) and IBS3 sites in 58 subgroup IIA sequences; mutual information (Chiu and Kolodziejczak, 1991), which measures the extent to which the base contents of one site can be predicted from those of the other one, is high (0.834). The use of EBS3–IBS3, rather than δ–δ′ (Jacquier and Jacquesson-Breuleux, 1991), to designate interactions between the intron and 3′ exon is made necessary by the existence of a distinct δ–δ′ pairing in subgroup IIB molecules (B) and improves the consistency of the group II nomen clature, by restricting Greek letters to tertiary interactions within the intron (Figure 3). (B) Subgroup IIB. Same as (A). Question marks refer to potential interactions, the existence of which is established in this work (see Figure 3). The table at the bottom shows that the base contents of the IBS3 and δ sites are not significantly correlated in 69 subgroup IIB sequences (mutual information 0.127).
While it might be concluded from the preceding argument that exon ligation is not generally guided in subgroup IIB introns, we now report that the first residue of the 3′ exon is actually base paired to the intron during exon ligation catalyzed by a self-splicing subgroup IIB molecule. However, the intron partner (EBS3) of the 3′ exon IBS3 site is not the δ nucleotide, which is itself engaged in base pairing with another intron residue (δ′). The two newly identified sites, EBS3 and δ′, happen to be facing each other within an internal RNA loop, the sequence and location of which are well conserved in subgroup IIB introns. We propose that this loop, by means of its specific fold, is primarily responsible for positioning the IBS3–EBS3 pair and EBS1–IBS1 helix in the appropriate conformation for exon ligation. The novel structural constraints uncovered in this work make it possible to build a three-dimensional model of a large section of the active center of group II introns. We further show that this model is supported by data from experiments in which hydroxyl radicals are used for the first time to probe the higher order structure of a group II ribozyme and its complexes with the exons.
Results
Identification of G293 as a potential exon-binding site
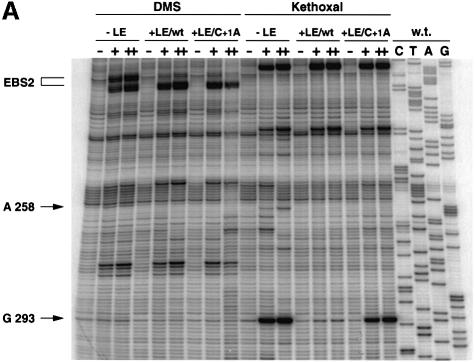
In order to uncover a possible binding site for the first base of the 3′ exon, we have chemically probed a group IIB self-splicing intron either alone or in the presence of wild-type and mutated versions of its ligated exons. Group II introns are normally excised as branched molecules called lariats, and the lariat form of intron Pl.LSU/2 from Pylaiella littoralis mitochondria was previously shown to form populations of molecules with a seemingly uniform conformation, which makes it suitable for direct chemical probing (Costa et al., 1997b, 1998; Costa and Michel, 1999). As shown in Figure 2A, probing the Pl.LSU/2 lariat with dimethylsulfate (DMS) in the presence of a 16mer oligoribonucleotide (LE/wt, Table I) that encompasses the last 13 residues of the 5′ exon and the first three nucleotides of the 3′ exon results in the expected footprinting of the A-rich EBS2 sequence and (to a lesser extent) of position A258, within EBS1 (Figure 3; both footprints are attributable to the 5′ exon; see Costa and Michel, 1999). In this case, comparison with another oligonucleotide with a base substitution (C to A) at the first position (+1) of the 3′ exon (LE/C+1A, Table I) revealed no difference that could be ascribed to that substitution. In contrast, when kethoxal, which specifically targets guanines with accessible N1 and N2 groups, is used as a probe, the strong protection that the LE/wt molecule confers to the otherwise (Figure 2A, lane –LE) very reactive G293 is no longer observed if this molecule is replaced by its LE/C+1A counterpart. That this footprint is specific to the 3′ exon was checked further by verifying (Figure 2B) that a previously described (Costa and Michel, 1999) unreactive 13mer analog of the 5′ exon (3′dE5, Table I) completely fails to protect position 293.
Fig. 2. Mapping by reverse transcription of DMS and kethoxal exon footprints on the lariat form of intron Pl.LSU/2. All modification reactions were at 30°C for 0 (–), 7 (+) or 15 (++) min. C, T, A and G are sequencing lanes (recall that reverse transcriptase stops one nucleotide before a modified base). (A) DMS and kethoxal probing of wild-type lariat either alone (–LE) or in the presence of 2 µM LE/wt or LE/C+1A (Table I) ligated exon molecules. (B) Kethoxal modification of wild-type and C252G intron lariats in the absence or presence of an inactive oligonucleotide analog of the 5′ exon (3′dE5, Table I and Costa and Michel, 1999). Saturation conditions (see text) were ensured by using 6 µM (wild-type lariat) or 30 µM (C252G lariat) 3′dE5.
Table I. Oligonucleotides for footprinting experiments and kinetic analysesa.
| Name | Sequence |
|---|---|
| rE5 | 5′-UGUUUAUUAAAAA |
| 3′dE5 | 5′-UGUUUAUUAAAA3′dAb |
| 2′dE5 | 5′-UGUUUAUUAAAA2′dAc |
| LE/wt | 5′-UGUUUAUUAAAAACAC |
| LE/C+1A | 5′-UGUUUAUUAAAAAAAC |
aUnless otherwise stated, sugars are riboses; the IBS1 sequence (Figure 3) is underlined.
bThe terminal 3′-OH group is replaced by a hydrogen.
cThe 2′-OH group of the 3′-terminal nucleotide is replaced by a hydrogen.
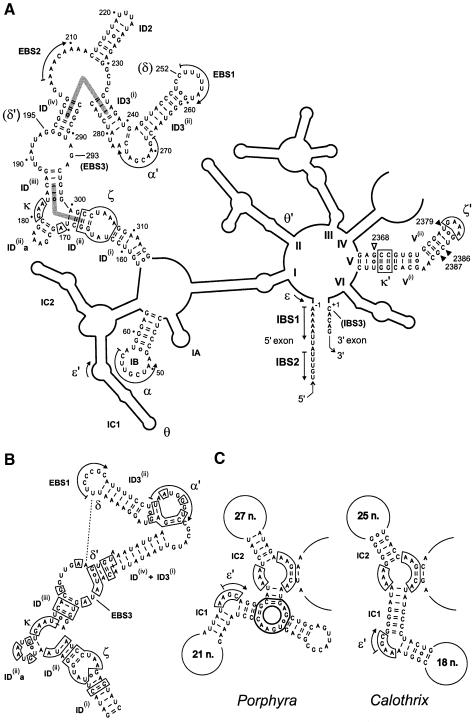
Fig. 3. (A) Secondary structure model of the Pl.LSU/2 intron, redrawn from Costa and Michel (1999) with minor modifications (as suggested by comparative analysis, G241 is paired with C276 rather than U279). Individual nucleotides are shown only in those sections of the molecule that are modeled in Figure 6. Greek letters designate tertiary interactions within the intron (Costa and Michel, 1995; Michel and Ferat, 1995; Costa et al., 1997a; Boudvillain and Pyle, 1998; this work). Pairs of helices proposed to stack end-to-end (see text) are indicated by gray bars. Open and filled inverted triangles indicate domain V residues whose reactivity to hydroxyl radicals is increased or decreased, respectively, upon binding of the 5′ exon. Note that numbering takes into account the open reading frame in domain IV, which was removed in the constructs used for in vitro experiments (Costa et al., 1997b). (B) Subdomain ID in intron Aae.LSU/5 from Agrocybe aegerita (DDBJ/EMBL/GenBank accession No. AF087656). Boxed nucleotides are in common with Pl.LSU/2. Other related introns lack bulges in stems ID3(ii) and ID(iv)+ID3(i) (M.Granlund, F.Michel and M.Norgren, submitted; F.Lang, personal communication). (C) Subdomain IC in introns Ppu.LSU/1 from Porphyra purpurea (Burger et al., 1999) and Cal.x1 from Calothrix PCC7601 (Ferat and Michel, 1993). Homologous nucleotides are boxed with thin lines. Heavy boxing: three-way junction specific to the Porphyra intron.
