Abstract
p53-mediated transcription activity is essential for cell cycle arrest, but its importance for apoptosis remains controversial. To address this question, we employed homologous recombination and LoxP/Cre-mediated deletion to produce mutant murine embryonic stem (ES) cells that express p53 with Gln and Ser in place of Leu25 and Trp26, respectively. p53Gln25Ser26 was stable but did not accumulate after DNA damage; the expression of p21/Waf1 and PERP was not induced, and p53-dependent repression of MAP4 expression was abolished. Therefore, p53Gln25Ser26 is completely deficient in transcriptional activation and repression activities. After DNA damage by UV radiation, p53Gln25Ser26 was phosphorylated at Ser18 but was not acetylated at C-terminal sites, and its DNA binding activity did not increase, further supporting a role for p53 acetylation in the activation of sequence-specific DNA binding activity. Most importantly, p53Gln25Ser26 mouse thymocytes and ES cells, like p53–/– cells, did not undergo DNA damage-induced apoptosis. We conclude that the transcriptional activities of p53 are required for p53-dependent apoptosis.
Keywords: acetylation/DNA damage/phosphorylation/stability/transactivation
Introduction
p53 is the most commonly mutated tumor suppressor gene in human cancers, and its role in tumor suppression is further highlighted by the creation of p53–/– mice, which are highly cancer prone and develop a large spectrum of tumors (Donehower et al., 1992; Jacks et al., 1994). It has become clear that p53 has at least two roles in preventing cancer: cell cycle arrest in G1, which allows time for the repair of DNA damage, or apoptosis, which eliminates cells with damaged genomes (Ko and Prives, 1996; Giaccia and Kastan, 1998; Prives and Hall, 1999). These roles are partly dependent on cell type, but both prevent the genome from accumulating mutations and transmitting these to daughter cells. Structural and functional analyses of p53 have shown that p53 is a transcription factor with a sequence-specific DNA binding domain in the central region and a transcriptional activation domain at the N-terminus (Ko and Prives, 1996). A number of genes, including WAF1, MDM2, GADD45, cyclin G and PERP, have been identified as direct transcriptional targets regulated by p53 (Ko and Prives, 1996; Attardi et al., 2000). Mdm2 is the product of a protooncogene that complexes with p53 to inhibit transcriptional activity and promote its degradation, thereby creating an autoregulatory feedback loop that regulates p53 expression and activity (Haupt et al., 1997; Kubbutat et al., 1997). PERP, a member of the PMP-22/gas3 family, might be involved in apoptosis (Attardi et al., 2000). In addition to its transcriptional activation activity, p53 can also negatively regulate the expression of genes, including the microtubule-associated protein MAP4 (Murphy et al., 1996; Zhao et al., 2000). It appears that the N-terminus of p53 is involved in both activation and repression of gene expres sion because two missense mutations (Leu22Trp23 to Gln22Ser23 mutations) at the N-terminus of human p53 appear to disrupt both the transcriptional activation and repression activities of p53 (Lin et al., 1994; Murphy et al., 1996; Roemer et al., 1996).
In response to DNA damage and other cellular stresses, the cellular levels of p53 protein are greatly increased, and the ability of p53 to bind specific DNA sequences is activated (Ko and Prives, 1996). p53 protein levels are regulated post-transcriptionally; thus, the accumulation of p53 following DNA damage results primarily from an increase in protein stability (Mosner et al., 1995). Accumulating evidence suggests, however, that phosphorylation may play important roles in regulating both the stability of p53 and its DNA binding activity (Meek, 1999). In this context, phosphorylation of human p53 at Ser15 and Ser20 may induce conformational changes in the N-terminus that disrupt Mdm2 binding and lead to its stabilization (Shieh et al., 1997, 1999; Dumaz and Meek, 1999; Unger et al., 1999), while phosphorylation-driven acetylation of the C-terminus may activate sequence-specific DNA binding (Gu and Roeder, 1997; Sakaguchi et al., 1998; Corbet et al., 1999).
It has become clear that the transcriptional activity of p53 is required for p53-dependent cell cycle arrest in G1; however, the mechanisms by which p53 induces apoptosis are not clear (Ko and Prives, 1996). In particular, it remains controversial as to whether p53 transcriptional activity is also necessary for p53-dependent apoptosis. Studies by Caelles et al. (1994) and Wagner et al. (1994) showed that apoptosis occurred in the presence of inhibitors of transcription or translation, suggesting that p53-dependent apoptosis can occur independently of its transcription activity. In addition, Haupt et al. (1995) found that the p53 mutant, p53Gln22Ser23, which is incapable of activating transcription, was still capable of inducing apoptosis in a transient transfection assay. In contrast, Sabbatini et al. (1995), Yonish-Rouach et al. (1995) and Attardi et al. (1996), using this same p53 mutant, concluded that the transcriptional activity of p53 is required for p53-dependent apoptosis. Different cell lines, experimental protocols, cell growth states or genetic backgrounds may have contributed to the conflicting conclusions. Therefore, to address this issue in a physiological context, we introduced missense mutations that encode Gln and Ser in place of Leu25 and Trp26 (corresponding to Leu22 and Trp23 of human p53) into the endogenous p53 gene of mouse embryonic stem (ES) cells, and from which thymocytes were derived. Consistent with the equivalent human mutations, p53Gln25Ser26 is completely deficient in transcriptional activation and repression activities. Analysis of the apoptotic responses to DNA damage of these mutant ES cells and of mouse thymocytes derived from the mutant ES cells indicates that the transcriptional activities of p53 are essential for the p53-dependent apoptotic response. In addition, our studies suggest that in vivo, several phosphorylation and acetylation events participate in regulating p53 transcriptional activity.
Results
Introduction of Leu25Trp26 to Gln25Ser26 missense mutations into the p53 gene in murine ES cells
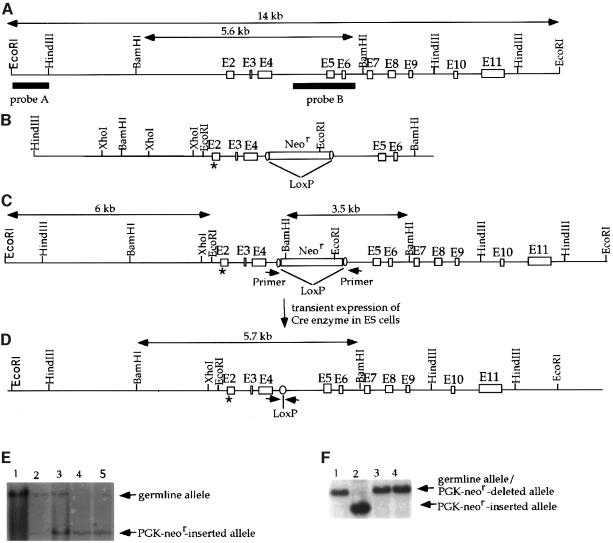
To introduce the mutations at residues 25–26 of mouse p53 (corresponding to residues 22–23 of human p53), a mouse p53 genomic fragment containing exons 2–6 was isolated from a mouse genomic library, subcloned into pBluescript SK and mapped by restriction enzyme digestions. Recombinant methods and site-directed mutagenesis were then employed to construct a ‘knock-in’ vector by mutating the nucleotides encoding leucine (Leu) at residue 25 and tryptophan (Trp) at residue 26 in exon 2 to encode Gln and Ser, respectively, and inserting a PGK-neomycin resistance gene (PGK-Neor) flanked by LoxP sites into intron 4 (Figure 1A–C). Homologous recombination between the endogenous p53 genomic loci of ES cells and the knock-in vector replaced the p53 germline exon 2 with sequences harboring the Gln25Ser26 mutations together with the neighboring PGK-Neor gene (Figure 1D). We then screened EcoRI-digested DNA from G418-resistant colonies with probe A (Figure 1) by Southern blot hybridization to detect a 6 kb mutant restriction fragment and the 14 kb germline fragment (Figure 1A, D and E). Subsequently, homozygous mutant ES cells, with both alleles mutated, were generated by selecting with higher concentrations of G418 as described previously (Xu et al., 1996). The existence of the Neor gene in the mutant alleles could affect the transcription through this locus. Therefore, the PGK-Neor gene flanked by two LoxP sites was excised from the genome of the double mutant ES cells through transient expression of the Cre enzyme, leaving two recombined LoxP sites in the genome of the mutant ES cells (Xu et al., 1996; Figure 1D and E). The ES cells with the PGK-Neor gene deleted from both alleles are referred to as p53Gln25Ser26 ES cells. p53 genomic DNA and mRNA from p53Gln25Ser26 ES cells were sequence analyzed to confirm that the Gln25 and Ser26 mutations, but no other mutations, were present in the p53 gene.
