Abstract
Background
The role played by calcium as a regulator of circadian rhythms is not well understood. The effect of the pharmacological inhibition of the ryanodine receptor (RyR), inositol 1,4,5-trisphosphate receptor (IP3R), and endoplasmic-reticulum Ca2+-ATPase (SERCA), as well as the intracellular Ca2+-chelator BAPTA-AM was explored on the 24-h rhythmicity of the liver-clock protein PER1 in an experimental model of circadian synchronization by light and restricted-feeding schedules.
Methods
Liver explants from Period1-luciferase (Per1-luc) transgenic rats with either free food access or with a restricted meal schedule were treated for several days with drugs to inhibit the activity of IP3Rs (2-APB), RyRs (ryanodine), or SERCA (thapsigargin) as well as to suppress intracellular calcium fluctuations (BAPTA-AM). The period of Per1-luc expression was measured during and after drug administration.
Results
Liver explants from rats fed ad libitum showed a lengthened period in response to all the drugs tested. The pharmacological treatments of the explants from meal-entrained rats induced the same pattern, with the exception of the ryanodine treatment which, unexpectedly, did not modify the Per1-luc period. All effects associated with drug application were reversed after washout, indicating that none of the pharmacological treatments was toxic to the liver cultures.
Conclusions
Our data suggest that Ca2+ mobilized from internal deposits modulates the molecular circadian clock in the liver of rats entrained by light and by restricted meal access.
Keywords: Food-entrainable oscillator, liver explants, clock proteins, intracellular calcium, SERCA, IP3R, RyR.
Background
In most species, numerous physiological processes are regulated by a timing system associated with the rotational movement (~24 h) of our planet. The circadian rhythms are anticipatory adjustments of metabolic and behavioral processes elicited by daily environmental fluctuations [1]. At the cellular level, circadian rhythms are maintained by a system of interlocking positive (CLOCK and BMAL) and negative (PER and CRY) feedback loops of transcription/translation driven by the core clock genes [2]. The circadian clock, in turn, communicates rhythmicity to a number of output pathways by controlling E-box-associated gene expression and the metabolic status of the cell [3].
Environmental factors that provide entraining cues (zeitgebers) communicate the passage of time to cellular circadian clocks. Two well-recognized zeitgebers are the alternating light/dark cycle and food availability. Photic stimuli entrain the main mammalian pacemaker, the suprachiasmatic nucleus (SCN). It is now well established that restricted feeding also synchronizes rhythmic processes through activation of a food-entrainable oscillator (FEO). The anatomical substrate for the FEO and its dependence on canonical clock gene function remain topics of debate [4,5]. However, two aspects are accepted: 1) the FEO is not completely dependent on the SCN [6]; 2) it requires food restriction (FR) with circadian entraining limits (~22-26 h) [7]. Activation of the FEO influences specific brain areas related to feeding and metabolic control [8] and peripheral tissues such as the liver, which displays a particularly rapid and robust phase shift in response to food entrainment in many physiological and metabolic hepatic functions [9-12]. FR and the associated FEO expression modify not only the phase but also the amplitude of diverse circadian rhythms. For example, notable changes in 24-h variations in the amplitude of free fatty acid concentrations, stomach content, and hepatic triacylglyceride levels have been reported in FR protocols [13-15]. However, it has not been reported if meal entrainment causes modifications in the free-running period of rhythmic parameters.
Compelling evidence for a link between the molecular clock and metabolic networks has been reviewed during light and food entrainment [16-19]. We suggest that intracellular calcium is a strong candidate to coordinate the circadian timing system and biochemical reactions due to its ubiquitous role as a metabolic regulator [20]. Fluctuations of cytoplasmic calcium codify a message that is interpreted in a spatio-temporal manner by metabolic and transcriptional factors. The handling of intracellular calcium is the result of a coordinated activation of ion channels, ATPases, and exchangers in organelles such as the endoplasmic reticulum, mitochondria, and nucleus [21]. Important elements involved in intracellular calcium dynamics include the calcium-release channels inositol 1,4,5-trisphosphate receptor (IP3R) and ryanodine receptor (RyR), as well as the sarco-endoplasmic reticulum calcium ATPase (SERCA). Although all of these proteins are expressed and widely studied in the liver [22], the precise regulatory role for calcium dynamics in the function of the molecular clock in the liver has not been described.