Disruption of the G293-C+1 pair impairs exon ligation
In the experimental set-up we chose to use (Scheme 1), ligated exons (LE) result when the reaction intermediate, a lariat molecule consisting of the branched intron with the 3′ exon still attached (IE3), is incubated with a saturating (Costa and Michel, 1999, and Table II) concentration of the 5′ exon (E5). However, additional products are generated, due to the reversibility of the transesterification reaction that constitutes the first step of the splicing pathway (left part of Scheme 1). Thus, a fraction of IE3•E5 complexes react to reconstitute transiently the precursor (E5IE3). Moreover, the lariat intron (I) produced during exon ligation is also ‘debranched’ by the 5′ exon to generate E5I, a linear intron–5′ exon molecule [Scheme 2, which can be investigated separately by mixing purified lariat intron molecules with E5, and which was shown by Chin and Pyle (1995) to reach an equilibrium]:
Scheme 1
Table II. Kinetic parameters of lariat debranching reactionsa.
| Molecules | Kdb (nM) | kobsc (per min) | Fraction of productd |
|---|---|---|---|
| EBS3 (293) | |||
| G (wt) | 420 ± 60 | 1.28 ± 0.06 (1.26e) | 0.186 ± 0.009 (0.172e) |
| A | 100 ± 20 | 1.84 ± 0.11 (1.93e) | 0.166 ± 0.010 (0.159e) |
| U | 690 ± 110 | 1.20 ± 0.10 (1.28e) | 0.153 ± 0.010 (0.189e) |
| C | 2200 ± 200 | 1.23 ± 0.06 (1.68e) | 0.138 ± 0.005 (0.143e) |
| δ–δ′ (252:195) | |||
| C:G (wt) | 420 ± 60 | 1.28 ± 0.06 | 0.186 ± 0.009 |
| U:A | 3200 ± 400 | 1.58 ± 0.10 | 0.144 ± 0.006 |
| C:Gf (wt) | 0.82 ± 0.14 | 0.169 ± 0.011 | 0.114 ± 0.007 |
| C:Af | 870 ± 60 | 0.198 ± 0.011 | 0.120 ± 0.003 |
aPurified lariat intron samples were used (Scheme 2). Values are for the rE5 5′ exon.
bSee Materials and methods and Costa and Michel (1999).
cObserved rate of lariat debranching (kdeb,I + kbr,I, Scheme 2) at a saturating concentration of 5′ exon.
dFinal fraction of debranched molecules at a saturating concentration of 5′ exon.
eValues in parentheses were obtained by quantitating all intron-containing products in reactions of IE3 lariat intermediate with the rE5 exon (Figure 4A) and extracting rate constants for intron debranching and branching (IE3 molecules had matched EBS3:IBS3 combinations).
fTemperature was 25°C instead of 40°C.
Scheme 2
Exon ligation is nevertheless observed to proceed to completion (Figure 4A) when the concentration of 5′ exon is much higher than that of IE3. Under such conditions, controls showed that rates for each individual step (Figure 4A) are independent of the initial intron concentration (Materials and methods), as should be the case for pseudo-first-order reactions.
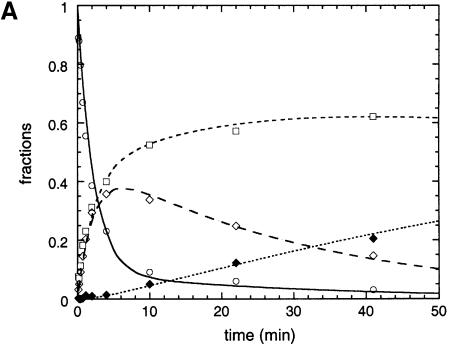
Fig. 4. Reactivity of IE3 lariat intermediate molecules with matched and mismatched base combinations at positions 293 (EBS3) and +1 (IBS3). (A) Example of reaction kinetics. Purified 293A:+1U IE3 molecules (20 nM) were reacted (Materials and methods) with 20 µM 2′dE5 exon (Table I). Experimental data (fractions of the initial molar concentration of IE3 as a function of time) were fitted with the help of the Kintecus software (J.Ianni) to curves generated from the set of linear differential equations corresponding to Schemes 1 and 2. Open circles and the continuous curve (—), IE3; open diamonds and long dashes (– – –), E5IE3; open squares and small dashes (---), lariat intron (I); filled diamonds and dotted curve (……), E5I. (B) Calculated rate constants of exon ligation (klig, Scheme 1) for IE3 molecules with diverse 293:+1 base combinations. Conditions as in (A) with 5 µM rE5 5′ exon (thick horizontal black bars) and 20 µM 2′dE5 (open bars) for all combinations except those including C293, for which 25 µM rE5 and 100 µM 2′dE5 were used (vertical lines connect black and open bars for the same mutant). Reactions with the rE5 exon of IE3 molecules carrying matched EBS3–IBS3 combinations were somewhat too rapid for their rate to be estimated precisely. (C) Relative rate of debranching over branching for IE3 molecules. The exon was 2′dE5. Ratios are expressed as kdeb,IE3/(kbr,IE3 + kdeb,IE3) (see Scheme 1) in order to allow comparison with final fractions of debranched lariat intron in Table II [note that the wild-type lariat intron is debranched more extensively (final fraction ∼0.38) by the 2′dE5 than the rE5 exon].
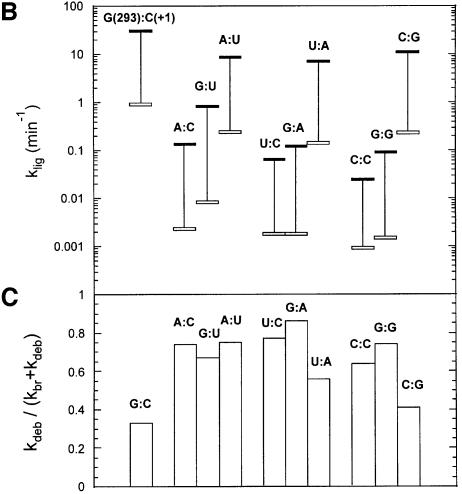
As shown in Figure 4B, the rate constant (klig) of exon ligation is much decreased by mutations at either position 293 or +1 (splicing of mutated molecules nevertheless remains faithful, as judged from the migration of the ligated exon product in denaturing gels; data not shown). Unfortunately, the reaction of the wild-type IE3 molecule with an all-ribose (rE5, Table I) 5′ exon is too rapid, even at pH 5.8, for its rate to be estimated accurately. However, ligation can be slowed down ∼50-fold by removing the 2′-OH group at exon position –1 (Podar et al., 1998; Dème et al., 1999) and, in this context, single base substitutions at positions 293 and +1 are seen to reduce klig further by 100- to 1000-fold (Figure 4B). Under these conditions, it also becomes apparent that efficient ligation is largely restored by combining those single substitutions so as to generate Watson–Crick combinations other than the wild-type one (as expected for a canonical base pair, the G293:U+1 wobble combination displays somewhat intermediate behavior). These experiments thus demonstrate both the existence of the 293(EBS3):+1(IBS3) pair and its importance for the second step of splicing.