Fig. 1. Construction of p53Gln25Ser26 ES cells. (A) The mouse germline p53 locus. Blank boxes represent the p53 exons and the two filled bars represent the two probes (A and B) used to detect the wild-type and mutant p53 alleles by Southern blot analysis. The germline 14 kb EcoRI and 5.6 kb BamHI fragments are indicated. (B) The targeting construct. The position of the mutations encoding Gln25 and Ser26 in place of Leu25 and Trp26 in exon 2 of the p53 gene is indicated by an asterisk. The PGK-Neor gene flanked by LoxP sites was inserted into an engineered SalI site within intron 4. (C) Targeted p53 locus. The sizes of the mutant EcoRI and BamHI fragments are indicated. The positions of the PCR primer sites that were used to screen for deletion of the PGK-Neor segment are indicated by arrowheads. (D) Mutant p53 allele with the PGK-Neor gene segment deleted. The size of the mutant BamHI fragment after deletion of the PGK-Neor gene is indicated; arrows show the new positions of the PCR primer sites. (E) Southern blot analysis of genomic DNA derived from wild-type (lane 1), heterozygous p53Gln25Ser26 mutant (lanes 2 and 3) and homozygous mutant (lanes 4 and 5) ES cells with the PGK-Neor gene inserted. Genomic DNA was digested with EcoRI and hybridized with probe A. The positions of the EcoRI restriction fragments from the germline alleles are indicated by an arrow. (F) Southern blotting analysis of genomic DNA derived from wild-type (lane 1), homozygous mutant ES cells with the PGK-Neor gene inserted (lane 2), and p53Gln25Ser26 ES cells with the PGK-Neor gene deleted (lanes 3 and 4). Genomic DNA was digested with BamHI and hybridized with probe B. The positions of the BamHI fragments derived from wild-type and mutant alleles after deletion of the PGK-Neor gene and of the mutant allele with the PGK-Neor gene inserted are indicated with arrows.
p53 induction and p53-dependent gene expression in mutant cells following DNA damage
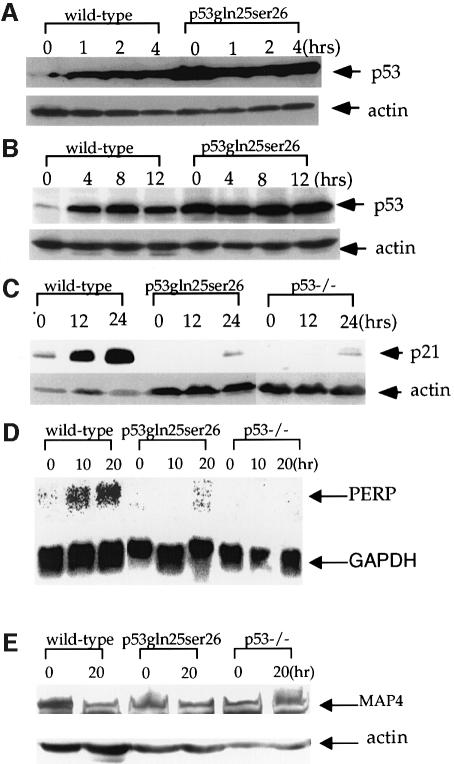
Previous studies have shown that the interaction between human p53 and Mdm2 is disrupted by mutations that change Leu22 to Gln and Trp23 to Ser, leading to p53 stability (Haupt et al., 1997; Kubbutat et al., 1997). Therefore, we analyzed the p53 protein levels in p53Gln25Ser26 ES cells with or without DNA damage to test the importance of the p53–Mdm2 interaction in regulating p53 expression in a physiological environment. The basal p53 protein level in p53Gln25Ser26 cells was much higher than in wild-type cells, and, unlike wild-type ES cells, which undergo significant p53 accumulation after ionizing radiation (IR) and UV treatment, p53Gln25Ser26 accumulation was not induced at any time following DNA damage (Figure 2A and B).
Fig. 2. Induction of p53 and p53-dependent gene expression in wild-type and p53Gln25Ser26 cells following DNA damage. Cell extracts were prepared from wild-type and p53Gln25Ser26 ES cells at the time points indicated after exposure to (A) 5 Gy γ-irradiation or (B) 60 J/m2 UV-C light, and the samples were processed for western immunoblot analysis as described in Materials and methods. The genotypes and time points are labeled on the top of the lane. p53 and actin are indicated on the right. (C) p21 protein induction in wild-type, p53Gln25Ser26 and p53–/– differentiated ES cells following 60 J/m2 UV treatment. The positions of p21 and actin are indicated by arrows. (D) PERP mRNA induction in wild-type, p53Gln25Ser26 and p53–/– differentiated ES cells following 60 J/m2 UV treatment. The positions of PERP and GAPDH mRNA are indicated by arrows. (E) Repression of MAP4 expression in wild-type, p53Gln25Ser26 and p53–/– ES cells following 60 J/m2 UV radiation. The positions of MAP4 protein and actin are indicated with arrows. The genotypes and times after DNA damage are given at the top of each panel.