The aim of this study was to explore the rhythmic properties of Per1-Luc in liver explants (mainly the period) exposed for several days to drugs that specifically alter calcium dynamics in the endoplasmic reticulum. Experiments were conducted using rats fed ad libitum (AL) or under a restricted feeding (FR) schedule. Our data reveal that targeted disruption of calcium dynamics affects circadian rhythms of Per1-luc expression in a manner that does not depend on the entrainment cue (meal vs. light), except for the case of ryanodine treatment which, surprisingly, did not alter Per1-luc rhythmicity in the FR group.
Methods
Animals
Male transgenic rats bearing a Period1 gene whose promoter region is linked to the coding sequence of the luciferase gene were used as experimental subjects (Per1-luc) [23]. Transgenic Per1-luc rats were bred and raised in the vivarium of the Biology Department at the University of Virginia under standard conditions (12:12 h light/dark with lights on as Zeitgeber Time 0 or ZT0) with ad libitum access to food and water. At 5-6 weeks of age (~200 g), animals were transferred to a light- and temperature-controlled cabinet for 5 days prior to food restriction. On day 6, rats were separated into two groups: one group continued with ad libitum feeding (AL), whereas a second group was given a restricted meal (FR from ZT4 to ZT6) for 10 days. All animals were sacrificed on day 15, and liver tissue was recovered for explant culture as described below. All procedures were done according to the National Institutes of Health Guidelines for the Care and Use of Animals, and the Guidelines of the Animal Care and Use Committee at the University of Virginia, USA.
Locomotor activity recording
Total locomotor activity was monitored with infra-red motion detectors (IR; Quorom RR-150) mounted ~13 cm above each cage. Each IR detector was modified such that a delay of 8-12 s occurred between detected events. Data were collected in 1-min bins (~5-7 detections/min) with ClockLab software (Actimetrics, USA), compressed into 5-min bins, and finally actograms were generated offline with the same software.
Tissue culture and bioluminiscence recording
Rats were sacrificed on day 15, and hepatic tissues were isolated and cultured as previously described [23]. Briefly, a fraction of one hepatic lobe was quickly removed and chilled in Hanks Balanced Salt Solution (HBSS) at ~4°C. Small explants of liver tissue (1-2 mm2) were dissected free from the larger fragment, placed on culture membranes (Millicell-CM; Millipore), and sealed in a glass-covered (Circle #0, Fisher Scientific), 3.5-ml petri dish (BD-Falcon) containing 1.2 ml of culture medium [serum-free, low sodium-bicarbonate, no phenol red, Dulbecco's modified Eagle's medium (Gibco) supplemented with 10 mM HEPES (pH 7.2), 2% B27 (Invitrogen), 0.1% Luciferin (Promega), and antibiotics (25 U/ml penicillin and 25 μg/ml streptomycin; Gibco). Tissue bioluminescence was measured at 1-min intervals with photomultiplier tubes (Hamamatsu Corporation, USA) housed in a light-tight chamber maintained at 36°C. Each explant was kept 1 day without any manipulation before the addition of drugs. Actions and final concentrations of drugs were as follows: BAPTA-AM (membrane-permeable Ca2+ chelator) 30 μM (Sigma, USA); thapsigargin (SERCA antagonist) 1 μM (Calbiochem, USA); aceto-methoxy 2-aminoethoxydiphenyl borate (2-APB; IP3R antagonist) 100 μM (Calbiochem, USA); ryanodine (RyR antagonist) 100 μM (Calbiochem, USA), and dimethyl sulfoxide (DMSO) 0.1% as vehicle. Drugs were applied continuously from day 2 to 6 (in a chronic manner). On day 7, drugs were removed, fresh medium was added, and bioluminescence was recorded for an additional 3-4 days (washout). The doses of all these well-characterized drugs were consistent with those employed in in vivo and ex vivo experimental systems [24-26].
