Abstract
Variations in the intein-mediated protein splicing mechanism are becoming more apparent as polymorphisms in conserved catalytic residues are identified. The conserved Ser or Cys at the intein N-terminus and the conserved intein penultimate His are absent in the KlbA family of inteins. These inteins were predicted to be inactive, since an N-terminal Ala cannot perform the initial reaction of the standard protein splicing pathway to yield the requisite N-terminal splice junction (thio)ester. Despite the presence of an N-terminal Ala and a penultimate Ser, the KlbA inteins splice efficiently using an alternative protein splicing mechanism. In this non-canonical pathway, the C-extein nucleophile attacks a peptide bond at the N-terminal splice junction rather than a (thio)ester bond, alleviating the need to form the initial (thio)ester at the N-terminal splice junction. The remainder of the two pathways is the same: branch resolution by Asn cyclization is followed by an acyl rearrangement to form a native peptide bond between the ligated exteins.
Keywords: catalytic mechanism/intein/KlbA/Methanococcus jannaschii/protein splicing
Introduction
Inteins are intervening sequences that are post-translationally spliced from precursor proteins. Upon excision of the intein, a native peptide bond is formed between the exteins (the protein fragments that formerly surrounded the intein). The intein splicing domain is composed of two regions that flank a second intein domain consisting of a small linker or a homing endonuclease (Belfort and Roberts, 1997; Duan et al., 1997; Hall et al., 1997; Klabunde et al., 1998; Perler, 1998). As first described in mobile introns, the embedded homing endonuclease can initiate intein gene mobility through double-strand break–repair pathways (Belfort and Roberts, 1997; Gimble, 2000). The intein splicing domain plus the first C-extein residue direct the self-catalytic protein splicing mechanism, which consists of four steps involving three conserved nucleophiles: (i) Ser, Thr or Cys at the intein N-terminus [although Thr has yet to be found at this position, it has been shown to substitute for Ser in one intein (Hodges et al., 1992)]; (ii) Asn or Gln at the intein C-terminus; and (iii) Ser, Thr or Cys at the beginning of the C-extein (Pietrokovski, 1994, 1998a,b; Dalgaard et al., 1997; Perler et al., 1997; Noren et al., 2000; Perler, 2000).
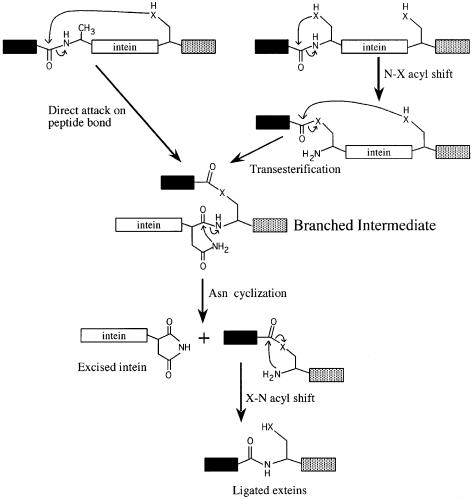
The mechanism of protein splicing (Figure 1) has been extensively reviewed (Noren et al., 2000). In step 1, the conserved Ser or Cys at the intein N-terminus undergoes an acyl rearrangement to form a (thio)ester at the N-terminal splice junction. This (thio)ester bond is then attacked by the Ser, Thr or Cys at the C-terminal splice junction, resulting in cleavage of the N-terminal splice junction, ligation of the exteins and formation of a branched (thio)ester intermediate. The branched intermediate is resolved when Asn or Gln cyclizes, breaking the peptide bond at the C-terminal splice junction. As a result of a second acyl rearrangement, the (thio)ester bond between the exteins is replaced by a normal peptide bond. Reactions at the N-terminal splice junction are facilitated by an oxyanion hole, which increases the electrophilicity of the carbonyl carbon of the scissile bond and includes several amino acids (aa) from intein block B (Pietrokovski, 1994, 1998b; Dalgaard et al., 1997; Duan et al., 1997; Kawasaki et al., 1997; Perler et al., 1997; Klabunde et al., 1998; Noren et al., 2000; Poland et al., 2000). The conserved intein penultimate His is thought to assist in Asn and Gln cyclization (Cooper et al., 1993; Xu and Perler, 1996; Duan et al., 1997; Klabunde et al., 1998; Chen et al., 2000; Noren et al., 2000; Poland et al., 2000). Each step in the standard protein splicing mechanism has been demonstrated to be required in inteins with consensus residues in conserved motifs (Noren et al., 2000).
Fig. 1. The standard and alternative self-catalytic protein splicing mechanisms. Inteins that begin with Ala can splice by a modified mechanism that was originally disproven for standard inteins with conserved nucleophiles. The two pathways differ in the reactions required to form the branched intermediate. In the alternative protein splicing mechanism (left), a direct nucleophilic attack on the N-terminal splice junction peptide bond by the C-extein nucleophile replaces the first two steps of the standard intein-mediated protein splicing pathway (right). Resolution of the branched intermediate and formation of a peptide bond between the exteins is the same in both mechanisms. The intein C-terminal Asn can be regenerated by hydrolysis of the succinimide ring. In some inteins, Asn is replaced by Gln, which can undergo a similar cyclization reaction (Pietrokovski, 1998a). The oxygen or the sulfur present in the side chain of Ser, Thr or Cys is represented by an ‘X’, the black box represents the N-extein and the stippled box represents the C-extein. All tetrahedral intermediates, assisting groups and proton transfer steps are omitted for clarity. The standard intein-mediated protein splicing mechanism and its proof have been recently reviewed in Noren et al. (2000).