Similarly to mouse embryonic fibroblasts, retinoic acid-induced differentiated ES cells undergo p53-dependent induction of p21 expression and cell cycle G1 arrest following DNA damage (Xu and Baltimore, 1996; Sabapathy et al., 1997; Aladjem et al., 1998). To confirm that p53Gln25Ser26 is indeed defective in transcriptional activity, we analyzed p53-dependent p21 expression in wild-type, p53Gln25Ser26 and p53–/– differentiated ES cells. As expected, p21 protein levels increased significantly by 24 h after UV treatment in wild-type cells, but little p21 protein was observed in both p53Gln25Ser26 and p53–/– cells with or without DNA damage (Figure 2C). The expression of PERP mRNA is also induced by p53 in mouse cells following DNA damage (Attardi et al., 2000). Therefore, we analyzed the p53-dependent expression of PERP mRNA in wild-type, p53Gln25Ser26 and p53–/– differentiated ES cells following UV radiation. While PERP mRNA is induced significantly 10 h after UV radiation, there is little PERP mRNA in p53Gln25Ser26 and p53–/– cells with or without DNA damage (Figure 2D). We conclude that p53Gln25Ser26 is deficient in transcriptional activation activity. Therefore, the high basal level of p53 in the mutant cells is likely due to the lack of Mdm2 in these cells, since the transcription of Mdm2 is activated by p53 and Mdm2–p53 interaction leads to p53 degradation (Ko and Prives, 1996).
Since human p53 with Gln22Ser23 mutations is defective in transcription repression activity (Murphy et al., 1996; Roemer et al., 1996), we also tested the p53-dependent repression of MAP4 expression in wild-type and mutant ES cells following UV irradiation because UV irradiation induces p53-dependent apoptosis. Consistent with previous findings, while MAP4 expression was repressed in wild-type cells 20 h after UV radiation, little repression of MAP4 expression was detected in either p53Gln25Ser26 or p53–/– cells (Figure 2E). Similar results were obtained when we analyzed the p53-dependent repression of MAP4 mRNA expression in wild-type and mutant cells following UV radiation (data not shown). Therefore, p53Gln25Ser26 is also defective in its transcriptional repression activity.
p53Gln25Ser26 is largely nuclear and binds to the p53 consensus binding site
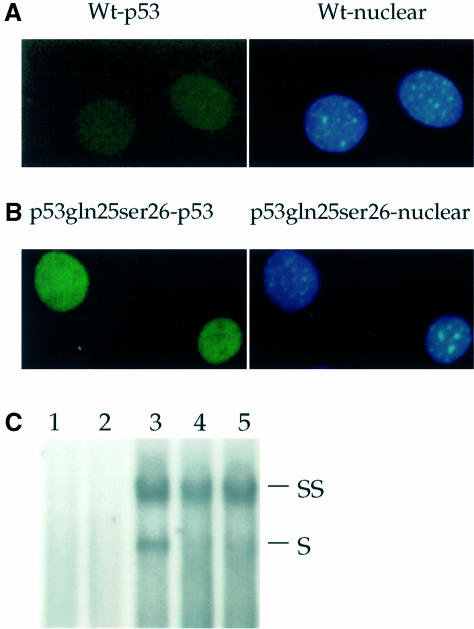
While the finding of high p53 protein levels but no p21 expression in p53Gln25Ser26 cells indicates that p53Gln25Ser26 is defective in transcriptional activity, it was necessary to rule out the possibility that p53Gln25Ser26 is cytoplasmic and thus could not activate p21 expression. Therefore, we examined the cellular localization of wild-type and p53Gln25Ser26 in differentiated ES cells by immunofluorescence staining. A low level of p53 was detected in the nuclei of wild-type cells (Figure 3A); however, p53 protein levels in p53Gln25Ser26 cells were higher than in wild-type cells, and a majority of p53Gln25Ser26 was localized in nuclei (Figure 3B). In addition, we tested the specific DNA binding activity of p53Gln25Ser26 with and without DNA damage (Figure 3C). Our data indicated that p53Gln25Ser26 can bind to the consensus/GADD45 p53-specific DNA binding site, but that the DNA binding activity was not induced by DNA damage (Figure 3C).
Fig. 3. Cellular localization and DNA binding of p53Gln25Ser26. Indirect immunofluorescence: wild-type (A) and p53Gln25Ser26 (B) differentiated ES cells were fixed and stained with PAb421 for p53 (left panels) and DAPI for DNA (right panels) as described in Materials and methods. (C) EMSA: nuclear extracts were prepared from wild-type, p53Gln25Ser26 and p53–/– differentiated ES cells before or 4 h after exposure to 60 J/m2 UV light. EMSA was performed using a 32P-labeled, double-stranded p53 consensus binding sequence as described in Materials and methods. Shown is an autoradiogram of the polyacrylamide gel. The lanes correspond to the nuclear p53-specific DNA binding activity from: 1, p53–/– differentiated ES cells; 2, wild-type differentiated ES cells with no treatment; 3, wild-type differentiated ES cells 4 h after exposure to UV; 4, differentiated p53Gln25Ser26 ES cells with no UV treatment; 5, differentiated p53Gln25Ser26 ES cells 4 h after exposure to UV. The positions of supershifted (SS) as well as specific (S) p53-specific DNA complexes are indicated. PAb421 antibody against p53 was used to supershift the p53 complexes.
Phosphorylation and acetylation of p53 in p53Gln25Ser26 cells following DNA damage
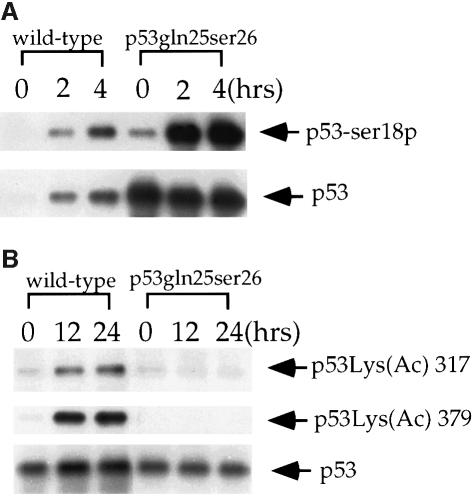
Acetylation of Lys382 and Lys320 of human p53 was recently reported to activate sequence-specific DNA binding activity, and phosphorylation of N-terminal residues was reported to modulate the degree of p53 acetylation (Gu and Roeder, 1997; Lambert et al., 1998; Sakaguchi et al., 1998). Therefore, we checked p53Gln25Ser26 phosphorylation at Ser18 (corresponding to human Ser15) and acetylation at the two C-terminal sites in an attempt to understand why specific DNA binding activity was not induced in p53Gln25Ser26 cells following DNA damage. Consistent with previously published results, phosphorylation of mouse p53 at Ser18 was undetectable in untreated cells but increased significantly in wild-type cells following UV treatment (Figure 4A). Similarly, phosphorylation of p53Gln25Ser26 at Ser18 was also induced by UV treatment, although a small amount of phosphorylation at Ser18 was observed in untreated p53Gln25Ser26 cells (Figure 4A). However, while acetylation of p53 at Lys317 and Lys379 (corresponding to human Lys320 and Lys382) was induced in wild-type cells following UV treatment, acetylation of these residues was essentially absent in p53Gln25Ser26 cells following the same UV treatment.