Data Analysis
Data analysis was performed using the Lumicycle Data Analysis Software (version 2.1, 2008) and OriginPro (version 7.0, 2002). Raw data were detrended and smoothed as described previously [27]. The period (τ) of Per1-luc expression was calculated using a χ-square periodogram analysis of 24-h cycles during drug treatments (days 3-6) and after washout (days 8-11) [28]. The periods during treatment and after washout were calculated separately, omitting the transient gap that resulted during the washout handling. These data have been excluded from the Figures to facilitate the analysis of representative traces.
Statistical Analysis
All results are presented as mean ± SEM. The effects of feeding condition (AL vs. FR) and culture treatment (DMSO vs. drug treatments vs. washout) on the Per1-luc period (τ) were examined with a two-way ANOVA followed by a Bonferroni post-hoc test (significance at p <0.01). Statistical analysis was performed using GraphPad Prism software (version 5.0, 2007).
Results
Food Anticipatory Activity during restricted meal
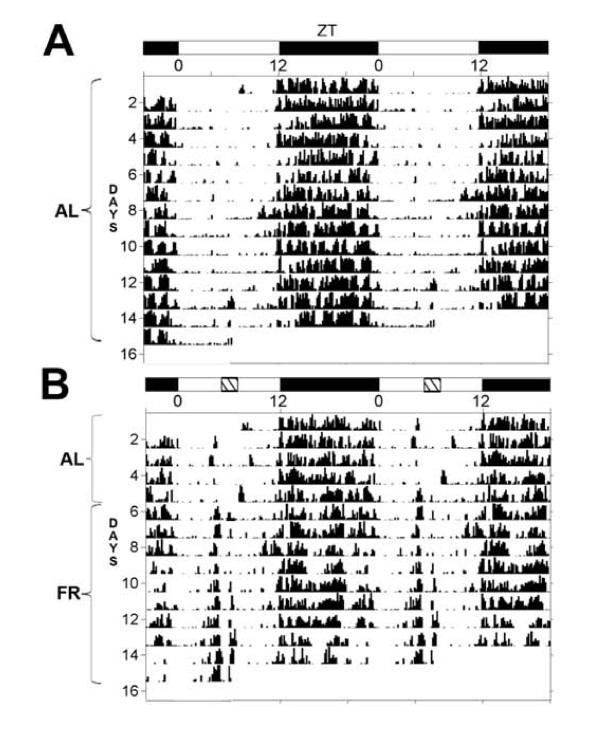
As expected, representative actograms of locomotor activity revealed the appearance of robust food anticipatory activity (FAA) when rats had access to a 2-h restricted meal for 10 days beginning on day 3 (Figure 1B). These data agree with previous reports and are accepted as evidence of FEO activation in Per1-luc transgenic rats [29].
Figure 1.
Representative double-plotted actograms of locomotor activity from Per1-luc transgenic rats fed ad-libitum or under restricted feeding in a 12:12 h light:dark cycle. In the first 5 days all rats were allowed free access to food; one group continued this condition from day 6 to day 15 (A) while another group (B) had a restricted feeding schedule with food available from ZT4 to ZT6 (indicated by the hatched box at the top of the figure inside the bar indicating the L:D cycle). These data are representative of 6 animals.
The meal-entrained phase of Per1-luc expression is maintained in liver explant cultures
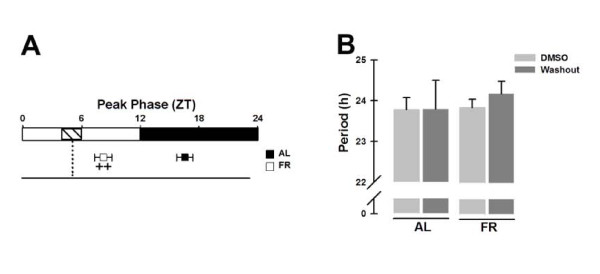
As expected [9,29], the liver cultures isolated from AL rats showed a Per1-luc peak phase during subjective night, whereas the explants from FR animals showed a Per1-luc peak phase close to the time of the former food access (peak phase: AL = ZT 15.7 ± 0.8 h vs. FR = ZT 7.7 ± 0.8 h; Bonferroni post-test p < 0.001; Figure 2A). The Per 1-luc period (τ) in all these conditions showed a constant value close to 24 h, indicating that DMSO is an appropriate reagent vehicle in this experimental protocol (Figure 2B).
Figure 2.