In the last 10 years, over 100 inteins have been identified in all three domains of life, including bacteriophage and a eukaryotic virus [see the Intein Registry in InBase at http://www.neb.com/neb/inteins.html (Perler, 2000)]. Polymorphism in catalytic residues (including residues incapable of participating in the requisite chemical reactions) has become more evident as more inteins have been sequenced (Dalgaard et al., 1997; Perler et al., 1997; Pietrokovski, 1998a,b; Perler, 2000). Three families of intein alleles, present in the KlbA protein (Dalgaard et al., 1997; Gorbalenya, 1998; Pietrokovski, 1998b), the DnaB helicase insertion site dnab-2 and the Snf2 helicase, have an Ala at position 1 instead of the conserved Ser, Thr or Cys (Table I) (Perler, 2000). In all inteins tested to date, splicing is blocked when a residue other than Ser, Thr or Cys is substituted at the intein N-terminus (Hodges et al., 1992; Cooper et al., 1993; Xu and Perler, 1996; Evans et al., 1999; Mathys et al., 1999; Wood et al., 1999). The Methanococcus jannaschii KlbA intein was the first intein identified with an N-terminal Ala and it was predicted to be inactive (Gorbalenya, 1998; Pietrokovski, 1998b). The 168 aa M.jannaschii KlbA intein also lacks a penultimate His, but retains other conserved intein residues. Subsequently, intein alleles were found in KlbA genes from several archaea: Pyrococcus furiosus (Maeder et al., 1999), P.horikoshii OT3 (Kawarabayasi et al., 1998) and P.abyssi (Perler, 2000). Unlike the M.jannaschii (168 aa) and P.abyssi (196 aa) KlbA inteins, the P.furiosus (522 aa) and P.horikoshii OT3 (520 aa) KlbA inteins contain a homing endonuclease domain. It is unlikely that the remainder of their signature motifs would be maintained if these inteins were inactive (unless they had all recently lost their N-terminal Ser or Cys). The function of KlbA is unknown, but it is part of the kilB operon (orfA), which is thought to be involved in type II secretory pathways (Dalgaard et al., 1997).
Table I. Inteins that begin with Ala.
| Inteina | Size (aa) | Insertion site | N-terminus | Block B | C-terminus |
|---|---|---|---|---|---|
| Pho KlbA | 520 | klba-a | GHDG/ALYDFS | GNEVILTRSHPLFA | NGILVSN/CMGT |
| Pab KlbA | 196 | klba-a | GHDG/ALYYFS | GKELVLTQDHPVFV | NGIVVSN/CMGT |
| Pfu KlbA | 522 | klba-a | GHDG/ALYDFS | GNEIILTRNHPLAF | NGILVSN/CMGT |
| Mja KlbA | 168 | klba-a | GHDG/ALAYDE | RREITLTHDHPVYI | EGFAVSN/CSGT |
| Mav DnaB | 337 | dnaB-b | GVGK/ALALDT | GTVIVADAAHQWLT | AMVPTHN/STLG |
| Mle DnaB | 145 | dnaB-b | GVGK/ALALDT | GTVIVADAQHQWPT | GMVPTHN/STLG |
| Dra Snf2 | 343 | snf2-a | GLGK/AQPLDA | GASVEADAEHLWNV | GYIVTHN/TLQT |
The number of amino acids in each intein is indicated, as is the insertion site in the extein (Pietrokovski, 1998b; Perler, 2000). Splice junctions are indicated with a slash (/). Amino acid sequences are listed using the single-letter code.
aDra, Deinococcus radiodurans; Mav, Mycobacterium avium; Mle, Mycobacterium leprae; Pab, P.abyssi; Pho, P.horikoshii OT3; Pfu, P.furiosus; DnaB, DnaB helicase; Snf2, Snf2 helicase. See the InBase database for complete intein motif sequences and references (Perler, 2000).
Dalgaard et al. (1997) suggested that the M.jannaschii KlbA intein could splice by a non-canonical pathway first described by Xu et al. (1994) in which the C-extein Ser, Thr or Cys directly attacks a peptide bond at the N-terminal splice junction, instead of a (thio)ester bond (Figure 1). This study examines whether KlbA inteins can splice in Escherichia coli and defines an alternative protein splicing pathway for the M.jannaschii KlbA intein in the absence of an intein N-terminal nucleophile.
Results
Splicing of KlbA inteins
The study of protein splicing has been facilitated by cloning intein genes into model precursors that allow easy purification and analysis based on size, antibody recognition and affinity tags. One such system, the maltose-binding protein–intein–paramyosin (MIP) precursor, consists of an intein inserted in-frame between the E.coli maltose-binding protein (MBP) and the ΔSal fragment of Dirofilaria immitis paramyosin, and has previously been used to study splicing of four inteins (Xu et al., 1993; Telenti et al., 1997; Chen et al., 2000). The M.jannaschii KlbA intein was cloned into the MIP system with a single native N-extein residue (Gly) and three native C-extein residues (Cys-Ser-Gly). Amino acids in the precursor are numbered as follows: (i) numbering of intein residues begins at its N-terminus with 1; (ii) numbering of C-extein residues begins at its N-terminus with +1 and includes the plus sign to denote C-extein residues; and (iii) the N-extein residue adjacent to the N-terminal splice site is numbered –1.
When expressed at either 37 or 15°C, the M.jannaschii KlbA intein MIP fusion protein spliced to completion in vivo, as indicated by the presence of spliced MBP–paramyosin (MP) (72 kDa) and free KlbA intein, and the absence of MIP precursor or single splice junction cleavage products (Figure 2). Although the predicted molecular mass of the M.jannaschii KlbA intein is 18 kDa, its apparent molecular mass in SDS–PAGE was 22 kDa. This difference in relative mobility may be due to intrinsic properties of the protein or to incomplete denaturation of a protein from an extreme thermophile. Aberrant mobility was also observed in the intein–paramyosin (IP) fragment (see below). The identity of MP was confirmed by western blot analysis with anti-MBP and anti-paramyosin sera (Xu et al., 1993), and the identity of both MP and the KlbA intein were confirmed by N-terminal amino acid sequence (data not shown). Splicing was proven by sequencing across the extein ligation site in MP. Factor Xa protease digestion of ligated MP generated 43 and 29 kDa fragments (Figure 2). The 43 kDa Factor Xa cleavage product yielded the predicted MBP N-terminal sequence and the 29 kDa product yielded a sequence that begins at the Factor Xa cleavage site in the N-extein (GTLEG), spans the splice junction (G/C) and continues into the C-extein for 15 aa. The ability to sequence through the splice junction by Edman degradation indicates that the exteins are connected by a standard peptide bond.