In contrast to their impairment of exon ligation, base substitutions at position 293 have only moderate (<5-fold) effects on the Kd for the 5′ exon and essentially no influence on the rate of production and final (equilibrium) fraction of debranched molecules when assayed in the context of the lariat intron [Scheme 2 and Table II; Kd was estimated from the dependence of the final fraction of debranched product on 5′ exon concentration, as explained in Costa and Michel (1999)]. Since Scheme 2 is related to the first step of splicing, the latter thus appears essentially unaffected. Nevertheless, all combinations other than G:C and C:G show somewhat elevated rates of debranching of the IE3 form (compared with branching, Figure 4C), which most probably reflects the fact that the EBS3–IBS3 base pair, which is shown by kethoxal modification to exist in the complex between the lariat intron and ligated exons (Figure 2A), is also part of the ground state of the wild-type IE3 molecule [disruption or weakening of another interaction that is believed to come into existence only after the first step of splicing was reported by Chanfreau and Jacquier (1996) to have the same consequences in the related Sc.a5γ intron from yeast mitochondria].
Statistical evidence for the EBS3–IBS3 and δ–δ′ pairings
Only one residue separates G293 from the 3′ branch of helix ID(iv), whose length and sequence tend to be rather well conserved in subgroup IIB introns (Figure 1B in Michel et al., 1989). It is therefore straightforward to align available subgroup IIB sequences locally and check whether the base homologous to nucleotide 293 is generally constrained to pair with the first nucleotide of the 3′ exon. As seen in Table III, this is clearly the case, since a Watson–Crick base pair could form in 53 out of 69 sequences (mutual information 0.593). Moreover, in five of the remaining sequences, a G:U pair could exist.
Table III. Statistical evidence for EBS3–IBS3 and δ–δ′ pairings in group IIB intronsa.
| EBS3 (293 and counterparts) | IBS3 |
|||
|---|---|---|---|---|
| A | C | G | U | |
| A | 1 | 1 | 2 | 18 |
| C | 2 | 0 | 1 | 0 |
| G | 0 | 18 | 0 | 5 |
| U |
16 |
2 |
1 |
2 |
| δ | δ′ (195 and counterparts) |
|||
| |
A |
C |
G |
U |
| A | 3 | 0 | 0 | 0 |
| C | 2 | 0 | 16 | 0 |
| G | 0 | 8 | 1 | 0 |
| U | 30 | 6 | 1 | 2 |
aThe same 69 sequences were analyzed as in Figure 1B.
Pairing of G293 with the first base of the 3′ exon implies that nucleotide 252 (δ), immediately 5′ of EBS1, lies relatively close to it during the ligation step. We therefore sought a possible partner for C252 among the bases that are part of the internal loop between helices ID(iii) and ID(iv). As seen in Table III, nucleotide 195, immediately 5′ of the 5′ branch of helix ID(iv), tends to co-vary with the base at position 252 (mutual information 0.548), and indeed seems capable of forming a canonical base pair with the latter [54 out of 69 sequences; the lack of A(δ):U(δ′) combinations could reflect the necessity to avoid extending the ID(iv) helix by base pairing between nucleotide 195 and the well-conserved A at position 292].
Disruption of the δ–δ′ pair interferes with exon binding
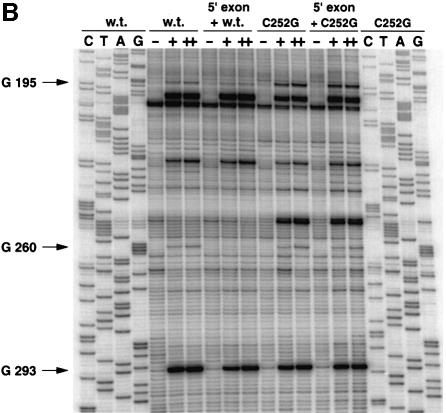
In order to assess the possible existence of a C252(δ)–G195(δ′) base pair in the Pl.LSU/2 intron, we generated molecules with U:A and G:C combinations at these two sites and compared them both with those carrying single substitutions and the wild type. While mutant precursor transcripts were sufficiently reactive to generate workable amounts of excised intron, all purified lariat intron molecules except U252–A195 proved unable to debranch significantly (Scheme 2) when confronted with 5 µM rE5 5′ exon at 40°C. Since closer examination of the kinetic parameters for the debranching reaction of the U:A lariat revealed a marked increase in Kd (Table II), we sought conditions that would allow tighter binding of the 5′ exon.
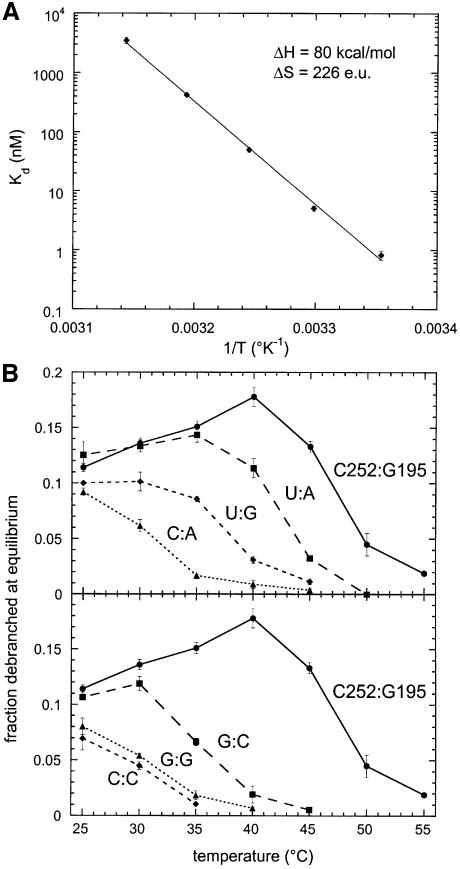
As seen in Figure 5A, not only does the Kd for dissociation of the 5′ exon from wild-type lariat molecules show the expected strong dependence on temperature, but the Van’t Hoff plot of log(Kd) versus 1/T appears linear between 25 and 45°C, which suggests that neither the lariat nor the 5′ exon undergo conformational changes liable to interfere with their mutual recognition within the temperature range investigated. The overall decrease in Kd between 40 and 25°C is ∼500-fold and, when assayed at the latter temperature in the presence of 5 µM rE5, all mutated lariats were found to debranch to a significant extent (Figure 5B), in keeping with the possibility that their Kd was now either below or in the same range as the concentration of 5′ exon. That this was the case was verified by determining the Kd for the 252C:195A lariat, which although ∼1000-fold higher than for the wild type, is nevertheless <5 µM at 25°C (Table II).