Per1-luc bioluminescence rhythms in liver explants treated with DMSO. (A) Peak phases (in ZT) from Per1-luc expression rhythms calculated from the first peak occurring at the beginning of the culture. The dashed rectangle inside the subjective light period indicates the former food-restricted schedule, and the vertical dotted line represents the former meal time of the FR rats. (B) Period from Per1-luc expression in liver cultures from AL (left bars) and FR (right bars) rats during and after (washout) DMSO treatment. All data plotted are presented as means ± SEM, n = 6. ++Significant difference between AL vs. FR group on the indicated treatment day (Bonferroni post hoc test, α = 0.05).
Chelation of intracellular calcium with BAPTA-AM affected liver Per1-luc rhythmicity
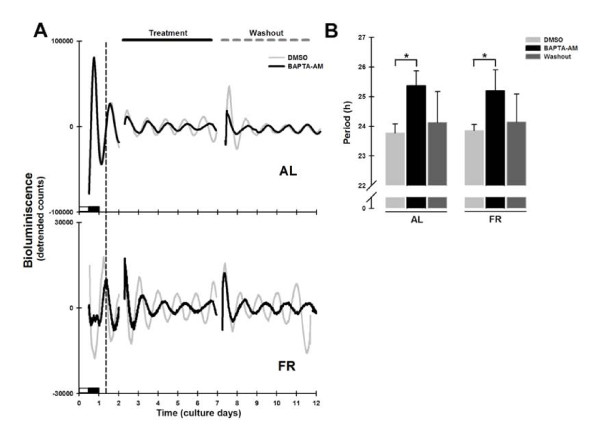
To determine the role of variations of cytosolic calcium in Per1-luc circadian expression, liver cultures were treated with BAPTA-AM. In previous work, the use of this chelator promoted damping of Per1-luc rhythm in suprachiasmatic nucleus explants [26]. Representative rhythms of Per1-luc expression in liver explants from AL and FR groups treated with BAPTA-AM are shown in Figure 3A. A significant τ lengthening (BAPTA-AM: AL = 25.4 ± 0.5 h and FR = 25.2 ± 0.7 h vs. DMSO: AL = 23.8 ± 0.3 h and FR = 23.9 ± 0.2 h; Bonferroni post-test p < 0.001; Figure 3B) was observed when BAPTA treatment was applied to the liver explants in both groups. The BAPTA-AM action was reversed after the change of culture medium (washout: AL = 24.3 ± 1.0 h and FR = 24.1 ± 0.9 h; Figure 3B).
Figure 3.
Per1-luc bioluminescence rhythms in liver explants treated with BAPTA-AM. (A) Representative daily records of Per1-luc expression in liver explants from AL (superior panel) and FR rats (inferior panel) during BAPTA-AM treatment (100 μM). The grey trace represents the DMSO control and the black trace the BAPTA-AM treated culture. The vertical dotted line represents the former meal time of the FR rats. The horizontal black line above the upper panel indicates the onset and duration of drug treatment which ended with washout of the drug on day 6, marked by the horizontal gray dashed line. The subjective night (or dark phase) is marked only on first day of culture. (B) Period from Per1-luc expression in liver cultures from AL (left bars) and FR (right bars) rats during BAPTA-AM treatment and after washout. All data plotted is presented as means ± SEM, n = 6. *Significant difference vs. the DMSO control and x means difference against washout (Bonferroni post hoc test, α = 0.01).
Thapsigargin altered the period of Per1-luc expression in AL and FR rats
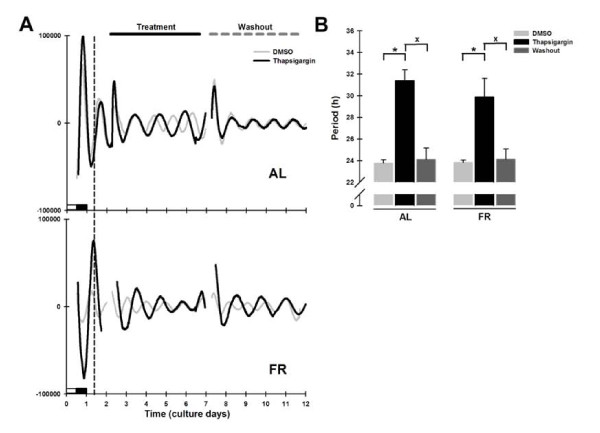
Thapsigargin has been widely used as a potent SERCA inhibitor in many types of cells and experimental conditions [30]. Thapsigargin treatment promoted a large increase in the period of Per1-luc expression in liver cultures from both AL and FR animals (Figure 4A). Quantification of these actions revealed a significantly lengthened period of Per1-luc expression (AL = 31.4 ± 1.0 h and FR = 29.9 ± 1.7 h vs. DMSO; Bonferroni post-test p < 0.001; Figure 4B). This effect was eliminated by the washout of the drug in both groups (AL = 24.1 ± 1.0 h and FR = 24.1 ± 0.9 h vs. DMSO; p = 0.58; Figure 4B).