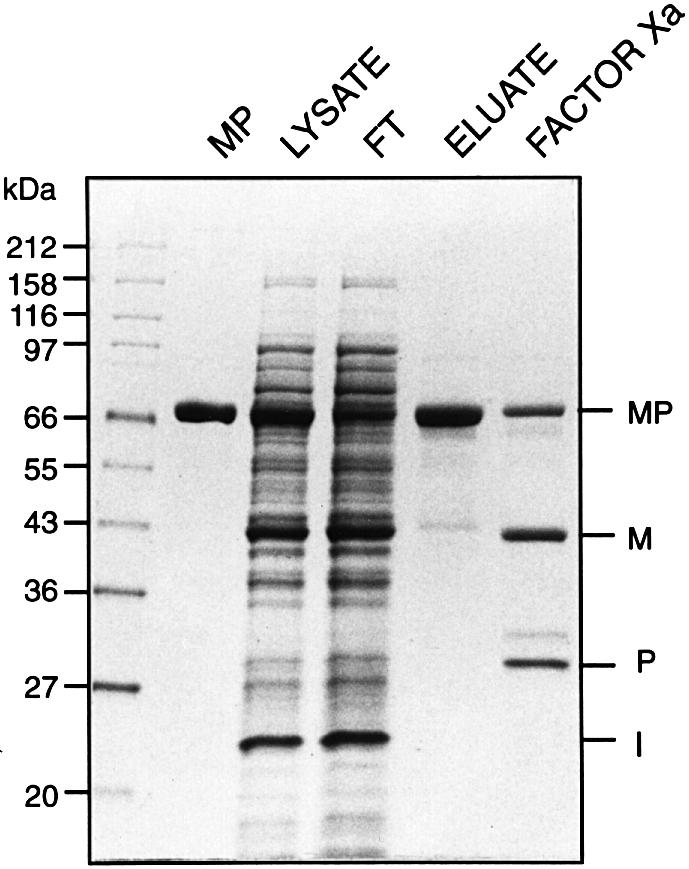
Fig. 2. Protein splicing of the M.jannaschii KlbA intein in MIP. Expression of the MIP precursor in E.coli results in production of spliced exteins (MP) plus free intein (I). MP lane: a control MBP::paramyosin fusion protein. Lysate lane: soluble cell extract from cultures induced for 2 h at 37°C. FT and eluate lanes: MP was purified by affinity chromatography over amylose resin to yield spliced MP in the eluate, while free intein remained in the flow through (FT). Only proteins with a MBP domain will bind to amylose columns. Various cell lysates contained differing amounts of a highly expressed protein that co-migrated with MBP, but did not bind to amylose resin. Factor Xa lane: the eluate treated with Factor Xa protease. The 30 kDa unlabeled band in the Factor Xa lane is the large subunit of the protease. SDS–polyacrylamide gels were stained with Coomassie Blue.
KlbA genes from several archaea, including all sequenced Thermococcales strains, contain allelic inteins inserted at the same site in the klbA gene (Dalgaard et al., 1997; Kawarabayasi et al., 1998; Pietrokovski, 1998b; Maeder et al., 1999; Perler, 2000). Primers for polymerase chain reaction (PCR) amplification were designed based on these sequences to examine more Thermococcales strains for the presence of klbA intein genes. A 2.1 kb fragment (similar in size to the expected product from the P.furiosus klbA intein) was amplified from Thermococcales strains GB-C, GI-J and GB-D (Jannasch et al., 1992; Southworth et al., 1996). DNA sequencing of the PCR products from strains GB-D, GI-J and GB-C indicated that these intein genes were inserted at the same position in the klbA gene as the other klbA inteins. The encoded residues from all three strains were identical to those of the P.furiosus KlbA intein at the N-terminal (G/ALYDFSVIQ) and C-terminal (NYVANGILVSN/C) splice junctions. Splicing of the Pyrococcus sp. GB-D KlbA intein in MIP was observed in the presence of 163 aa of native N-extein and 30 aa of native C-extein (data not shown). Since proteolytic degradation of the precursor made it difficult to identify precursor and products, no further characterization was attempted with the Pyrococcus sp. GB-D KlbA intein.
Mutation of the M.jannaschii KlbA intein N-terminal Ala1 and C-extein Cys+1
Both protein splicing pathways depicted in Figure 1 predict that the C-extein nucleophile (Cys+1) cleaves the N-terminal splice junction to form a branched intermediate. In the standard pathway, Cys+1 attacks a (thio)ester bond, whereas in the alternative pathway, it attacks an amide bond. In either scenario, mutation of Cys+1 should block N-terminal splice junction cleavage and branched intermediate formation, but not C-terminal splice junction cleavage by Asn cyclization. To test this hypothesis, Cys+1 in the M.jannaschii KlbA intein MIP precursor was mutated to Ala. As predicted, both N-terminal splice junction cleavage and branch intermediate formation were blocked in vivo and after overnight incubation in vitro of amylose purified protein (Figure 3; Table II). Overnight incubation resulted in C-terminal splice junction cleavage, yielding MBP–intein (MI) and paramyosin. Similar results were obtained with a Cys+1Ser mutant, indicating that Ser cannot substitute for Cys in this context (Table II).
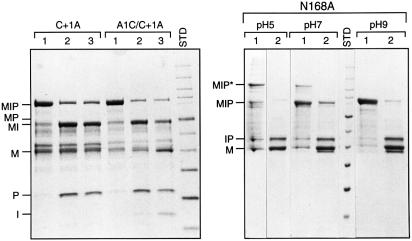
Fig. 3. Mutation of Ala1, Asn168 and Cys+1 in MIP. Left panel: mutation of Cys+1 blocks splicing and formation of the DTT-cleavable branched thioester intermediate, while substitution of the intein N-terminal Ala with Cys (in the absence of Cys+1) results in generation of a DTT-cleavable thioester bond at the N-terminal splice junction. MBP-containing proteins from soluble lysates of the Cys+1Ala (C+1A) MIP single mutant or the Ala1Cys Cys/1Ala (A1C/C+1A) MIP double mutant were purified over amylose resin (lane 1) and then incubated overnight at room temperature in the absence (lane 2) or presence (lane 3) of 50 mM DTT. SDS–polyacrylamide gels were stained with Coomassie Blue. Right panel: mutation of Asn168 to Ala blocks splicing and C-terminal splice junction cleavage, resulting in accumulation of the branched intermediate (MIP*), especially at lower pH. MBP-containing proteins from Asn168Ala samples were purified over amylose resin, after which the pH was adjusted to the indicated values (lane 1). The samples were then treated with 20 mM DTT for 20 min (lane 2). The thioester bond in the branched intermediate was rapidly cleaved by 20 mM DTT to form MBP and IP. Aggregation was observed in the absence of DTT in the sample buffer (lane 1), especially at lower pH. At higher pH, varying proportions of IP showed aberrant migration patterns, migrating slightly faster than MBP. SDS–polyacrylamide gels were stained with Coomassie Blue.