Fig. 5. Reactivity of lariat intron molecules with matched and mismatched base combinations at positions 252 (δ) and 195 (δ′). (A) Van’t Hoff plot of Kd for dissociation of the rE5 5′ exon from the wild-type lariat (252C:195G). Kd was estimated from the dependence of the final fraction of debranched product on rE5 concentration (Costa and Michel, 1999). (B) Final fraction of debranched molecules at 5 µM rE5 as a function of temperature.
In order to characterize the other mutated molecules, we resorted to the simple test of determining the final fraction of debranched lariat as a function of temperature for a constant 5 µM concentration of 5′ exon: as the temperature is raised, this fraction is seen (Figure 5B) to decrease progressively to undetectable levels, with the temperature at mid-transition presumably corresponding to that at which Kd becomes approximately equal to 5 µM. By this criterion, the C:A combination appears less fit to bind the exon than U:G, which fares, in turn, less well than U:A and, finally, C:G. While this order is of course the expected one for a Watson–Crick pair, exchanging G and C results in a molecule that loses the ability to bind the exon at ∼35°C, rather than at ∼47°C for the wild type. However, whatever the reason for the poorer performance of the G:C combination, it nevertheless is seen to offer some significant measure of compensation over C:C and G:G mismatches, which provides a further indication of a canonical interaction between positions 252(δ) and 195(δ′).
Is the δ–δ′ pair part of the ground state of the intron?
The inability of molecules lacking the δ–δ′ pair to bind their 5′ exon tightly strongly suggests that one function of this interaction is to constrain the structure of the ID3 loop in such a way that the EBS1–IBS1 helix can be stabilized. In order to determine whether formation of the δ–δ′ pair is facilitated by binding of the exon, we probed the wild-type and a C252G mutant lariat with kethoxal in the presence and absence of the unreactive 3′dE5 5′ exon (Figure 2B). In the C252G mutant molecule, the G at position 195 reacts with kethoxal in both the absence and presence of the exon (whose binding was verified to be complete by DMS modification; data not shown). By comparison, G195 is significantly (by a factor of ∼4.5) protected in the wild-type lariat, where it can pair with the C at 252 and, even though its reactivity is reduced further (∼3-fold) upon binding of the exon, this strongly suggests that the δ–δ′ pair exists in the absence of the EBS–IBS pairings (another residue that responds to the presence of the exon is G260, which, although assumed to base pair with U250, is not fully protected unless both the δ–δ′ and IBS1–EBS1 interactions are present). Finally, we did not note any difference in the extents of protection from kethoxal modification afforded to G195 by the 5′ exon and ligated exons (data not shown). Therefore, the EBS3–IBS3 and δ–δ′ base pairs must co-exist in the ground state of the complex between the intron lariat and its exons.
Three-dimensional architecture of domains ID and V
Subdomain ID has a central role in organizing the active center of group II introns because it includes not only the three exon-binding sites [EBS1, EBS2 and (this work) EBS3], but also two important receptor sites (Figure 3A) for the small and well-conserved domain V, which is regarded as being involved in catalysis. We now detail how a three-dimensional model of the complex formed by the ligated exons, subdomain ID and domain V can be built by combining all available data.
Besides confirming the widespread occurrence of the δ–δ′ and EBS3–IBS3 interactions, comparative analysis of available subgroup IIB sequences reveals two other major constraints on the architecture of subdomain ID. A number of subgroup IIB introns lack the EBS2–IBS2 pairing and, in most of these molecules, stems ID(iv) and ID3(i) are fused together (see Figure 3B). This shows that these two helices, which are almost always contiguous when distinct, actually stack end-to-end. A second pair of helices, which are most likely to be coaxial, are ID(ii) and ID(iii). These two stems are part of a three-way junction motif, H(elix)1-A–H(elix)2-GAA–H(elix)3-, which recurs at different locations in self-splicing introns. As shown in Figure 3C, this motif may be replaced by a single continuous helix, equivalent to H1 + H3, and, since the substitution occurs in closely related molecules, it is quite unlikely to change the relative geometry of the sections distal to H1 and H3 (in the example provided, the loops on either side of H1–H3 are well-conserved, important components of domain I).
The ID(ii)–ID(ii)a–ID(iii) junction is also of interest as a binding site for domain V. Not only does chemical modification of the adenines of the internal loop interfere with binding of domain V by domain I (Jestin et al., 1997), but these three bases are footprinted by a separate domain V molecule (Konforti et al., 1998b; Costa and Michel, 1999). Based on nucleotide analog interference experiments, Boudvillain and Pyle (1998) suggested that the section of domain V that is contacted by these adenines consists of base pairs 4 and 5, and named this novel interaction κ–κ′ (Figure 3A). They further proposed that the structure of the κ three-way junction is similar to that of a GAAA terminal loop in interaction with a helical receptor (Pley et al., 1994), and we have followed this suggestion, which is consistent with the coaxial stacking of the ID(ii) and ID(iii) helices. Basal of helix ID(ii) is the ζ receptor for the terminal loop of domain V (Costa and Michel, 1995). The structure of the complex between this 11 nucleotide motif and a GAAA loop has been determined at atomic resolution (Cate et al., 1996), so that the entire complex between domain V and its ζ and κ receptors can be built as a single piece within which the relative layout (angle and distance) of the two domain V helices is reasonably well specified.
In modeling the ligated exons, we have assumed that the nucleotides 3′ and 5′ of the reactive phosphate lie in helical continuity with respect to one another. This arrangement, which is almost certain to prevail in subgroup IIA introns, which have a continuous IGS, is also consistent with hydrolytic cleavage of continuous double helices by the yeast Sc.a5γ intron (Jacquier and Jacquesson-Breuleux, 1991). G293 was therefore placed on top of U253 and the helical stack was extended further by placing the EBS1–IBS1 pairing in the continuity of the ID3(ii) helix, as strongly suggested by the fact that the EBS1 sequence abuts the 3′ branch of ID3(ii) in a majority of group II introns. Coupling of this particular architecture, which allows a Watson–Crick pair to form between C252 (δ) and G195 (δ′), with the stacking of helices ID3(i) and ID(iv) makes it necessary for the backbone to undergo a complete reversal at the junction of helices ID3(i) and ID3(ii) and essentially sets the geometry of the distal section of subdomain ID, including the α–α′ pairing.
Because the precise structure of the ID(iii)–ID(iv) internal loop remains unknown, assembly of the proximal and distal sections of subdomain ID must rest on compatibility with the overall architecture of the intron. A major constraint is the need for the ID(i) helix and the stem (IB) that supports the α sequence to come together, since they emerge from the same multibranched internal loop. The only way that we succeeded in meeting this requirement was by arranging the two pieces as shown in Figure 6A.