Figure 4.
Per1-luc bioluminescence rhythms in liver explants treated with thapsigargin. (A) Representative daily records of Per1-luc expression in liver explants from AL (superior panel) and FR (inferior panel) rats during thapsigargin treatment (100 μM). The grey trace represents the DMSO control and the black trace the thapsigargin-treated culture. The vertical dotted line represents the former meal time of the FR rats. The horizontal black line above the upper panel indicates the onset and duration of drug treatment which ended with washout of the drug on day 6, marked by the horizontal gray dashed line. The subjective night (or dark phase) is marked only on the first day of culture. (B) Period from Per1-luc expression in liver cultures from AL (left bars) and FR (right bars) rats during thapsigargin treatment and after washout. All data plotted are presented as means ± SEM, n = 6. *Significant difference vs. the DMSO control, and x means difference against washout (Bonferroni post hoc test, α = 0.01).
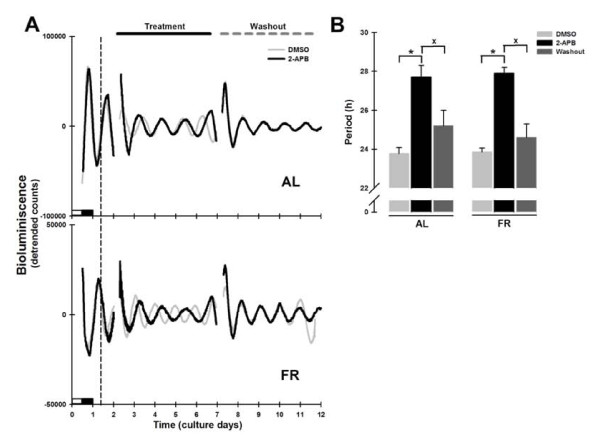
2-APB altered the period of Per1-luc expression in liver explants from AL and FR rats
2-APB is commonly used as an inhibitor of IP3Rs [25,31]. As was the case with BAPTA-AM and thapsigargin, exposure of liver explants from both AL and FR rats to 2-APB significantly lengthened the period of Per1-luc expression (Figure 5A; AL = 27.7 ± 0.6 h and FR = 27.9 ± 0.3 h vs. DMSO; Bonferroni post-test p < 0.001; Figure 5B). As with BAPTA-AM and thapsigargin treatments, this effect was abolished when the drug was removed (AL = 25.2 ± 0.8 h and FR = 24.6 ± 0.7 h; p = 0.65; Figure 5B).
Figure 5.
Per1-luc bioluminescence rhythms in liver explants treated with 2-APB. (A) Representative daily records of Per1-luc expression in liver explants from AL (superior panel) and FR (inferior panel) rats during 2-APB treatment (100 μM). The grey trace represents the DMSO control and the black trace the 2-APB treated culture. The vertical dotted line represents the former meal time of the FR rats. The horizontal black line above the upper panel indicates the onset and duration of drug treatment which ended with washout of the drug on day 6, marked by the horizontal grey dashed line. The subjective night (or dark phase) is marked only on the first day of culture. (B) Period from Per1-luc expression in liver cultures from AL (left bars) and FR (right bars) rats during 2-APB treatment and after washout. All data plotted are presented as means ± SEM, n = 6. *Significant difference vs. the DMSO control, and x means difference against washout (Bonferroni post hoc test, α = 0.01).