Table II. The effects of mutation on splicing and cleavage.
| Residue | Mutation | MIP precursor | Spliced MP | C-terminal cleavage | N-terminal cleavage | DTT cleavage |
|---|---|---|---|---|---|---|
| MIP | none | – | >90% | – | – | n.d. |
| Ala1 | Cys, Ser | – | >90% | + | – | n.d. |
| Gly | + | – | >90% | – | – | |
| Thr91 | Ala | + | >90% | – | – | n.d. |
| Thr93 | Ala | + | ++ | +++ | – | – |
| His94 | Ala | + | >90% | – | – | n.d. |
| Asp95 | Ala | – | >90% | + | – | n.d. |
| His96 | Ala | >90% | – | + | – | – |
| Ser167 | His | – | ++ | +++ | – | – |
| Asn168 | Ala | >90% | – | – | – | >90% |
| Cys+1 | Ala,Ser | >90% | – | + | – | – |
| Ala1/Cys+1 | Ser/Ala | >90% | – | + | – | – |
| Cys/Ala | >90% | – | + | – | + | |
| Ser+2 | Ala | +++ | ++ | – | – | + |
Quantification of the molar percentages of M.jannaschii KlbA intein MIP precursor and products after induction at 37°C or overnight treatment with 50 mM DTT in single mutants or the Ala1/Cys+1 double mutant. The Ser+2Ala mutant spliced slowly, with >90% spliced product formed after overnight incubation in vitro at room temperature.
–, <5%; +, 5–25%; ++, 26–50%; +++, 51–75%; n.d., not determined due to >90% in vivo splicing.
Dithiothreitol (DTT) has been used to demonstrate both linear and branched thioester intermediates from steps 1 and 2 of the protein splicing pathway. It specifically cleaves thioester bonds in proteins under mild conditions, but not oxygen esters or amide bonds, and it drives the formation of the thioester by product elimination (Chong et al., 1996; Xu and Perler, 1996). This technique could not be applied to the wild-type M.jannaschii KlbA intein because no expression conditions were found that allowed purification of the MIP precursor. In the Cys+1Ala and Cys+1Ser mutants, DTT failed to cleave the mutated precursors (Figure 3; Table II).
The effect of substitutions at the M.jannaschii KlbA intein N-terminus depended on the amino acid present (Table II). Mutation of Ala1 to Gly blocked splicing and thiol induced cleavage. ‘Reversion’ of Ala1 to Ser or Cys yielded >90% spliced product. Since Cys1 did not interfere with splicing, the Ala1Cys mutant was examined to see if Cys1 was capable of undergoing the acyl shift that standard inteins use as the first step in the splicing pathway. In order to examine this question, Cys+1 had to be mutated to eliminate production of MBP and IP resulting from DTT cleavage of the thioester in the branched intermediate. The products formed by the Ala1Cys plus Cys+1Ala double mutant were similar to the Cys+1Ala single mutant, with MIP predominating in vivo and C-terminal splice junction cleavage predominating after overnight incubation in vitro (Figure 3; Table II). Incubation with 50 mM DTT resulted in N-terminal splice junction cleavage to release MBP. These results suggest that, in the absence of Cys+1 and splicing, Cys1 can undergo the standard intein acyl shift. An Ala1Ser/Cys+1Ala double mutant yielded the same products as the Ala1Cys/Cys+1Ala double mutant, with the exception that DTT treatment did not result in cleavage (Table II). This latter result was expected, since DTT does not cleave amide bonds or potential oxygen esters under these conditions (Chong et al., 1996; Xu and Perler, 1996).
Mutation of M.jannaschii KlbA intein block B residues
Mutation and structural data suggest that residues in intein block B facilitate reactions at the N-terminal splice junction (Duan et al., 1997; Kawasaki et al., 1997; Perler et al., 1997; Klabunde et al., 1998; Pietrokovski, 1998b; Noren et al., 2000; Poland et al., 2000). Residues in intein block B were examined to see if they also facilitated attack by the C-extein nucleophile in the M.jannaschii KlbA intein. The most highly conserved residues in block B are a Thr and a His separated by 2 aa (Table I) (Pietrokovski, 1994, 1998b; Dalgaard et al., 1997; Perler et al., 1997; Perler, 2000). The M.jannaschii KlbA intein has two possible Thr-x-x-His motifs beginning at Thr91 and Thr93. Ala scanning was performed on the following block B residues: Thr91, Thr93, His94, Asp95 and His96 (Figure 4; Table II). Mutation of Thr91 and His94 had no observable effect on splicing, while mutation of Thr93, Asp95 and His96 inhibited splicing to varying degrees. Substitution of Thr93 resulted in mostly C-terminal splice junction cleavage and a small amount of spliced product. In the Asp95Ala sample, >90% of the precursor spliced, but a small amount of C-terminal splice junction cleavage (MI + paramyosin) occurred. The His96 mutation had the largest effect on splicing, with >90% of the MIP precursor remaining intact in vivo and after overnight incubation in vitro. Although expression of the His96Ala precursor at 37°C did not result in C-terminal splice junction cleavage, expression at 15°C yielded approximately equal amounts of precursor and C-terminal splice junction cleavage products (data not shown). Overnight incubation with 50 mM DTT had no effect on the His96Ala mutant, indicating that this precursor was unable to form the branched thioester intermediate.
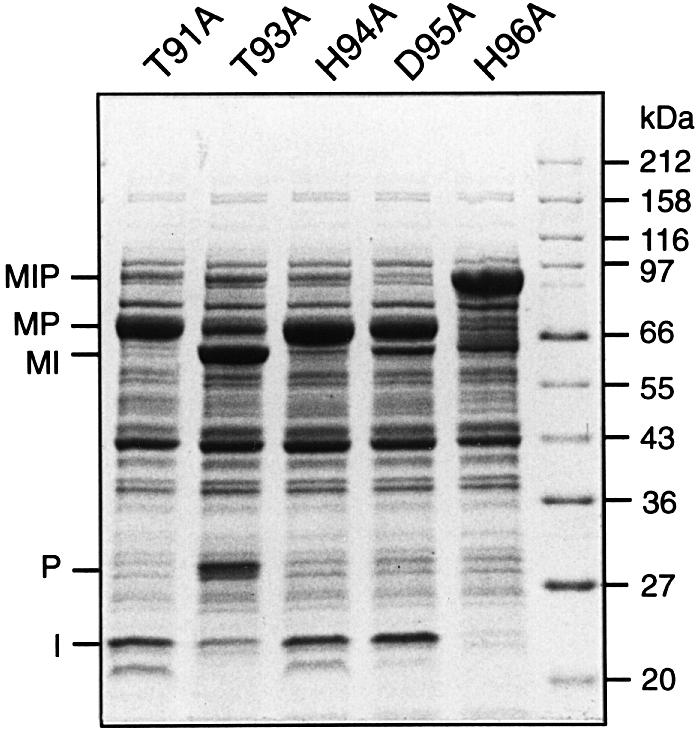
Fig. 4. Mutation of residues in the M.jannaschii KlbA intein block B. Ala scanning of residues in intein block B demonstrates that Thr93 and His96 are important for splicing, while Asp95 plays only a minor role. SDS–polyacrylamide gel of soluble cell lysates induced at 37°C was stained with Coomassie Blue. Lanes are labeled with the block B mutation present in MIP, using the single-letter amino acid code.