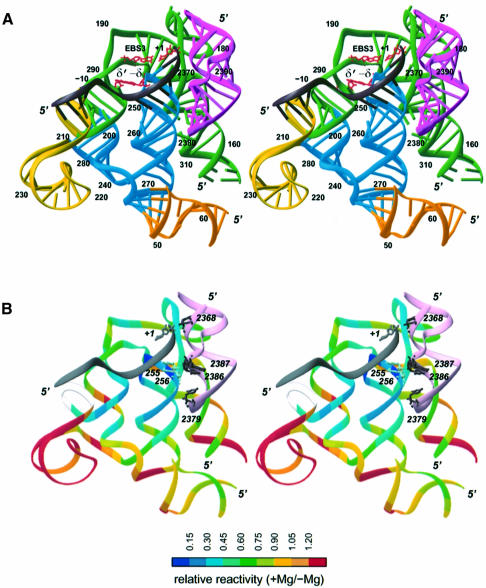
Fig. 6. Three-dimensional model of the complex between subdomain ID, domain V and the ligated exons. (A) Coloring by domains: dark gray, exons; green, basal section of ID [ID(i)–ID(iv)]; blue, ID3; yellow, ID2 stem and basal loop; orange, IB; magenta, domain V. Numbering is according to Figure 3A. The EBS3–IBS3(+1) and δ–δ′ base pairs are shown in red. (B) Coloring (except for domain V and the exons) is according to the extent of protection from hydroxyl radical cleavage in the presence of magnesium (values from Figure 7B; values could not be obtained for positions 202–207, shown in off-white, see legend to Figure 7C). Nucleotides shown in domain V (pink) are those whose reactivity changes upon binding of the 5′ exon. Figure prepared with Ribbons (Carson, 1991).
Hydroxyl radical footprinting of the intron by the 5′ exon
A striking feature of the model depicted in Figure 6A is the extensive contact surface of domain V and the EBS1–IBS1 helix. Since interlocking of these two structures was not part of the premises of the modeling process, but rather came as a consequence of ensuring a globally correct architecture, we have sought to verify its existence experimentally. Because the surfaces predicted to face one another consist exclusively of backbone components, we chose to probe the complex between the lariat intron and the 5′ exon with Fe(II)-EDTA-generated hydroxyl radicals, which primarily target accessible riboses (Balasubramanian et al., 1998). As previously observed on other large self-assembling RNAs with a compact structure (Latham and Cech, 1989), many sections of the molecule acquire some degree of protection from the probe in the presence of a concentration of magnesium sufficient to ensure higher order folding (Figure 7A). As also expected, a number of these protected segments happen to coincide with sites known to be involved in tertiary interactions (Figure 7B). More generally, reactivities determined experimentally in the presence of the 5′ exon were found to be in good agreement with the modeled structure (Figure 6B), for all of the most highly accessible nucleotides happen to lie at the surface, while protection reaches a maximum at the tip of the ID3 stem, which is predicted by our modeling of the intron–exon complex to be the most buried section of subdomain ID. In addition, despite the fact that only about a third of the Pl.LSU/2 ribozyme was modeled, there are relatively few segments whose protection cannot be accounted for by the current structure.
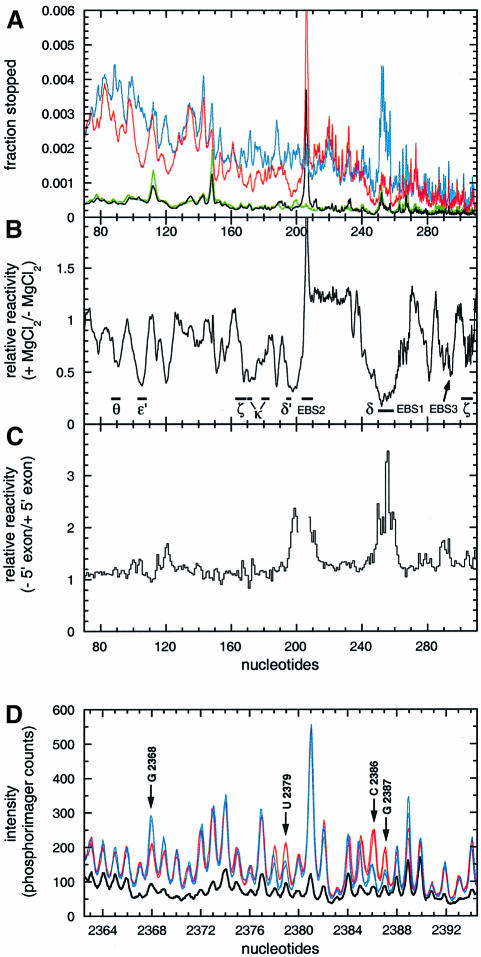
Fig. 7. Quantitation of wild-type lariat intron reactivity to hydroxyl radicals. Nucleotide numbering is according to Figure 3A. Detection of events was by reverse transcription with labeled primers (Materials and methods). (A) Scans of primer extensions over subdomains IC and ID. The lariat intron was exposed to hydroxyl radicals in the presence of the 3′dE5 5′ exon (6 µM) at either 10 mM MgCl2 (red curve, f+Mg) or 0 mM MgCl2 (blue curve, f–Mg). Black and green curves, mock-treated samples with (b+Mg) and without (b–Mg) magnesium, respectively. Ordinates, fraction of primer extensions ending 3′ of nucleotide x (relative to all longer molecules). (B) Reactivity in the presence of magnesium relative to that in its absence [(f+Mg – b+Mg)/(f–Mg – b–Mg)]. (C) Footprints of the 3′dE5 5′ exon on subdomains IC and ID. Ordinates, ratio of the fraction of molecules ending 3′ of nucleotide x in the absence of exon over that in the presence of 3′dE5. Figures could not be obtained over positions 202–207 due to a strong stop in magnesium-containing lanes. (D) Footprints of the 3′dE5 5′ exon on domain V. The lariat was exposed to hydroxyl radicals in the absence (red scans) or presence (blue scans) of 6 µM 3′dE5. For each condition, two scans corresponding to two independent experiments were super imposed to show the reproducibility of 5′ exon footprints. Thick black scan, mock-treated sample.
When patterns of protection in the presence and absence of the 5′ exon were compared over subdomains IC and ID (Figure 7C), significant changes were found to be confined essentially to nucleotides that are either part of the exon-binding sites or in their vicinity. Since becoming part of a double-stranded helix fails by itself to offer protection from free radicals (Celander and Cech, 1990), the strong additional shielding of EBS1 seen in the presence of the exon must reflect the fact that formation of the EBS1–IBS1 helix not only confers a rigid structure to the ID3 terminal loop, but pushes the EBS1 segment into close contact with some other intron component, which, according to the model, should be domain V. By surveying the entire molecule, we found that aside from the EBS sites and their vicinity, the only nucleotides whose reactivity to hydroxyl radicals responds to the presence of the exon are indeed all part of domain V (Figure 7D). Residues U2379, C2386 and G2387 (Figures 3 and 6B), which, in the modeled structure, face the two EBS1 riboses (255 and 256, Figure 7C) that are most highly protected by the exon, are themselves shielded (by up to 2.5-fold) in its presence, while G2368, whose ribose was placed next to the reactive phosphate, is reproducibly more reactive when probed in the presence of the 5′ exon than in its absence.