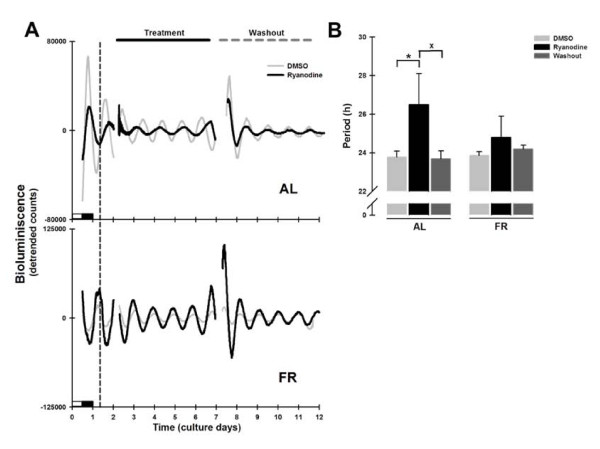
Ryanodine lengthened the period of Per1-luc expression in liver explants from AL but not FR rats
The open probability of the RyR increases in response to ryanodine in the nM range, a state of subconductance is adopted when ryanodine is between 1 and 10 μM, and the channel is inhibited at higher concentration (80-100 μM) [32]. Surprisingly, treatment of the liver cultures with an inhibitory concentration of ryanodine only affected the Per1-luc period of the AL group (ryanodine AL = 26.5 ± 1.6 h, AL = 23.8 ± 0.3 h vs. DMSO; Bonferroni post-test p < 0.001; Figure 6). The period was longer than that observed for DMSO during treatment, but returned to basal values after washout (washout AL = 23.7 ± 0.4 h, AL = 23.8 ± 0.3 h vs. DMSO; p = 0.32; Figure 6B).
Figure 6.
Per1-luc bioluminescence rhythms in liver explants treated with ryanodine. (A) Representative daily records of Per1-luc expression in liver explants from AL (superior panel) and FR (inferior panel) rats during ryanodine treatment (100 μM). The grey trace represents the DMSO control and the black trace the ryanodine treated culture. The vertical dotted line represents the former meal time of the FR rats. The horizontal black line above the upper panel indicates the onset and duration of drug treatment which ended with washout of the drug on day 6, marked by the horizontal grey dashed line. The subjective night (or dark phase) is marked only on the first day of culture. (B) Period of Per1-luc expression in liver cultures from AL (left bars) and FR (right bars) rats during ryanodine treatment and after washout. All data plotted are presented as means ± SEM, n = 6. *Significant difference vs. the DMSO control, and x means difference against washout (Bonferroni post hoc test, α = 0.01).
Discussion
We tested the hypothesis that intracellular calcium plays a role in the regulation of the rhythmicity of hepatic Per1-luc under ex vivo conditions. Our data clearly show that the IP3R, RyR, and SERCA inhibitors and intracellular calcium fluctuations altered the daily expression of Per1-luc in liver explants. Furthermore, the pharmacological action of ryanodine was dependent upon feeding condition.
It has been reported that intracellular calcium plays diverse roles as a component of the timing system by regulating the entrainment process [25,33], clock gene expression [34,35], and output signaling [36]. Diurnal fluctuations of cytoplasmic calcium [37] as well of RyR and IP3R have been observed in the SCN [38,39]. Supporting the importance of intracellular calcium dynamics in the rhythmicity of the SCN, calcium buffering (with concentrations over 40 μM of BAPTA-AM) or low calcium levels (Ca2+-free media) promoted damping of Per1-luc expression in a dose-dependent manner in SCN explants [26]. In NIH3T3 and rat1-fibroblast cell cultures, thapsigargin and calimycin (a calcium ionophore) increased Per1 expression during the first hours of treatment [35,40].
Fluctuations of cytoplasmic calcium codify a message that is interpreted in a spatio-temporal manner by metabolic and transcription factors, thus regulating a variety of cellular processes associated with cell proliferation, apoptosis, biochemical control, and gene expression. This fine-tuned handling of intracellular calcium is the result of a coordinated activation of ion channels, metabolic pumps, and exchangers in organelles such as the endoplasmic reticulum, mitochondria, and nucleus [21]. A role for calcium in the regulation of circadian rhythms and activation of clock-gene proteins has already been reported in protozoa (Euglena gracilis and Paramecium multimicronucleatum), mollusks (Bulla gouldiana) [41-43], plants (Arabidopsis thaliana) [44], insects (Drosophila melanogaster) [45], and in the mammalian SCN [24,37]. However, the precise regulatory role for calcium dynamics in the function of the molecular clock in the liver has not been described.