Mutation of the M.jannaschii KlbA intein C-terminal Asn
Asn cyclization is required for resolution of the branched intermediate by C-terminal splice junction cleavage in both protein splicing pathways (Figure 1). As predicted, mutation of Asn168 to Ala blocked splicing and C-terminal splice junction cleavage (Figure 3; Table II). Unlike other Cys+1 inteins (Hirata et al., 1990; Kane et al., 1990; Davis et al., 1991, 1994; Chong et al., 1998), small amounts of a slowly migrating branched intermediate (MIP*) were occasionally observed with the M.jannaschii KlbA intein at neutral pH. Several lines of evidence indicated that this slowly migrating protein was the branched intermediate: (i) it reacted with anti-MBP and anti-paramyosin sera (data not shown); (ii) at pH 5, 60% of MIP* was converted to MBP + IP in 5 min by 20 mM DTT, with 100% cleavage at 20 min; and (iii) the expected pair of residues from the two predicted N-termini were present in each cycle of Edman degradation. The equilibrium between the MIP precursor and the MIP* branched intermediate could be shifted towards MIP* in the Asn168Ala mutant by decreasing the pH to 5, or towards MIP by increasing the pH to 9 (Figure 3). Although the equilibrium favored the precursor at pH 9, product elimination by DTT was still able drive thioester formation and cleavage.
Mutation of Ser167 and Ser+2
The KlbA inteins have a penultimate Ser rather than the conserved His. Splicing of most previously examined inteins that naturally lack a penultimate His was more efficient in E.coli when the penultimate position was ‘reverted’ to His (Huang et al., 1994; Scott et al., 1999; Chen et al., 2000). In the M.jannaschii KlbA intein, substitution of Ser167 with His inhibited splicing, yielding approximately equal amounts of spliced MP and C-terminal splice junction cleavage products (Table II).
Proximal extein residues affect splicing of many inteins. Mutation of Ser+2 to Ala slowed splicing, requiring incubation in vitro at room temperature to complete the splicing reaction (Table II and data not shown).
Discussion
When we first attempted to decipher the intein-mediated protein splicing mechanism, we examined two equally plausible pathways leading to the branched intermediate (Figure 1) (Xu et al., 1994; Shao et al., 1996; Xu and Perler, 1996). In a reaction similar to that used by serine and cysteine proteases, the C-extein nucleophile could directly attack a peptide bond at the N-terminal splice junction. Alternatively, the C-extein nucleophile could attack a (thio)ester previously formed by an acyl rearrangement of the intein N-terminal Ser, Thr or Cys. This latter pathway provided a role for the conserved intein N-terminal nucleophile. Subsequent splicing studies demonstrated an absolute requirement for a Ser, Thr or Cys at the N-terminus of every intein tested (Hodges et al., 1992; Cooper et al., 1993; Xu and Perler, 1996; Evans et al., 1999; Mathys et al., 1999; Wood et al., 1999). The presence of an N-terminal splice junction (thio)ester was proven by several biochemical and biophysical approaches (Chong et al., 1996; Shao et al., 1996; Xu and Perler, 1996; Noren et al., 2000). Taken together, all of the previous data support the four-step protein splicing mechanism requiring a linear (thio)ester intermediate and ruled out the alternative pathway involving a direct attack on the peptide bond at the N-terminal splice junction. Due to the difference in functionality, Ala1 cannot form the initial linear (thio)ester intermediate, so inteins that begin with Ala are unable to splice by the consensus protein splicing mechanism. However, this study demonstrates that at least one family of inteins beginning with Ala can splice. In fact, splicing of the M.jannaschii KlbA intein was so efficient that <5% precursor remained when the wild-type intein was expressed in a model precursor under all experimental conditions (as assayed in SDS–PAGE gels stained with Coomassie Blue). Many standard inteins fail to splice completely in similar situations. Splicing was also observed with the larger Pyrococcus sp. GB-D KlbA intein, which has a homing endonuclease domain, suggesting that splicing is not affected by the type of intein central domain (linker or endonuclease).
All of the mutagenesis data indicate that the mechanism of protein splicing in the M.jannaschii KlbA intein is the same as standard inteins except that the C-extein nucleophile (Cys+1) attacks a peptide bond at the N-terminal splice junction instead of a (thio)ester bond. As in other inteins, the M.jannaschii KlbA intein C-terminal Asn168 is required for branch resolution by C-terminal splice junction cleavage. In the absence of Cys+1, no DTT cleavable thioester is formed, no N-terminal splice junction cleavage occurs and no branched intermediate is formed. The M.jannaschii KlbA intein branched intermediate is identical to standard branched intermediates, having the same two N-termini (the N-extein and the intein), the same (thio)ester linkage between the N-extein and the remainder of the precursor, and the same branch point (the +1 residue at the beginning of the C-extein). The pH characteristics of the equilibrium between the precursor and the branched intermediate were similar in the M.jannaschii KlbA intein Asn168Ala mutant and the Pyrococcus sp. GB-D DNA polymerase intein (Xu et al., 1993). The observation that this equilibrium is shifted towards the branched intermediate at lower pH suggests that protonation of the leaving group in the tetrahedral intermediate is the rate-limiting step in branch formation. The M.jannaschii KlbA intein has apparently retained the capacity to form a linear thioester if Cys is present at its N-terminus and splicing is blocked by mutation of Cys+1 to Ala. However, the slow rate and low yield of DTT cleavage products from this Ala1Cys Cys+1Ala mutant suggest that splicing probably proceeds by the alternative pathway in the Ala1Cys precursor. The presence of a Ser as the second residue in the C-extein (Ser+2) also raised the possibility that this residue might somehow substitute for the missing N-terminal nucleophile. However, mutation of Ser+2 to Ala did not block splicing.