Discussion
The experiments reported herein demonstrate the existence of two novel base–base interactions, δ–δ′ and EBS3–IBS3, which the statistical analyses presented in Table III show to be characteristic of one of the two major subdivisions of group II self-splicing introns. Although these two base pairs involve nucleotides in the same internal loop [ID(iii)–ID(iv)] of the ribozyme secondary structure, their functional significance is quite different. As indicated by kethoxal modification of the lariat intron, the δ–δ′ interaction, which is most probably present prior to exon binding, persists in intron–exon complexes, and kinetic analyses show that its role is to facilitate base pairing between the 5′ exon and the intron. On the other hand, base pairing of EBS3 with its 3′ exon partner, which we have shown to be essential for efficient exon ligation, corresponds to one particular conformation of the intron in which the 3′ splice site, rather than the 2′-OH group responsible for branch formation, is positioned in the active site (Chanfreau and Jacquier, 1996); this readily accounts for the fact that mutations that prevent formation of the EBS3–IBS3 pair promote debranching of IE3 lariat intermediate molecules (Figure 4C).
While the δ–δ′ and EBS3–IBS3 interactions appear confined to members of group IIB, their existence has structural implications for both subclasses of group II introns. By bringing together domain V and the EBS1–IBS1 helix, the model in Figure 6 and the hydroxyl radical protection data that support it provide us for the first time with a structural framework within which the interplay of two key components of the group II ribozyme may be analyzed. Because it is the most conserved section of group II introns in terms of sequence, domain V had long been suspected to play a central role in the splicing reaction. This was confirmed when it was found that substitution of many of its bases, 2′-OH and phosphate groups interferes with function in such a way that in set-ups in which this domain is brought in as a separate piece, activity cannot be restored by increasing its concentration (Chanfreau and Jacquier, 1994; Peebles et al., 1995; Abramovitz et al., 1996; Konforti et al., 1998a). By these criteria, G2368 (Figures 3 and 6B) may well be regarded as of utmost importance, because in the related a5γ intron neither substitution of the guanine with A, C or U nor replacement of the phosphate group by an Rp phosphorothioate appears compatible with efficient catalysis (Chanfreau and Jacquier, 1994; Peebles et al., 1995). It would therefore seem particularly fitting to have this residue sitting next to the reactive phosphate at the junction of the two exons, as is the case in our model, had it not been shown by Konforti et al. (1998a) that the guanine moieties essential for ‘catalysis’ are N7 and O6, which happen to be located (Figure 6) on the ‘wrong’ side with respect to the exons (this supposes that the Hoogsteen face of G2368 is directly, rather than indirectly involved in the reaction). However, the contradiction may be only apparent, because G2368 is strikingly insensitive to the nature of the base facing it. Both G:C and G:A occur repeatedly as substitutes for the dominant G:U combination in natural group II sequences (Michel et al., 1989; F.Michel, in preparation) and, at least in the Sc.a5γ intron, G:A appears to react just as well as the G:U wild type (Peebles et al., 1995). This suggests that despite G2368 being situated in the middle of a helix, it may not be paired with U2397 in the active state of the ribozyme, in which case its precise location cannot be guessed. In fact, the enhanced susceptibility of G2368 to hydroxyl radicals upon binding of the 5′ exon (Figure 7D) points to some rearrangement either of this nucleotide or of one of its immediate neighbors.
It is only recently that domain V was shown not only to participate in catalysis, but also to promote binding of the 5′ exon. Estimates of the concentration of exon necessary to protect the EBS1 and EBS2 bases from chemical modification, as well as kinetically determined Kd values, indicate that the affinity of the Pl.LSU/2 intron for its 5′ exon is decreased by ∼100-fold upon removal of the last three base pairs of the distal helix of domain V (Costa and Michel, 1999). This somewhat unanticipated finding is readily accounted for in the context of the present work, since it is precisely the distal section [V(ii)] of domain V that happens to be protected from hydroxyl radicals by the 5′ exon (Figure 7D) and, according to the model in Figure 6, would be liable to stabilize the EBS1–IBS1 pairing by contacting the middle part of the EBS1 segment. An obvious advantage of having one of the intron–exon pairings lying alongside domain V would be to improve discrimination against mismatched substrates. Distortion of that part of the backbone of EBS1 that contacts domain V could not only further compromise substrate binding (i.e. beyond the energetic penalty associated with the mismatch itself), but might also affect catalysis if the resulting deformation were to extend to the active site, which we believe to be located at the other point of contact between domain V and the EBS1–IBS1 helix. The fact is that the related Sc.a5γ molecule has been shown to be rather intolerant of mismatched substrates: replacement of base pair 4 of the IBS1–EBS1 helix—the one closest to domain V according to our modeling—by a G:A mismatch not only increases Km by 240-fold (which is not too different from the expected effect of a single mismatch on duplex stability; Kierzek et al., 1999), but also reduces kcat by a factor of 60 (Xiang et al., 1998).
Somewhat surprisingly, mismatches in the EBS2–IBS2 pairing, which is situated further away from the active site, can be just as detrimental to binding and catalysis as those in the EBS1–IBS1 helix (Xiang et al., 1998). As suggested in Figure 6, the G-rich (Michel et al., 1989) shallow groove of helix ID(iv) could make extensive contacts to the backbone of the EBS2 segment of group IIB introns, thus playing much the same role in substrate discrimination as the shallow groove of helix V(ii). Our choice of having the EBS2 rather than the IBS2 strand facing the ribozyme reflects the fact that group II introns, which use both RNA and (during transposition) DNA substrates in vivo, appear insensitive in vitro to the presence of deoxyriboses in the 5′ exon, except at positions –1 and –2 (Griffin et al., 1995): those two riboses are the only ones to come near intron residues in our model. Assuming that discrimination is indeed ensured by helix ID(iv), effects on catalysis could be accounted for by invoking the δ–δ′ and EBS3–IBS3 base pairs, which connect the base of the ID(iv) stem to the active site. It will be of interest to investigate whether subgroup IIA introns, in which the EBS2 sequence is part of a terminal loop, are equally intolerant of mismatched EBS2–IBS2 pairings.
Finally, it remains to be determined whether in the Sc.a5γ subgroup IIB intron, which has been the prevalent in vitro model system for group II self-splicing until now, the residue that base pairs with the first nucleotide of the 3′ exon (an A) is the uridine counterpart of G293, as should be expected from our data, or, rather, the uridine located at the δ site, immediately 5′ of EBS1, as proposed by Jacquier and Jacquesson-Breuleux (1991). Although these authors were partly able to redirect cleavage at the 3′ site by mutating the δ site, they took care to stress that their data pointed to the first base of the 3′ exon being involved in yet another interaction with an unknown intron partner. In fact, the a5γ molecule is unique among group II introns in that its EBS1 segment appears to extend over seven rather than six nucleotides; this could allow the U at the δ site to pair either with the (adenine) counterpart of G195 (our δ′ site) or, by stacking on top of EBS1, with the first adenine of the 3′ exon. The matter is less anecdotal than it might seem, because the same sort of flexibility may well have existed at the time at which an ancestral group II molecule switched from direct to indirect guiding of exon ligation, or vice versa. Group II introns are proving surprisingly diverse in terms of the structural devices they use to carry out such essential steps as the selection of a proper 3′ splice site (this work) or the necessary rearrangement of part of their active center after the first step of splicing (Chanfreau and Jacquier, 1996; Costa et al., 1997a). The evolutionary versatility implied by this diversity makes it perhaps more credible that group II introns gave rise to the spliceosomal machinery, but also less likely that we will ever be able to retrace the steps they took in doing so.