Fig. 4. UV-induced phosphorylation and acetylation of p53 in wild-type and p53Gln25Ser26 cells. (A) Western immunoblot analysis showing the phosphorylation of mouse p53 at Ser18 in wild-type and p53Gln25Ser26 ES cells before and 2 or 4 h after exposure to 60 J/m2 UV light. The immunoblot was probed with affinity purified antibodies specific for murine p53 phosphorylated at Ser18 (p53-Ser18P, top strip); the blot was then stripped and probed with PAb240 to detect the total p53 signal (bottom strip). (B) Western immunoblot showing the acetylation of mouse p53 at Lys317 and Lys379 (corresponding to human Lys320 and Lys382) in differentiated wild-type and p53Gln25Ser26 ES cells before and 18 or 24 h after exposure to 60 J/m2 UV light. The blot was probed with affinity purified antibodies specific for mouse p53 acetylated at Lys317 (corresponding to Lys320 of human p53; top strip), stripped and reprobed with antibodies specific for mouse p53 acetylated at Lys379 (corresponding to Lys382 of human p53; central strip), and finally stripped and reprobed with PAb240. The genotypes are given above the panels, the positions of p53 are indicated with arrows.
p53-dependent apoptosis in p53Gln25Ser26 ES cells following UV irradiation
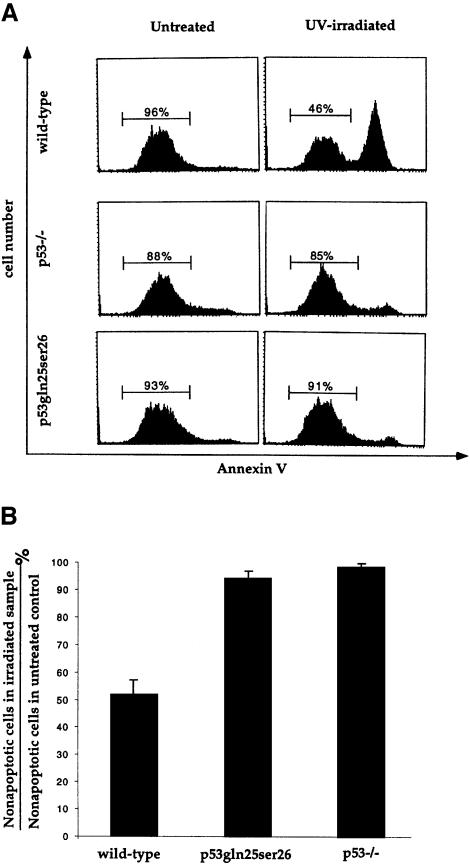
Since p53Gln25Ser26 is completely defective in transcriptional activity, p53Gln25Ser26 cells provide a physiological system with which to test whether the transcriptional activity of p53 is required for p53-dependent apoptosis. ES cells undergo p53-dependent apoptosis following UV treatment (Sabapathy et al., 1997; Corbet et al., 1999). Therefore, we compared UV-induced apoptosis in wild-type, p53Gln25Ser26 and p53–/– ES cells after UV treatment. To identify the cells undergoing apoptosis, the cells were stained with annexin V and analyzed by flow cytometry as described previously (Koopman et al., 1994; Martin et al., 1995). Around 50% of wild-type ES cells underwent apoptosis by 12 h after exposure to 60 J/m2 UV irradiation, but a very small fraction of control p53–/– ES cells underwent apoptosis after this exposure, consistent with previous findings that ES cells undergo p53-dependent apoptosis following UV treatment (Figure 5A and B). Similarly to p53–/– ES cells, only a small fraction of p53Gln25Ser26 ES cells became apoptotic after the same UV treatment, indicating that p53-dependent apoptosis was completely abolished in p53Gln25Ser26 ES cells (Figure 5A and B). Therefore, these data show that the transcriptional activity of p53 is required for p53-dependent apoptosis.
Fig. 5. Induction of apoptosis in wild-type, p53–/– and p53Gln25Ser26 ES cells by UV treatment. (A) Flow cytometric analysis of wild-type, p53–/– and p53Gln25Ser26 ES cells harvested 12 h after exposure to 60 J/m2 UV irradiation. Cell number is plotted as a function of the intensity of staining for annexin V; cells stained positive with annexin V antibodies are apoptotic. The percentages of non-apoptotic cells are indicated. (B) The percentile ratio of non-apoptotic cells in irradiated wild-type, p53Gln25Ser26 and p53–/– ES cells relative to non-apoptotic cells in unirradiated controls. The mean and standard deviation from three independent experiments is given.
p53-dependent apoptosis in p53Gln25Ser26 mouse thymocytes following IR
Mouse thymocytes also undergo p53-dependent apoptosis following IR exposure (Clarke et al., 1993; Lowe et al., 1993). To further confirm our findings that the transcriptional activity of p53 is required for p53-dependent apoptosis, we employed the RAG2-deficient blastocyst complementation approach to derive thymocytes from the mutant ES cells; these were then analyzed for p53-dependent apoptosis as described previously (Lowe et al., 1993; Xu et al., 1996). Briefly, p53Gln25Ser26 ES cells were injected into RAG2–/– blastocysts, which were subsequently implanted into a foster mother. Some offspring from such implants are chimeric with the ES cells contributing to all cell lineages. Because thymocyte development is blocked at the CD4–CD8– stage in RAG2–/– mice, which usually constitutes ∼1–5% of the normal thymus cellularity, all CD4+CD8+ double positive as well as CD4+ or CD8+ single positive thymocytes (>80% thymus cellularity) in the chimeric mice will be derived from the injected mutant ES cells (Figure 6A). The thymocytes harboring p53Gln25Ser26 mutations recovered from the RAG2–/– chimeric mice, as well as wild-type and p53–/– thymocytes, were treated with increasing doses of IR and analyzed for apoptotic cells 10 h later. To prevent contamination of CD4–CD8– thymocytes derived from RAG2–/– blastocysts in the chimeric mice, we analyzed only the CD4+ thymocytes since CD4+CD8+ thymocytes are the ones undergoing p53-dependent apoptosis following IR (Clarke et al., 1993; Lowe et al., 1993). Identification by annexin V staining of thymocytes undergoing apoptosis was performed as described previously (Koopman et al., 1994; Martin et al., 1995). An increasing fraction (50–70%) of apoptotic cells was observed when wild-type thymocytes were exposed to 5, 10 or 20 Gy of IR; in contrast, little IR-induced apoptosis was observed in p53–/– and p53Gln25Ser26 thymocytes after exposure to 5, 10 or 20 Gy (Figure 6B). In conclusion, these data also show that the transcriptional activity of p53 is required for p53-dependent apoptotic function.
Fig. 6. Induction of apoptosis in wild-type, p53Gln25Ser26 and p53–/– thymocytes by ionizing radiation. (A) Thymocytes were harvested from wild-type, p53–/– mice and p53Gln25Ser26–RAG2–/– chimeric mice, stained for CD4 and CD8, and analyzed by flow cytometry as described in Materials and methods. Cells residing in the lymphocyte gate were analyzed and the percentage of total cells in a particular gate is indicated. (B) The mean value of the percentile ratio of non-apoptotic CD4+ thymocyte number in wild-type, p53Gln25Ser26 and p53–/– thymocytes treated with 5, 10 and 20 Gy of IR to the non-apoptotic thymocyte number from untreated controls from three independent experiments is given. Error bars show the standard deviation.