The liver is a complex organ formed by several cellular types. Even the most prevalent population, that of hepatocytes, varies with anatomical localization and metabolic performance (periportal and pericentral cells). So far, it is not known if the molecular clock in the liver is equally distributed throughout the entire organ, though our preliminary data in experiments done in vivo suggest zone-specific expression of PER1 protein in the hepatic parenchyma (data not shown). The subcellular distribution of the endoplasmic reticulum also changes within the hepatocytes, and potentially the intracellular location of the IP3R, RyR, and SERCA. Different isoforms of calcium-release channels are situated in regions of the endoplasmic reticulum next to the plasma membrane (IP3R type II), or around the nucleus (IP3R type I and RyR) [22]. The heterogeneity of these calcium-handling proteins might underlie a complex interaction between the molecular clock, the intracellular calcium dynamics, and eventually the circadian control of liver metabolic activity. Also, the calcium content of the endoplasmic reticulum is important for the regulation of metabolic pathways, the cell cycle, and cancer progression [46].
The treatments of liver explants from AL and FR rats with BAPTA, 2-APB, and thapsigargin resulted in a lengthening in the Per1-luc period. However, the alteration in the period varied from ~1.5 h (with BAPTA-AM) to ~4 h (with 2-APB) and ~7 h (with thapsigargin). Our data strongly suggest that in the liver there exist different types of interactions between the intracellular calcium dynamics and the molecular clock, ranging from mild (related to cytoplasmic calcium levels, with BAPTA-AM), to medium (related to the calcium pool released by IP3R, as with 2-apb), and strong (related to the calcium content within the endoplasmic reticulum, as observed with thapsigargin treatment). Therefore, it is well known that these interactions between the handling of intracellular calcium and the molecular clock of the liver are independent of the type of circadian synchronization (by light or by meal) and the feeding protocol (AL or FR). In contrast, the period of Per1-luc expression was lengthened during RyR inhibition only in explants from AL animals. No effect was observed when ryanodine was added to explants from FR animals. This fact implies that during the restricted feeding schedule, the period of Per1-luc expression is no longer dependent on the calcium released by the RyR. In a different set of experiments, we observed that the 24-h rhythmicity of [3H]-ryanodine binding to liver microsomal membranes in the daytime FR protocol showed an increase in amplitude, without change in phase (data not shown). This result suggests that the rhythmic properties of the RyR change as a consequence of FR schedule/FEO expression. In addition, the damping rate and the relative amplitude of the Per1-luc rhythm were not changed by the treatments (data not shown).
The changes in Per1-luc period rhythmicity associated with drug treatments could not be interpreted in terms of "after-effects". An "after-effect" is a transient change in the free-running period of an oscillator after a defined period of prior entrainment [47].
The influence of intracellular calcium on liver Per1-luc rhythmicity could be explained by transcriptional regulation of the mRNA for this clock gene or by modulation of the phosphorylation status of PER1 protein. For example, a hypo-phosphorylated state of PER1 delays its degradation and slows the rest of the circadian network [48]. As shown previously by Oh-hashi et al. [35], a significant increase in Per1 and Per2 transcription (also Cry1 as discussed in this report) occurred 1 h after NIH3T3 cells were treated with 0.5 μM thapsigargin. A reduction in cytoplasmic calcium availability is expected with all pharmacological treatments, since BAPTA-AM buffers transient elevations of free intracellular calcium concentration, thapsigargin promotes a depletion of the internal calcium deposits, and the inhibition of IP3R and RyR hinders the release of calcium. The decrease in intracellular calcium could change Per1-luc rhythmicity by modifying the function of other clock genes/proteins such as Cry1 or the casein kinase I epsilon (CKIε), which acts as a negative transcriptional regulator of Per1 expression or as a PER1-phosphorylating element that promotes proteosomal degradation [48]. Elevation of intracellular calcium concentration and the AMP/ATP ratio activate the 5'-AMP kinase (AMPK), which has been proposed as a metabolic sensor capable of coordinating the circadian clockwork of peripheral oscillators through phosphorylation of Cry1 and CKIε, allowing a rapid degradation of Per1 expression and shortening of the period [33,49]. It has been suggested that CamKII (calcium/calmodulin-dependent protein kinase II) could mediate the coupling between intracellular calcium levels and the circadian clock, since it promotes the expression of Per1 in the SCN in a calcium-dependent process [27,50]. For example, a role for this calcium-sensitive protein in regulating the circadian oscillations driven by CLOCK/CYCLE has been reported in reconstituted tissue cultures from D. melanogaster [51]. In addition, there is evidence that buffering intracellular calcium can lengthen the in vivo locomotor activity of the fruit fly [50]. These data correlate with our results showing that a decrease in intracellular calcium has a slowing effect on the period.