Materials and methods
Kinetic analyses
DNA constructs and mutagenesis, as well as the synthesis and purification of intron transcripts internally labeled with 32P are described in Costa and Michel (1999). Kinetic analyses were carried out essentially as previously reported (Costa and Michel, 1999). Briefly, purified and renatured lariat intron or IE3 samples were incubated with a concentration of 13 nucleotide 5′ exon substitute in excess of ribozyme, after which all intron-containing products were electrophoresed and quantitated with a PhosphorImager (Molecular Dynamics). Ionic conditions were 1 M NH4Cl, 10 mM MgCl2 and either 40 mM MES pH 5.8 (for rate measurements) or 40 mM HEPES pH 7.0 (in Figure 5B, as well as for determinations of Kd, see Table II). Unless otherwise stated, the temperature was 40°C. Ribozyme concentration was set routinely at 20 nM after it had been checked that rates did not vary within a wide range of initial concentrations (2–50 nM). As shown in Table II, indirectly estimating the rate constants of debranching of lariat intron molecules and the final fraction of debranched product from reactions of IE3 molecules yields essentially the same values as direct measurements of intron debranching (Scheme 2) using purified lariat intron samples. This is consistent with the assumption that the final fraction of debranched intron reflects an equilibrium between debranching and branching (Chin and Pyle, 1995). Consequently, measuring the fraction of debranched product as a function of the concentration of 5′ exon allows access to Kd rather than Km (for equations and experimental procedures see Costa and Michel, 1999).
Chemical probing and reverse transcription
Modification reactions contained 13 nM lariat destined to be reverse transcribed in 50 µl of 40 mM Na–HEPES pH 7.6 at 37°C, 100 mM NH4Cl, 0.02% (w/v) SDS and either 10 mM MgCl2 (for footprinting experiments with the 3′dE5 oligonucleotide, Figure 2B) or 20 mM CaCl2 (to prevent reverse self-splicing in footprinting experiments with the ligated exons, Figure 2A; substitution of magnesium by calcium does not appear to perturb the overall tertiary structure of the intron, since it has no detectable effect on the pattern of modification by DMS; M.Costa, unpublished data). Base-specific probing reactions were performed at 30°C for 7 or 15 min in the presence of either DMS diluted to 1:600 or kethoxal diluted to 1.5 mg/ml. Hydroxyl radical probing conditions were 1 mM (NH4)2Fe(SO4)2, 2 mM Na2EDTA pH 8.0, 5 mM dithiothreitol (DTT) and 0.1% H2O2, for 5 min at 23°C in either the magnesium buffer detailed above (which ensures probing of the native tertiary structure) or in 40 mM Na–HEPES pH 7.6 at 37°C (which only allows stabilization of the secondary structure). Chemical probing reactions were stopped with 50 µl of either 0.7 M β-mercaptoethanol (DMS probing), 60 mM potassium borate pH 7.5 (kethoxal probing) or 20 mM thiourea (hydroxyl radical probing). All reactions were subsequently precipitated with 10 µg of bulk yeast tRNA, 10 µl of 3 M sodium acetate and 330 µl of ethanol, and pellets were washed with 70% ethanol prior to drying. Modified RNAs were resuspended in water (DMS- or hydroxyl radical-modified samples) or in 25 mM K+ borate pH 7.5 (kethoxal-modified samples) at a final concentration of 0.08 µM prior to reverse transcription.
In order to ensure that populations of molecules being probed were correctly and uniformly folded, RNA samples were renatured by addition of buffer at a temperature close to the melting range for tertiary structure (Costa et al., 1998), followed by slow cooling to the probing temperature and a further 15 min incubation. For footprinting experiments, appropriate concentrations of ligated exons or 5′ exon analogs in 1× concentrated buffer were added to renatured lariat molecules and incubation was continued for at least 20 min to ensure probing under equilibrium binding conditions.
Reverse transcription of modified or mock-treated RNA molecules with 5′-32P-labeled complementary DNA primers and subsequent treatment of the samples were performed as described in Costa et al. (1998). After denaturation by heating at 80°C for 10 min, samples were loaded onto denaturing polyacrylamide gels. Fixed and dried gels were quantitated with a PhosphorImager (Molecular Dynamics). In all experiments, the intron was fully scanned (from positions 7 to 2424) at least once by reverse transcription with the appropriate set of primers (importantly, the pattern of chemical modification revealed by reverse transcription was verified to be the same for differently located primers). For unknown reasons, some YG dinucleotides consistently give rise to a double stop after treatment with kethoxal (e.g. Figure 2B).
Comparative sequence analyses and modeling
The sequence alignments that formed the basis for Table III and the tables in Figure 1 are available from the authors. In generating the tables, introns inserted at homologous sites in related genomes were counted only once. Modeling was carried out with the program MANIP, as described by its authors (Massire and Westhof, 1998). Atomic coordinates of the model, refined with NUCLIN-NUCLSQ (Westhof et al., 1985) and used for generating Figure 6, have been deposited in the Protein Data Bank.
Acknowledgments
Acknowledgements
We are extremely grateful to Harry Noller in whose laboratory at the Center for Molecular Biology of RNA most of the experiments were carried out and who provided two of us (M.C. and F.M.) with generous and warm support throughout this work. We also thank Jay Nix for helping us to draw Figure 6, Alain Jacquier and Dan Herschlag for valuable critical reading of the manuscript, and James Murray for invaluable help with oligoribonucleotide synthesis. This work was supported by an NIH grant (#GM17129) to Harry Noller and a grant from the Programme Physique et Chimie du Vivant of the CNRS to F.M. and E.W. M.C. acknowledges a long-term fellowship from the Human Frontier Science Program.
Note added in proof
A Pl.LSU/2 precursor transcript missing intron nucleotides 171–182 was found to react to completion at 20 mM MgCl2, 1 M NH4Cl, 40 mM Tris pH 7.5 and 45°C with a half-time similar to its wild-type counterpart. Thus, fusion of stems ID(ii) and ID(iii) (Figure 3) into a single continuous helix is compatible with group II ribozyme function.