Mutation of Thr93 and His96 in block B inhibited splicing and thiol cleavage at the N-terminal splice junction, but not C-terminal splice junction cleavage (His96Ala exhibited C-terminal splice junction cleavage when expressed at 15°C). This suggests that these residues facilitate reactions at the M.jannaschii KlbA intein N-terminal splice junction. This is consistent with mutagenesis and structural data from standard inteins (Duan et al., 1997; Hall et al., 1997; Kawasaki et al., 1997; Klabunde et al., 1998; Poland et al., 2000). The crystal structure of the Mycobacterium xenopi gyrase A intein shows the equivalent residues from block B (Thr72, His75) in position to assist in nucleophilic displacement reactions at the N-terminal splice junction (Klabunde et al., 1998). Similar results were observed with the Saccharomyces cerevisiae VMA intein (Duan et al., 1997; Poland et al., 2000). Although the residue equivalent to Asp95 in the M.xenopi gyrase A and S.cerevisiae VMA inteins was in position to facilitate reactions at the N-terminal splice junction, mutation of Asp95 to Ala in the M.jannaschii KlbA intein had little effect.
In previous studies of inteins that naturally lack a penultimate His, splicing of three out of four inteins improved when the penultimate position was ‘reverted’ to His, indicating that these inteins had not fully adapted to the loss of this conserved residue (Huang et al., 1994; Wu et al., 1998; Scott et al., 1999; Chen et al., 2000). In the one exception, substitution of the native M.jannaschii RNA polymerase A′ intein penultimate residue by His resulted in N-terminal splice junction cleavage, suggesting that His now inhibited Asn cyclization (Chen et al., 2000). The M.jannaschii KlbA intein is the second example of an intein in which splicing is reduced when the penultimate residue is reverted to His. However, this time a penultimate His causes Asn cyclization prior to branch formation. It is possible that, in order to balance a potentially less efficient or slower rate of branch formation in the M.jannaschii KlbA intein, an equally inefficient Asn cyclization step is preferred, which may be provided by the absence of a penultimate His.
The KlbA inteins have overcome the barriers to direct nucleophilic attack on the peptide bond at the N-terminal splice junction that are present in previously studied inteins with Ser or Cys at their N-terminus. It is unclear why other inteins cannot perform similar reactions, since the block B oxyanion hole is still available to facilitate direct attack on the N-terminal splice junction. Possibly, (thio)ester formation may be necessary in standard inteins to align the C-extein nucleophile, to remove steric hindrance or to induce a conformational shift that results in an increase in the nucleophilicity of Cys+1. The crystal structure of a S.cerevisiae VMA intein precursor (Poland et al., 2000) has helped to resolve this question by revealing that Cys+1 is too far away to attack directly either a peptide or a thioester bond at the N-terminal splice junction, leading the authors to suggest that inteins must undergo a conformational shift to allow attack by the Cys+1 nucleophile. We propose that in the M.jannaschii KlbA intein and other Ala1 inteins, Cys+1 is already in position to attack the N-terminal splice junction amide bond. Substitution of Ala1 by the more flexible Gly blocks branch formation, supporting the hypothesis that the M.jannaschii KlbA intein is already aligned for optimal attack at the N-terminal splice junction. We are presently working on establishing conditions for structural analysis of M.jannaschii KlbA intein precursors and intermediates, which will help resolve these questions. In conclusion, the robust nature of the intramolecular protein splicing reaction is becoming increasingly evident as new variations in the mechanism dictated by nucleophile and assisting group polymorphisms are identified.
Materials and methods
Cloning and mutagenesis of KlbA inteins
All clones were sequenced by the NEB Core facility and all enzymes were obtained from New England BioLabs (Beverly, MA) and used as described by the manufacturer. The M.jannaschii KlbA intein was cloned by PCR from M.jannaschii genomic DNA (supplied by Gary Olsen and David Graham, University of Illinois) using the primers 5′-GATGCA CTCGAGGGAGCTTTAGCTTATGATGAACCTATTTACTTAAGCG ATGGGAAT and 5′-GGAATTGAGGCCTGAACAGTTTGAGACAGCAAAACCTTCGTTTTT. PCR mixtures contained Vent DNA polymerase (2 units), Vent Polymerase Buffer (New England BioLabs), 200 µM dNTP, 10 µM each primer and 100 ng genomic DNA in a 100 µl reaction and amplification was carried out using a Perkin–Elmer Cetus thermal cycler 480 at 94°C for 30 s, 50°C for 30 s and 72°C for 30 s, for 20 cycles. The M.xenopi gyrase A intein in pMXP (Southworth et al., 1999) was replaced with the M.jannaschii KlbA intein PCR product using XhoI and StuI sites, resulting in pMIP(WT).
The splice junction mutants (Ala1Cys, Ala1Ser, Ala1Gly, Ser167His, Asn168Ala, Cys+1Ser, Cys+1Ala, Ser+2Ala and double mutants Ala1Cys/Cys+1Ala, Ala1Ser/Cys+1Ala) were constructed by PCR as described above, except pMIP(WT) was used as the template, and appropriate forward and reverse primers were used to introduce the desired mutation. Block B mutants (Thr91Ala, Thr93Ala, His94Ala, Asp95Ala and His96Ala) were obtained in a similar manner after the introduction of a silent AgeI site (QuikChange site-directed mutagenesis kit; Stratagene, La Jolla, CA) downstream of block B in the intein. Reverse primers with the desired mutations were used in an amplification reaction and cloned by shuffling the XhoI–AgeI fragment into the wild-type intein.
Pyrococcus sp. GB-D (Jannasch et al., 1992) and Thermococcales strains GI-J and GB-C (Southworth et al., 1996) were screened for KlbA inteins using primers designed from conserved sequences in known klbA genes. A region of the klbA gene spanning the intein insertion site was amplified using the forward primer 5′-CTTCCTGATGGAAGT AGAGTCAATG (489 nt from the 5′ end of the intein) and reverse primer 5′-CATTATCCTGGGAACGTTCATTGGAGG (84 nt from the 3′ end of the intein). PCR was performed as above, except for use of 50 ng of genomic DNA and Vent DNA polymerase, Exo– (4 units) and extension at 72°C for 132 s, 30 cycles. Agarose gel purified PCR products were directly sequenced. The Pyrococcus sp. GB-D KlbA intein was cloned into MIP by PCR using the primers 5′-GATGCAAC TAGTGGTCTTCCTGATGGAAGTAGAGTCAATG and 5′-GGAA TTGAGGCCTGTCATTATCCTGGGAACGTTCATTGGAGG, which contain partial native extein sequences. Amplification was as described above. The PCR products were digested with SpeI and StuI and cloned into pMP1 (pMXP with the intein replaced by a cassette containing SpeI and SphI restriction sites inserted between XhoI and StuI), which was also digested with SpeI and StuI.