Discussion
The p53 tumor suppressor gene protects vertebrate organisms from cancer through at least two mechanisms. As initially demonstrated by Kastan et al. (1992), p53 activates a G1 checkpoint in response to DNA damage that, in principle, provides time for repair of the DNA damage. In some circumstances, however, p53 activates apoptosis, and this response is also believed to play an important role in tumor suppression (Levine, 1997). Although the mechanism by which p53 activates G1 arrest is relatively well characterized and involves primarily transcriptional activation of the cyclin-dependent kinase inhibitor p21 (el-Deiry et al., 1994; Brugarolas et al., 1995; Deng et al., 1995), p53-mediated activation of apoptotic pathways is not well understood. Furthermore, conflicting evidence has been presented as to whether the activation of apoptosis by p53 is a transcriptionally dependent or independent event. For example, several groups reported that p53Gln22Ser23, which is completely deficient in the ability to activate or repress transcription, still induced apoptosis in a transient transfection assay, suggesting that p53-dependent apoptosis is independent of transcriptional activity (Caelles et al., 1994; Wagner et al., 1994; Haupt et al., 1995). In contrast, Sabbatini et al. (1995), Attardi et al. (1996) and Yonish-Rouach et al. (1995) using the same p53 mutant, reported that the transcriptional activity of p53 was required for p53-dependent apoptosis. The apparent discrepancies between these conclusions may result from differences in cell types, experimental protocols, cell growth states and/or genetic backgrounds. Moreover, most cell lines used in these studies were tumor lines that may harbor unknown genetic alterations. To avoid these technical problems, we employed ES cells to study the dependency of p53-induced apoptosis on transcriptional activity in a physiological context. This approach has several advantages. First, only primary cells with defined genetic backgrounds are used, which minimizes genetic variability and the possibility that inadvertant mutants may contribute to phenotype. Secondly, expression of the mutant p53 is driven by its own promoter and regulatory elements; thus, the problem of deregulated expression of p53, which is typical of transient transfections and non-homologous, stable transformations is avoided. Finally, ES cells are pluripotent stem cells that contribute to all cell types in mice. Therefore, primary cell types, such as the thymocytes used in this study, are easily derived from the mutant ES cells. These advantages now enable one to address the effects of p53 mutations on p53-dependent apoptosis in mutiple cell types under physiologically relevant conditions.
The p53 protein is a transcription factor, and analyses employing constructed mutants have suggested that transcriptional activation by p53 is critical for the induction of apoptosis (Gottlieb and Oren, 1998). Furthermore, several p53 target genes have been identified that are known to play a role in apoptosis. Bax, a pro-apoptotic member of the Bcl-2 family, has p53 binding sites in its promoter; thus, direct activation by p53 could provide a link with the apoptotic machinery (Miyashita and Reed, 1995). Nevertheless, the requirement for Bax in p53-dependent cell death is only partial, and Bax is fully dispensable for the p53-dependent cell death of thymocytes in response to γ-irradiation (Knudson et al., 1995). These results suggest that Bax induction may be relevant to p53-induced apoptosis only in certain cellular contexts. Other potential apoptosis target genes such as KILLER/DR5 and other PIGs (p53 inducible genes) have been described, but it remains to be seen whether these play critical roles in p53-dependent apoptosis (Polyak et al., 1997; Wu et al., 1999). Nevertheless, a recently identified p53 target gene, PERP, which is specifically induced upon DNA damage during apoptosis, provides a potentially compelling demonstration of a candidate effector in the p53 transcriptionally dependent apoptotic pathway (Attardi et al., 2000). The transcriptional activation of PERP by p53 appears crucial for PERP’s ability to induce cell death, and PERP apparently functions only to induce apoptosis and not cell cycle arrest. PERP is a new member of the PMP-22/gas3 family of tetraspan transmembrane proteins that have been implicated in cell growth regulation and apoptosis (Naef and Suter, 1999). A second gene, Pw1yPeg3, that is also specifically induced during apoptosis was recently reported (Relaix et al., 2000). Interestingly, Pw1yPeg3 cooperates with Siah1a, another p53-inducible gene, to induce apoptosis. Furthermore, the induction of Pw1yPeg3 during apoptosis requires activation of both p53 and c-myc expression. These data strongly suggest that Pw1yPeg3, like PERP, may be a critical downstream effector of the p53-mediated cell death pathway.
Although a growing number of p53-induced genes are implicated in the DNA damage-induced apoptotic pathway, it remains unclear whether any of them is directly involved in p53-dependent apoptosis and whether p53 also induces apoptosis through mechanisms that are independent of transcriptional activation. One formal hypothesis is that p53 may repress the transcription of certain genes required for cell survival. In support of this notion, it was shown that p53-mediated repression of MAP4 expression might be involved in p53-dependent apoptosis (Murphy et al., 1996). Importantly, the findings we report here demonstrated convincingly that changing Leu25 and Trp26 of murine p53 to Gln and Ser, respectively, simultaneously disrupts the transcriptional activation and repression activity of p53 in vivo as well as its apoptotic function. Therefore, in murine ES cells and thymocytes, the induction of apoptosis in response to DNA damage requires the p53-dependent transcriptional activation and/or repression of certain gene products. However, the relative contributions of p53 transcriptional activation activity and repression activity to apoptosis remain to be determined. In addition, it remains possible that in certain cells or conditions, apoptosis can be induced through the accumulation of p53 by mechanisms that do not require transcriptional activity.
Studies from several laboratories have begun to elucidate the steps leading to p53 activation. It is now clear that several sites in p53, including Ser15, become phosphorylated in response to DNA damage-inducing agents (Meek, 1999). In vitro, Ser15 can be phosphorylated by DNA-PK (Lees-Miller et al., 1992) and the related protein kinase ATM (Banin et al., 1998; Canman et al., 1998), and in vivo, efficient phosphorylation of Ser15 after cells have been exposed to IR requires a functional ATM gene (Siliciano et al., 1997). These results, coupled with the observation that p53 accumulation is delayed in ATM-deficient cells after exposure to IR (Kastan et al., 1992; Khanna and Lavin, 1993; Xu and Baltimore, 1996), suggest that phosphorylation of Ser15 may be important for stabilizing p53 (Shieh et al., 1999). Phosphorylation of the N-terminal serines 15, 33 and 37 has also been proposed to permit subsequent modification of the C-terminal lysine residues through the recruitment of p300/CBP/PCAF (Sakaguchi et al., 1997; Lambert et al., 1998). Our finding that p53Gln25Ser26 is not acetylated in response to UV light is consistent with the notion that the N-terminus of p53 is involved in recruitment of the histone acetylases and that acetylation of p53 at the C-terminus activates the specific DNA binding activity of p53 (Gu and Roeder, 1997; Sakaguchi et al., 1998). Our findings also suggest that interaction of p53 with components of the transcriptional apparatus may be a further requirement for C-terminal acetylation. Our study shows clearly that phosphorylation of Ser15 in response to DNA damage, which still occurs on the transcriptionally inactivated mutant p53, alone is not sufficient to promote acetylation of the C-terminal residues.
Materials and methods
Construction of the targeting vector
Leu25 and Trp26 of p53 are encoded by exon 2 of the mouse p53 gene (Bienz et al., 1984) (Figure 1A). A mouse p53 genomic DNA fragment containing exon 2 was isolated and mutations changing the nucleotides encoding Leu25 and Trp26 to Gln25 and Ser26 were introduced into p53 exon 2 by site-directed mutagenesis using a kit as recommended by the manufacturer (Stratagene). The p53 genomic DNA containing the mutated exon 2 was then used to construct the targeting vector by inserting the PGK-Neor gene flanked by two LoxP sites into the unique SalI site in intron 4 of the cloned p53 genomic DNA (Figure 1A and C). The thymidine kinase (TK) gene was inserted at one end of the p53 genomic DNA to allow negative selection for random integration. To ensure that the mutated exon replaced the germline exon, an EcoRI site was introduced into intron 1 for diagnostic purposes by site-directed mutagenesis (Figure 1A–C).