References
- Abramovitz D.L., Friedman,R.A. and Pyle,A.M. (1996) Catalytic role of 2′-hydroxyl groups within a group II intron active site. Science, 271, 1410–1413. [DOI] [PubMed] [Google Scholar]
- Balasubramanian B., Pogozelski,W.K. and Tullius,T.D. (1998) DNA strand breaking by the hydroxyl radical is governed by the accessible surface areas of the hydrogen atoms of the DNA backbone. Proc. Natl Acad. Sci. USA, 95, 9738–9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudvillain M. and Pyle,A.M. (1998) Defining functional groups, core structural features and inter-domain tertiary contacts essential for group II intron self-splicing: a NAIM analysis. EMBO J., 17, 7091–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G., Saint-Louis,D., Gray,M.W. and Lang,B.F. (1999) Complete sequence of the mitochondrial DNA of the red alga Porphyra purpurea. Cyanobacterial introns and shared ancestry of red and green algae. Plant Cell, 11, 1675–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson M. (1991) Ribbons 2.0. J. Appl. Crystallogr., 24, 958–961. [Google Scholar]
- Cate J.H., Gooding,A.R., Podell,E., Zhou,K., Golden,B.L., Kundrot,C.E., Cech,T.R. and Doudna,J.A. (1996) Crystal structure of a group I ribozyme domain: principles of RNA packing. Science, 273, 1678–1685. [DOI] [PubMed] [Google Scholar]
- Celander D.W. and Cech,T.R. (1990) Iron(II)-ethylenediaminetetraacetic acid catalyzed cleavage of RNA and DNA oligonucleotides: similar reactivity toward single- and double-stranded forms. Biochemistry, 29, 1355–1361. [DOI] [PubMed] [Google Scholar]
- Chanfreau G. and Jacquier,A. (1994) Catalytic site components common to both splicing steps of a group II intron. Science, 266, 1383–1387. [DOI] [PubMed] [Google Scholar]
- Chanfreau G. and Jacquier,A. (1996) An RNA conformational change between the two chemical steps of group II self-splicing. EMBO J., 15, 3466–3476. [PMC free article] [PubMed] [Google Scholar]
- Chin K. and Pyle,A.M. (1995) Branch-point attack in group II introns is a highly reversible transesterification, providing a potential proofreading mechanism for 5′-splice site selection. RNA, 1, 391–406. [PMC free article] [PubMed] [Google Scholar]
- Chiu D.K. and Kolodziejczak,T. (1991) Inferring consensus structure from nucleic acid sequences. Comput. Appl. Biosci., 7, 347–532. [DOI] [PubMed] [Google Scholar]
- Costa M. and Michel,F. (1995) Frequent use of the same tertiary motif by self-folding RNAs. EMBO J., 14, 1276–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M. and Michel,F. (1999) Tight binding of the 5′ exon to domain I of a group II self-splicing intron requires completion of the intron active site. EMBO J., 18, 1025–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M., Dème,E., Jacquier,A. and Michel,F. (1997a) Multiple tertiary interactions involving domain II of group II self-splicing introns. J. Mol. Biol., 267, 520–536. [DOI] [PubMed] [Google Scholar]
- Costa M., Fontaine,J.M., Loiseaux-de Goer,S. and Michel,F. (1997b) A group II self-splicing intron from the brown alga Pylaiella littoralis is active at unusually low magnesium concentrations and forms populations of molecules with a uniform conformation. J. Mol. Biol., 274, 353–364. [DOI] [PubMed] [Google Scholar]
- Costa M., Christian,E.L. and Michel,F. (1998) Differential chemical probing of a group II self-splicing intron identifies bases involved in tertiary interactions and supports an alternative secondary structure model of domain V. RNA, 4, 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dème E., Nolte,A. and Jacquier,A. (1999) Unexpected metal ion requirements specific for catalysis of the branching reaction in a group II intron. Biochemistry, 38, 3157–3167. [DOI] [PubMed] [Google Scholar]
- Ferat J.L. and Michel,F. (1993) Group II self-splicing introns in bacteria. Nature, 364, 358–361. [DOI] [PubMed] [Google Scholar]
- Griffin E.A.J., Qin,Z., Michels,W.J.J. and Pyle,A.M. (1995) Group II intron ribozymes that cleave DNA and RNA linkages with similar efficiency and lack contacts with substrate 2′-hydroxyl groups. Chem. Biol., 2, 761–770. [DOI] [PubMed] [Google Scholar]
- Guo H., Zimmerly,S., Perlman,P.S. and Lambowitz,A.M. (1997) Group II intron endonucleases use both RNA and protein subunits for recognition of specific sequences in double-stranded DNA. EMBO J., 16, 6835–6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier A. (1996) Group II introns: elaborate ribozymes. Biochimie, 78, 474–487. [DOI] [PubMed] [Google Scholar]
- Jacquier A. and Jacquesson-Breuleux,N. (1991) Splice site selection and role of the lariat in a group II intron. J. Mol. Biol., 219, 415–428. [DOI] [PubMed] [Google Scholar]
- Jacquier A. and Michel,F. (1987) Multiple exon-binding sites in class II self-splicing introns. Cell, 50, 17–29. [DOI] [PubMed] [Google Scholar]
- Jestin J.L., Deme,E. and Jacquier,A. (1997) Identification of structural elements critical for inter-domain interactions in a group II self-splicing intron. EMBO J., 16, 2945–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierzek R., Burkard,M.E. and Turner,D.H. (1999) Thermodynamics of single mismatches in RNA duplexes. Biochemistry, 38, 14214–14223. [DOI] [PubMed] [Google Scholar]
- Konforti B.B., Abramovitz,D.L., Duarte,C.M., Karpeisky,A., Beigelman,L. and Pyle,A.M. (1998a) Ribozyme catalysis from the major groove of group II intron domain 5. Mol. Cell, 1, 433–441. [DOI] [PubMed] [Google Scholar]
- Konforti B.B., Liu,Q. and Pyle,A.M. (1998b) A map of the binding site for catalytic domain 5 in the core of a group II intron ribozyme. EMBO J., 17, 7105–7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham J.A. and Cech,T.R. (1989) Defining the inside and outside of a catalytic RNA molecule. Science, 245, 276–282. [DOI] [PubMed] [Google Scholar]
- Massire C. and Westhof,E. (1998) MANIP: an interactive tool for modelling RNA. J. Mol. Graph. Model., 16, 197–205, 255–257. [DOI] [PubMed] [Google Scholar]
- Michel F. and Ferat,J.L. (1995) Structure and activities of group II introns. Annu. Rev. Biochem., 64, 435–461. [DOI] [PubMed] [Google Scholar]
- Michel F. and Jacquier,A. (1987) Long-range intron–exon and intron–intron pairings involved in self-splicing of class II catalytic introns. Cold Spring Harbor Symp. Quant. Biol., 52, 201–212. [DOI] [PubMed] [Google Scholar]
- Michel F., Umesono,K. and Ozeki,H. (1989) Comparative and functional anatomy of group II catalytic introns—a review. Gene, 82, 5–30. [DOI] [PubMed] [Google Scholar]
- Mohr G., Smith,D., Belfort,M. and Lambowitz,A.M. (2000) Rules for DNA target-site recognition by a lactococcal group II intron enable retargeting of the intron to specific DNA sequences. Genes Dev., 14, 559–573. [PMC free article] [PubMed] [Google Scholar]
- Peebles C.L., Zhang,M., Perlman,P.S. and Franzen,J.S. (1995) Catalytically critical nucleotides in domain 5 of a group II intron. Proc. Natl Acad. Sci. USA, 92, 4422–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pley H.W., Flaherty,K.M. and McKay,D.B. (1994) Model for an RNA tertiary interaction from the structure of an intermolecular complex between a GAAA tetraloop and an RNA helix. Nature, 372, 111–113. [DOI] [PubMed] [Google Scholar]
- Podar M., Perlman,P.S. and Padgett,R.A. (1998) The two steps of group II intron self-splicing are mechanistically distinguishable. RNA, 4, 890–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E., Dumas,P. and Moras,D. (1985) Crystallographic refinement of yeast aspartic acid transfer RNA. J. Mol. Biol., 184, 119–145. [DOI] [PubMed] [Google Scholar]
- Xiang Q., Qin,P.Z., Michels,W.J., Freeland,K. and Pyle,A.M. (1998) Sequence specificity of a group II intron ribozyme: multiple mechanisms for promoting unusually high discrimination against mismatched targets. Biochemistry, 37, 3839–3849. [DOI] [PubMed] [Google Scholar]