Expression, purification and protein characterization
All fusions were expressed in ER2683 cells by induction with isopropyl β-d-thiogalactoside for 2 h at 37 or 15°C overnight, purified over amylose resin (New England BioLabs) at pH 7.5 and treated with DTT at room temperature as previously described (Southworth et al., 1999). Purified protein was digested with Factor Xa protease as described by the manufacturer (New England BioLabs). Relative molecular masses were calculated in comparison with a broad range protein marker (New England BioLabs).
Soluble lysates and purified protein were boiled for 5 min in sample buffer plus DTT (New England BioLabs), loaded on to a 10–20% SDS–polyacrylamide gel (Invitrogen, Carlsbad, CA) and either stained with Coomassie Blue or transferred to nitrocellulose for western blot analysis with an anti-MBP or anti-paramyosin antibody as described previously (Xu et al., 1993). The mobility of IP in SDS–PAGE was dependent on the pH of the sample: it ran at its predicted molecular mass (51 kDa) at pH 5–6, at an apparent molecular mass of 41 kDa at pH 8–9 and at a combination of both at neutral pH. Increasing the time that the sample was boiled prior to loading resulted in an increased proportion of IP at 51 kDa, suggesting that the more quickly migrating form was not fully denatured. The M.jannaschii KlbA intein Asn168Ala mutant was also examined on 10–20% SDS–polyacrylamide gels in sample buffer (New England BioLabs) with 0.1 mM DTT or without DTT. The absence of DTT resulted in aggregation, especially at lower pH. DTT was added to either the crude lysate or purified proteins in concentrations ranging from 10 to 50 mM to study N-terminal splice junction cleavage. In pH studies, samples were adjusted to the appropriate pH by adding sodium phosphate buffer to a final concentration of 0.5 M.
Coomassie Blue-stained gels were digitized with a Microtek Scanmaker III and the signals quantified with NIH Image 1.51 software as described previously (Chen et al., 2000). The values for at least two independent experiments were averaged for each sample. Protein sequencing was performed as described previously (Chen et al., 2000). Briefly, protein samples were subjected to electrophoresis on Tris–glycine polyacrylamide gels (Invitrogen) and transferred to ProBlott polyvinylidene fluoride membranes (PE Biosystems, Foster City, CA). The membranes were stained with Coomassie Blue R-250; bands were excised and each was individually subjected to sequential Edman degradation. An Applied Biosystems 610A Data System was used for data acquisition and analysis.
Acknowledgments
Acknowledgements
We thank Gary Olsen and David Graham, University of Illinois, for kindly providing us with M.jannaschii genomic DNA. We thank Don Comb for support and encouragement, and Eric Adam, Bill Jack, Karen Noren, Steve Smith, Isaac Cann, Virginia Cornish and Chris Noren for helpful discussions and/or reading of the manuscript.
References
- Belfort M. and Roberts,R.J. (1997) Homing endonucleases: keeping the house in order. Nucleic Acids Res., 25, 3379–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Benner,J. and Perler,F.B. (2000) Protein splicing in the absence of an intein penultimate histidine. J. Biol. Chem., 275, 20431–20435. [DOI] [PubMed] [Google Scholar]
- Chong S., Shao,Y., Paulus,H., Benner,J., Perler,F.B. and Xu,M.Q. (1996) Protein splicing involving the Saccharomyces cerevisiae VMA intein. The steps in the splicing pathway, side reactions leading to protein cleavage, and establishment of an in vitro splicing system. J. Biol. Chem., 271, 22159–22168. [DOI] [PubMed] [Google Scholar]
- Chong S., Williams,K.S., Wotkowicz,C. and Xu,M.Q. (1998) Modulation of protein splicing of the Saccharomyces cerevisiae vacuolar membrane ATPase intein. J. Biol. Chem., 273, 10567–10577. [DOI] [PubMed] [Google Scholar]
- Cooper A.A., Chen,Y., Lindorfer,M.A. and Stevens,T.H. (1993) Protein splicing of the yeast TFP1 intervening protein sequence: a model for self-excision. EMBO J., 12, 2575–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard J.Z., Moser,M.J., Hughey,R. and Mian,I.S. (1997) Statistical modeling, phylogenetic analysis and structure prediction of a protein splicing domain common to inteins and hedgehog proteins. J. Comput. Biol., 4, 193–214. [DOI] [PubMed] [Google Scholar]
- Davis E.O., Sedgwick,S.G. and Colston,M.J. (1991) Novel structure of the recA locus of Mycobacterium tuberculosis implies processing of the gene product. J. Bacteriol., 173, 5653–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis E.O., Thangaraj,J.S., Brooks,P.C. and Colston,M.J. (1994) Evidence of selection for protein introns in the RecAs of pathogenic mycobacteria. EMBO J., 13, 699–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X., Gimble,F.S. and Quiocho,F.A. (1997) Crystal structure of PI-SceI, a homing endonuclease with protein splicing activity. Cell, 89, 555–564. [DOI] [PubMed] [Google Scholar]
- Evans T.C. Jr, Benner,J. and Xu,M.Q. (1999) The in vitro ligation of bacterially expressed proteins using an intein from Methanobacterium thermoautotrophicum. J. Biol. Chem., 274, 3923–3926. [DOI] [PubMed] [Google Scholar]
- Gimble F.S. (2000) Invasion of a multitude of genetic niches by mobile endonuclease genes. FEMS Microbiol. Lett., 185, 99–107. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E. (1998) Non-canonical inteins. Nucleic Acids Res., 26, 1741–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.M., Porter,J.A., Young,K.E., Koonin,E.V., Beachy,P.A. and Leahy,D.J. (1997) Crystal structure of a Hedgehog autoprocessing domain: homology between Hedgehog and self-splicing proteins. Cell, 91, 85–97. [DOI] [PubMed] [Google Scholar]
- Hirata R., Ohsumi,Y., Nakano,A., Kawasaki,H., Suzuki,K. and Anraku,Y. (1990) Molecular structure of a gene, VMA1, encoding the catalytic subunit of H+-translocating adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J. Biol. Chem., 265, 6726–6733. [PubMed] [Google Scholar]