Generation of homozygous p53Gln25Ser26 mutant ES cells
The targeting construct was linearized with XbaI and electroporated into 20 × 106 J-1 ES cells as described (Xu et al., 1996), and transfectants were selected with G418 (0.3 mg/ml) and gancyclovir (1 µM). Homologous recombination events were confirmed by Southern blotting of EcoRI-digested cell DNAs and hybridization with probe A, which revealed a 14 kb germline fragment from wild-type cells and a 6.5 kb fragment from homologous recombinants (Figure 1A–C). To generate homozygous mutant ES cells, heterozygous mutant ES cells were cultured under increasing concentrations of G418 as described (Xu et al., 1996). ES cell colonies surviving 4.8 mg/ml G418 were expanded and screened by Southern blotting as described above. To delete the PGK-neor gene from both alleles of the homozygous mutant ES cells, 20 µg of a circular plasmid that drives expression of Cre enzyme and a gene for puromycin resistance was transiently transfected into the PGK-neor-inserted homozygous mutant ES cells as described (Xu et al., 1996). Transfectants were plated, selected with 2 µM puromycin for 2 days, and then cultured in normal ES medium. Surviving ES cell colonies were screened for the Cre-mediated deletion by PCR using the primers indicated in Figure 1C. Positive ES cells identified by PCR were subcloned and subsequently confirmed by Southern blotting after BamHI digestion and hybridization with probe B, which reveals a 5.6 kb germline fragment, a 4.8 kb PGK-neor-inserted fragment or a 5.7 kb PGK-neor-deleted fragment.
Culture and differentiation of ES cells and irradiation treatment
Before irradiation treatment, ES cells were cultured without a feeder layer in Dulbecco’s modified Eagle’s medium (DMEM) medium supplemented with 15% fetal calf serum, glutamine, non-essential amino acids, antibiotics, 100 µM β-mercaptoethanol and recombinant LIF. ES cells were irradiated with 5 Gy γ-ray or exposed to 60 J/m2 UV light and harvested at various time points following treatment. Differentiation of ES cells in vitro with retinoic acid was performed as described (Aladjem et al., 1998). Briefly, subconfluent ES cell cultures were trypsinized, and individual ES cells were plated onto gelatinized 10 cm plates at a density of 2 million cells/plate in ES cell culture medium supplemented with 3 × 10–7 M retinoic acid but without LIF and the feeder layer. Most cells in the culture were differentiated after 4–5 days of treatment with retinoic acid. Differentiated ES cells were exposed to 60 J/m2 UV radiation.
Western blot analysis of p53, p21 and MAP4
Extracts from 4 × 105 cells per sample was separated by SDS–PAGE and transferred to nitrocellulose membranes. Membranes were blocked with 5% dry milk and probed with a monoclonal antibody against p53 (Pab240 from Santa Cruz) or polyclonal antibody against p21 (Santa Cruz) or rat monoclonal antibody against MAP4 (gift from Dr M.Murphy). Membranes were subsequently incubated with horseradish peroxide-conjugated secondary antibody, developed with enhanced chemiluminescence PLUS (ECL PLUS, Amersham), scanned with a Storm system (Molecular Dynamics) and quantitated with the ImageQuant program (Molecular Dynamics). Protein images were also developed by exposing the immunoblot to X-ray film. To insure that equal amounts of protein had been applied in each lane, the membrane was stripped and probed with a polyclonal antibody against actin (Santa Cruz), developed with ECL Plus and quantitated as described.
Northern blot analysis
Total RNA was isolated using TRI Reagent following the manufacturer’s instructions (Sigma). Northern blot analyses were performed as described using 20 µg of RNA in each lane (Xu et al., 1993). RNA was transferred to a nylon membrane and probed with a 3.1 kb HindIII fragment of the mouse MAP4 cDNA (gift from Dr M.Murphy) or a 200 bp mouse PERP cDNA. The 200 bp mouse PERP cDNA fragment was amplified from cDNA generated from UV-treated differentiated ES cells with primers PERP5 and PERP3, and confirmed by DNA sequencing. The sequence of the primers was as follows: PERP5 5′-atgctgcgctgcggcctggcct-3′; PERP3 5′-tagttggggaggcagcagaagaa-3′. The PCR was carried out in a final volume of 100 µl containing 150 ng of each primer, 1× PCR buffer, 0.2 mM dNTP and 2 U of Vent polymerase. The PCR went for 33 cycles, each consisting of 1 min at 94°C, 1.5 min at 60°C and 1 min at 72°C. The final reaction step was followed by an extension at 72°C for 10 min.
Phosphorylation- and acetylation-specific mouse p53 antibodies
PAbSer(P)15 rabbit polyclonal antibody specific for mouse p53 phosphorylated at Ser18 (corresponding to human Ser15) has been described previously (Sakaguchi et al., 1998; Jimenez et al., 1999). PAbLys(Ac)317m and PAbLys(Ac)379m antibodies specific for mouse p53 acetylated at Lys317 or Lys379 were prepared similarly. Briefly, rabbits were immunized with the acetylated mouse p53 peptide Ac-310–322(317Ac)C (i.e. Ac-SASPPQKK(Ac)KPLDGC-NH2) or Ac-374–386(379Ac)C (i.e. Ac-TSRHKK(Ac)TMVKKVGC-NH2), and acetylation site-specific antibodies were affinity purified by use of the corresponding SulfoLinked acetylated peptides. The purified antibodies were then passed through a column coupled with the respective unacetylated peptide to deplete antibodies that react with unacetylated mouse p53. The specificity of each antibody was confirmed by ELISA and immunoblot assays. Detection of the phosphorylated and acetylated p53 in irradiated and untreated cells was performed as described previously (Sakaguchi et al., 1998).
Immunofluorescence analysis
Differentiated wild-type and p53Gln25Ser26 ES cells were plated onto coverslips and 24 h after plating, the cells were washed once with PB (100 mM PIPES pH 6.8, 2 mM MgCl2, 1 mM EGTA), fixed in 3.7% formalin in PB for 20 min and extracted with 0.5% NP-40 in PB for 10 min. The treated cells were rinsed with phosphate-buffered saline (PBS) three times, blocked with 10% goat serum in PBS for 1 h and stained with a mouse monoclonal antibody against p53 (PAb421, Oncogene Research Products). After rinsing with PBS four times, cells were stained with fluorochrome-conjugated secondary antibody (Jackson ImmunoResearch Laboratories), rinsed with PBS, counter-stained for DNA and examined with an Olympus IX70 microscope fitted with appropriate fluorescence filters.
Electrophoretic mobility shift assays
Differentiated ES cells were exposed to 60 J/m2 UV and nuclear extracts were prepared at 4 h after irradiation as described (Sakaguchi et al., 1998). The electrophoretic mobility shift assay (EMSA) was performed by incubating 20 µg of nuclear extract in 25 µl DNA binding buffer [5% glycerol, 25 mM Tris–HCl pH 7.4, 50 mM KCl, 1 µg poly(dI-dC), 1 mM dithiothreitol (DTT), 0.5 µg anti-p53 antibody (PAb421, Oncogene Research Products), 1 ng 32P-labeled double-stranded p53 binding site (Santa Cruz)] for 30 min at room temperature, followed by separation on a 5% non-denaturing Tris–glycine gel.
Analysis of DNA damage-induced apoptosis in mouse ES cells and thymocytes
ES cells were plated in 6-well plates at a density of 2–3 × 105 cells per well, exposed to 40 or 60 J/m2 UV radiation, and harvested 12 h after UV treatment. Apoptotic cells were identified by staining with annexin V–FITC (PharMingen) as recommended by the manufacturer. Annexin V is a very sensitive probe to identify cells undergoing apoptosis (Koopman et al., 1994; Martin et al., 1995). Thymocytes were recovered from wild-type, p53–/– and p53Gln25Ser26–RAG2–/– chimeric mice, and IR-induced apoptosis assays were performed as described (Lowe et al., 1993). Thymocytes were resuspended in tissue culture medium (supplemented with 5% fetal bovine serum and 25 mM HEPES pH 7.4) at a density of 1 × 106 cells/ml and exposed to 5, 10 or 20 Gy of IR. Treated thymocytes and untreated controls were plated into wells of 24-well plates (1 ml/well) and incubated at 37°C. The percentage of apoptotic cells within the CD4+CD8+ and CD4+ thymocyte population was determined at 10 h after IR by staining with PE-conjugated anti-CD4 antibody and annexin V–FITC (both from PharMingen).
Acknowledgments
Acknowledgements
We thank Dr Maureen Murphy for the MAP4 cDNA and antibody and John Earle for blastocyst injection. This work was partially supported by an NIH grant (CA77563) and grants from the American Cancer Society and DOD Breast Cancer Research Program to Y.X. C.W.A. was supported in part by National Institutes of Health Grant GM52825 at Brookhaven National Laboratory under contract with the US Department of Energy.
References
- Aladjem M.I., Spike,B.T., Rodewald,L.W., Hope,T.J., Klemm,M., Jaenisch,R. and Wahl,G.M. (1998) ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr. Biol., 8, 145–155. [DOI] [PubMed] [Google Scholar]
- Attardi L.D., Lowe,S.W., Brugarolas,J. and Jacks,T. (1996) Transcriptional activation by p53, but not induction of the p21 gene, is essential for oncogene-mediated apoptosis. EMBO J., 15, 3702–3712. [PMC free article] [PubMed] [Google Scholar]
- Attardi L.D., Reczek,E.E., Cosmas,C., Demicco,E.G., McCurrach,M.E., Lowe,S.W. and Jacks,T. (2000) PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev., 14, 704–718. [PMC free article] [PubMed] [Google Scholar]
- Banin S. et al. (1998) Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science, 281, 1674–1677. [DOI] [PubMed] [Google Scholar]
- Bienz B., Zakut-Houri, R., Givol,D. and Oren,M. (1984) Analysis of the gene coding for the murine cellular tumour antigen p53. EMBO J., 3, 2179–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugarolas J., Chandrasekaran,C., Gordon,J.I., Beach,D., Jacks,T. and Hannon,G.J. (1995) Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature, 377, 552–557. [DOI] [PubMed] [Google Scholar]
- Caelles C., Helmberg,A. and Karin,M. (1994) p53-dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature, 370, 220–223. [DOI] [PubMed] [Google Scholar]
- Canman C.E., Lim,D.S., Cimprich,K.A., Taya,Y., Tamai,K., Sakaguchi,K., Appella,E., Kastan,M.B. and Siliciano,J.D. (1998) Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science, 281, 1677–1679. [DOI] [PubMed] [Google Scholar]
- Clarke A.R., Purdie,C.A., Harrison,D.J., Morris,R.G., Bird,C.C., Hooper,M.L. and Wyllie,A.H. (1993) Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature, 362, 849–852. [DOI] [PubMed] [Google Scholar]
- Corbet S.W., Clarke,A.R., Gledhill,S. and Wyllie,A.H. (1999) P53-dependent and -independent links between DNA-damage, apoptosis and mutation frequency in ES cells. Oncogene, 18, 1537–1544. [DOI] [PubMed] [Google Scholar]
- Deng C., Zhang,P., Harper,J.W., Elledge,S.J. and Leder,P. (1995) Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell, 82, 675–684. [DOI] [PubMed] [Google Scholar]
- Donehower L.A., Harvey,M., Slagle,B.L., McArthur,M.J., Montgomery,C.A.,Jr, Butel,J.S. and Bradley,A. (1992) Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature, 356, 215–221. [DOI] [PubMed] [Google Scholar]
- Dumaz N. and Meek,D.W. (1999) Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J., 18, 7002–7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry W.S. et al. (1994) WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res., 54, 1169–1174. [PubMed] [Google Scholar]
- Giaccia A.J. and Kastan,M.B. (1998) The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev., 12, 2973–2983. [DOI] [PubMed] [Google Scholar]
- Gottlieb T.M. and Oren,M. (1998) p53 and apoptosis. Semin. Cancer Biol., 8, 359–368. [DOI] [PubMed] [Google Scholar]
- Gu W. and Roeder,R.G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell, 90, 595–606. [DOI] [PubMed] [Google Scholar]
- Haupt Y., Rowan,S., Shaulian,E., Vousden,K.H. and Oren,M. (1995) Induction of apoptosis in HeLa cells by trans-activation-deficient p53. Genes Dev., 9, 2170–2183. [DOI] [PubMed] [Google Scholar]
- Haupt Y., Maya,R., Kazaz,A. and Oren,M. (1997) Mdm2 promotes the rapid degradation of p53. Nature, 387, 296–299. [DOI] [PubMed] [Google Scholar]
- Jacks T., Remington,L., Williams,B.O., Schmitt,E.M., Halachmi,S., Bronson,R.T. and Weinberg,R.A. (1994) Tumor spectrum analysis in p53-mutant mice. Curr. Biol., 4, 1–7. [DOI] [PubMed] [Google Scholar]
- Jimenez G.S. et al. (1999) DNA-dependent protein kinase is not required for the p53-dependent response to DNA damage. Nature, 400, 81–83. [DOI] [PubMed] [Google Scholar]
- Kastan M.B., Zhan,Q., el-Deiry,W.S., Carrier,F., Jacks,T., Walsh,W.V., Plunkett,B.S., Vogelstein,B. and Fornace,A.J.,Jr (1992) A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell, 71, 587–597. [DOI] [PubMed] [Google Scholar]
- Khanna K.K. and Lavin,M.F. (1993) Ionizing radiation and UV induction of p53 protein by different pathways in ataxia-telangiectasia cells. Oncogene, 8, 3307–3312. [PubMed] [Google Scholar]
- Knudson C.M., Tung,K.S., Tourtellotte,W.G., Brown,G.A. and Korsmeyer,S.J. (1995) Bax-deficient mice with lymphoid hyperplasia and male germ cell death. Science, 270, 96–99. [DOI] [PubMed] [Google Scholar]