- Hodges R.A., Perler,F.B., Noren,C.J. and Jack,W.E. (1992) Protein splicing removes intervening sequences in an archaea DNA polymerase. Nucleic Acids Res., 20, 6153–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang,S., Chen,L., Lemieux,C., Otis,C., Turmel,M. and Liu,X.Q. (1994) The Chlamydomonas chloroplast clpP gene contains translated large insertion sequences and is essential for cell growth. Mol. Gen. Genet., 244, 151–159. [DOI] [PubMed] [Google Scholar]
- Jannasch H.W., Wirsen,C.O., Molyneaux,S.J. and Langworthy,T.A. (1992) Comparative physiological studies on hyperthermophilic Archaea isolated from deep sea hot vents with emphasis on Pyrococcus strain GB-D. Appl. Environ. Microbiol., 58, 3472–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane P.M., Yamashiro,C.T., Wolczyk,D.F., Neff,N., Goebl,M. and Stevens,T.H. (1990) Protein splicing converts the yeast TFP1 gene product to the 69-kD subunit of the vacuolar H+-adenosine triphosphatase. Science, 250, 651–657. [DOI] [PubMed] [Google Scholar]
- Kawarabayasi Y. et al. (1998) Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3 (supplement). DNA Res., 5, 147–155. [DOI] [PubMed] [Google Scholar]
- Kawasaki M., Nogami,S., Satow,Y., Ohya,Y. and Anraku,Y. (1997) Identification of three core regions essential for protein splicing of the yeast Vma1 protozyme. J. Biol. Chem., 272, 15668–15674. [DOI] [PubMed] [Google Scholar]
- Klabunde T., Sharma,S., Telenti,A., Jacobs,W.R.,Jr and Sacchettini,J.C. (1998) Crystal structure of GyrA intein from Mycobacterium xenopi reveals structural basis of protein splicing. Nature Struct. Biol., 5, 31–36. [DOI] [PubMed] [Google Scholar]
- Maeder D.L., Weiss,R.B., Dunn,D.M., Cherry,J.L., Gonzalez,J.M., DiRuggiero,J. and Robb,F.T. (1999) Divergence of the hyperthermophilic archaea Pyrococcus furiosus and P. horikoshii inferred from complete genomic sequences. Genetics, 152, 1299–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathys S., Evans,T.C., Chute,I.C., Wu,H., Chong,S., Benner,J., Liu,X.Q. and Xu,M.Q. (1999) Characterization of a self-splicing mini-intein and its conversion into autocatalytic N- and C-terminal cleavage elements: facile production of protein building blocks for protein ligation. Gene, 231, 1–13. [DOI] [PubMed] [Google Scholar]
- Noren C.J., Wang,J. and Perler,F.B. (2000) Dissecting the chemistry of protein splicing and its applications. Angew. Chem. Int. Ed., 39, 450–466. [PubMed] [Google Scholar]
- Perler F.B. (1998) Protein splicing of inteins and hedgehog autoproteolysis: structure, function, and evolution. Cell, 92, 1–4. [DOI] [PubMed] [Google Scholar]
- Perler F.B. (2000) InBase, the intein database. Nucleic Acids Res., 28, 344–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perler F.B., Olsen,G.J. and Adam,E. (1997) Compilation and analysis of intein sequences. Nucleic Acids Res., 25, 1087–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrokovski S. (1994) Conserved sequence features of inteins (protein introns) and their use in identifying new inteins and related proteins. Protein Sci., 3, 2340–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrokovski S. (1998a) Identification of a virus intein and a possible variation in the protein-splicing reaction. Curr. Biol., 8, R634–635. [DOI] [PubMed] [Google Scholar]
- Pietrokovski S. (1998b) Modular organization of inteins and C-terminal autocatalytic domains. Protein Sci., 7, 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland B.W., Xu,M.Q. and Quiocho,F.A. (2000) Structural insights into the protein splicing mechanism of PI-SceI. J. Biol. Chem., 275, 16408–16413. [DOI] [PubMed] [Google Scholar]
- Scott C.P., Abel-Santos,E., Wall,M., Wahnon,D.C. and Benkovic,S.J. (1999) Production of cyclic peptides and proteins in vivo. Proc. Natl Acad. Sci. USA, 96, 13638–13643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y., Xu,M.-Q. and Paulus,H. (1996) Protein splicing: evidence for an N–O acyl rearrangement as the initial step in the splicing process. Biochemistry, 35, 3810–3815. [DOI] [PubMed] [Google Scholar]
- Southworth M.W., Kong,H., Kucera,R.B., Ware,J., Jannasch,H.W. and Perler,F.B. (1996) Cloning of thermostable DNA polymerases from hyperthermophilic marine Archaea with emphasis on Thermococcus sp. 9° N-7 and mutations affecting 3′–5′ exonuclease activity. Proc. Natl Acad. Sci. USA, 93, 5281–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southworth M.W., Amaya,K., Evans,T.C., Xu,M.Q. and Perler,F.B. (1999) Purification of proteins fused to either the amino or carboxy terminus of the Mycobacterium xenopi gyrase A intein. Biotechniques, 27, 110–120. [DOI] [PubMed] [Google Scholar]
- Telenti A., Southworth,M., Alcaide,F., Daugelat,S., Jacobs,W.R.,Jr and Perler,F.B. (1997) The Mycobacterium xenopi GyrA protein splicing element: characterization of a minimal intein. J. Bacteriol., 179, 6378–6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D.W., Wu,W., Belfort,G., Derbyshire,V. and Belfort,M. (1999) A genetic system yields self-cleaving inteins for bioseparations. Nature Biotechnol., 17, 889–892. [DOI] [PubMed] [Google Scholar]
- Wu H., Hu,Z. and Liu,X.Q. (1998) Protein trans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803. Proc. Natl Acad. Sci. USA, 95, 9226–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M. and Perler,F.B. (1996) The mechanism of protein splicing and its modulation by mutation. EMBO J., 15, 5146–5153. [PMC free article] [PubMed] [Google Scholar]
- Xu M., Southworth,M.W., Mersha,F.B., Hornstra,L.J. and Perler,F.B. (1993) In vitro protein splicing of purified precursor and the identification of a branched intermediate. Cell, 75, 1371–1377. [DOI] [PubMed] [Google Scholar]
- Xu M., Comb,D.G., Paulus,H., Noren,C.J., Shao,Y. and Perler,F.B. (1994) Protein splicing: an analysis of the branched intermediate and its resolution by succinimide formation. EMBO J., 13, 5517–5522. [DOI] [PMC free article] [PubMed] [Google Scholar]