- Ko L.J. and Prives,C. (1996) p53: puzzle and paradigm. Genes Dev., 10, 1054–1072. [DOI] [PubMed] [Google Scholar]
- Koopman G., Reutelingsperger,C.P., Kuijten,G.A., Keehnen,R.M., Pals,S.T. and van Oers,M.H. (1994) Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood, 84, 1415–1420. [PubMed] [Google Scholar]
- Kubbutat M.H., Jones,S.N. and Vousden,K.H. (1997) Regulation of p53 stability by Mdm2. Nature, 387, 299–303. [DOI] [PubMed] [Google Scholar]
- Lambert P.F., Kashanchi,F., Radonovich,M.F., Shiekhattar,R. and Brady,J.N. (1998) Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem., 273, 33048–33053. [DOI] [PubMed] [Google Scholar]
- Lees-Miller S.P., Sakaguchi,K., Ullrich,S.J., Appella,E. and Anderson,C.W. (1992) Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol. Cell. Biol., 12, 5041–5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A.J. (1997) p53, the cellular gatekeeper for growth and division. Cell, 88, 323–331. [DOI] [PubMed] [Google Scholar]
- Lin J., Chen,J., Elenbaas,B. and Levine A.J. (1994) Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev., 8, 1235–1246. [DOI] [PubMed] [Google Scholar]
- Lowe S.W., Schmitt,E.M., Smith,S.W., Osborne,B.A. and Jacks,T. (1993) p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature, 362, 847–849. [DOI] [PubMed] [Google Scholar]
- Martin S.J., Reutelingsperger,C.P., McGahon,A.J., Rader,J.A., van Schie,R.C., LaFace,D.M. and Green,D.R. (1995) Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J. Exp. Med., 182, 1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek D.W. (1999) Mechanisms of switching on p53: a role for covalent modification? Oncogene, 18, 7666–7675. [DOI] [PubMed] [Google Scholar]
- Miyashita T. and Reed,J.C. (1995) Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell, 80, 293–299. [DOI] [PubMed] [Google Scholar]
- Mosner J., Mummenbrauer,T., Bauer,C., Sczakiel,G., Grosse,F. and Deppert,W. (1995) Negative feedback regulation of wild-type p53 biosynthesis. EMBO J., 14, 4442–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M., Hinman,A. and Levine,A.J. (1996) Wild-type p53 negatively regulates the expression of a microtubule-associated protein. Genes Dev., 10, 2971–2980. [DOI] [PubMed] [Google Scholar]
- Naef R. and Suter,U. (1999) Impaired intracellular trafficking is a common disease mechanism of PMP22 point mutations in peripheral neuropathies. Neurobiol. Dis., 6, 1–14. [DOI] [PubMed] [Google Scholar]
- Polyak K., Xia,Y., Zweier,J.L., Kinzler,K.W. and Vogelstein,B. (1997) A model for p53-induced apoptosis. Nature, 389, 300–305. [DOI] [PubMed] [Google Scholar]
- Prives C. and Hall,P.A. (1999) The p53 pathway. J. Pathol., 187, 112–126. [DOI] [PubMed] [Google Scholar]
- Relaix F., Wei,X., Li,W., Pan,J., Lin,Y., Bowtell,D.D., Sassoon,D.A. and Wu,X. (2000) Pw1/Peg3 is a potential cell death mediator and cooperates with Siah1a in p53-mediated apoptosis. Proc. Natl Acad. Sci. USA, 97, 2105–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer K. and Mueller-Lantzsch,N. (1996) p53 transactivation domain mutant Q22, S23 is impaired for repression of promoters and mediation of apoptosis. Oncogene, 12, 2069–2079. [PubMed] [Google Scholar]
- Sabapathy K., Klemm,M., Jaenisch,R. and Wagner,E.F. (1997) Regulation of ES cell differentiation by functional and conformational modulation of p53. EMBO J., 16, 6217–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbatini P., Lin,J., Levine,A.J. and White,E. (1995) Essential role for p53-mediated transcription in E1A-induced apoptosis. Genes Dev., 9, 2184–2192. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K., Sakamoto,H., Lewis,M.S., Anderson,C.W., Erickson,J.W., Appella,E. and Xie,D. (1997) Phosphorylation of serine 392 stabilizes the tetramer formation of tumor suppressor protein p53. Biochemistry, 36, 10117–10124. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K., Herrera,J.E., Saito,S., Miki,T., Bustin,M., Vassilev,A., Anderson,C.W. and Appella,E. (1998) DNA damage activates p53 through a phosphorylation–acetylation cascade. Genes Dev., 12, 2831–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh S.Y., Ikeda,M., Taya,Y. and Prives,C. (1997) DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell, 91, 325–334. [DOI] [PubMed] [Google Scholar]
- Shieh S.Y., Taya,Y. and Prives,C. (1999) DNA damage-inducible phosphorylation of p53 at N-terminal sites including a novel site, Ser20, requires tetramerization. EMBO J., 18, 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano J.D., Canman,C.E., Taya,Y., Sakaguchi,K., Appella,E. and Kastan,M.B. (1997) DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev., 11, 3471–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger T., Juven-Gershon,T., Moallem,E., Berger,M., Vogt Sionov,R., Lozano,G., Oren,M. and Haupt,Y. (1999) Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J., 18, 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A.J., Kokontis,J.M. and Hay,N. (1994) Myc-mediated apoptosis requires wild-type p53 in a manner independent of cell cycle arrest and the ability of p53 to induce p21waf1/cip1. Genes Dev., 8, 2817–2830. [DOI] [PubMed] [Google Scholar]
- Wu G.S., Burns,T.F., McDonald,E.R.,III., Meng,R.D., Kao,G., Muschel,R., Yen,T. and el-Deiry,W.S. (1999) Induction of the TRAIL receptor KILLER/DR5 in p53-dependent apoptosis but not growth arrest. Oncogene, 18, 6411–6418. [DOI] [PubMed] [Google Scholar]
- Xu Y. and Baltimore,D. (1996) Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes Dev., 10, 2401–2410. [DOI] [PubMed] [Google Scholar]
- Xu Y., Baldassare,M., Fisher,P., Rathbun,G., Oltz,E.M., Yancopoulos,G.D., Jessell,T.M. and Alt,F.W. (1993) LH-2: A LIM/homeodomain gene expressed in developing lymphocytes and neural cells. Proc. Natl Acad. Sci. USA, 90, 227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Davidson,L., Alt.,F.W. and Baltimore,D. (1996) Deletion of the Igκ light chain intronic enhancer/matrix attachment region impairs but does not abolish VκJκ rearrangement. Immunity, 4, 1–10. [DOI] [PubMed] [Google Scholar]
- Yonish-Rouach E., Deguin,V., Zaitchouk,T., Breugnot,C., Mishal,Z., Jenkins,J.R. and May,E. (1995) Transcriptional activation plays a role in the induction of apoptosis by transiently transfected wild-type p53. Oncogene, 11, 2197–2205. [PubMed] [Google Scholar]
- Zhao R., Gish,K., Murphy,M., Yin,Y., Notterman,D., Hoffman,W.H., Tom,E., Mack,D.H. and Levine,A.J. (2000) Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev., 14, 981–993. [PMC free article] [PubMed] [Google Scholar]