Conclusion
Our data support the notion that intracellular calcium dynamics can act as a link between the internal metabolic cues and the circadian clock in the liver. Several drugs that interfere with endoplasmic reticulum-calcium homeostasis slowed the period of the hepatic clock gene Per1. We conclude that intracellular calcium dynamics are a key element in the mechanisms that coordinate proper function of the hepatic clock, acting through modification of the timing system in the liver, and in some cases, also according to the feeding condition.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
ABR and MDM designed and conducted the study, analyzed the data, and wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Adrián Báez-Ruiz, Email: adrienalin@hotmail.com.
Mauricio Díaz-Muñoz, Email: mdiaz@comunidad.unam.mx.
Acknowledgements
We thank Dr. Michael Menaker for allowing us to develop the present work in his laboratory, for the supply of Per1-luc transgenic rats, and for helpful discussions. We also thank Dr. Michael Sellix for his support in writing this manuscript and for his technical expertise. The skilled technical assistance of Naomi Ihara and Denise T. Holmes is also acknowledged. We thank Dr. Chris Colwell (UCLA) and Dr. Raúl Aguilar-Roblero (UNAM) for critical comments and suggestions, as well as Dr. Dorothy Pless and Dr. Michael Jeziorski for their assistance with the manuscript.
This study was supported by CONACYT (México, U-49047). A.B-R is a Ph.D. student from the program in Biomedical Sciences of the Universidad Nacional Autónoma de México.
References
- Menaker M, Takahashi JS, Eskin A. The physiology of circadian pacemakers. Ann Rev Physiol. 1978;40:501–526. doi: 10.1146/annurev.ph.40.030178.002441. [DOI] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15:R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- Yagita K, Yamaguchi S, Tamanini F, van der Horst GT, Hoeijmakers JH, Yasui A, Loros JJ, Dunlap JC, Okamura H. Dimerization and nuclear entry of mPer proteins in mammalian cells. Genes Develop. 2000;14:1353–1363. [PMC free article] [PubMed] [Google Scholar]
- Landry GJ, Yamakawa GR, Webb IC, Mear RJ, Mistleberger RE. The dorsomedial hypothalamic nucleus is not necessary for the expression of the circadian food-anticipatory activity in rats. J Biol Rhythms. 2007;22:467–478. doi: 10.1177/0748730407307804. [DOI] [PubMed] [Google Scholar]
- Fuller PM, Lu J, Saper CB. Differential rescue of light-and food-entrainable circadian rhythms. Science. 2008;320:1074–1077. doi: 10.1126/science.1153277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezuk P, Mohawk JA, Yoshikawa T, Sellix MT, Menaker M. Circadian organization is governed by extra-SCN pacemakers. J Biol Rhythms. 2010;25:432–442. doi: 10.1177/0748730410385204. [DOI] [PubMed] [Google Scholar]
- Davidson AJ, Castañon C, Stephan FK. Daily oscillations in liver function: diurnal vs circadian rhythmicity. Liver Int. 2004;24:179–186. doi: 10.1111/j.1478-3231.2004.00917.x. [DOI] [PubMed] [Google Scholar]
- Ángeles-Castellanos M, Mendoza J, Díaz-Muñoz M, Escobar C. Food entrainment modifies the c-Fos expression pattern in brain stem nuclei of rats. Am J Physiol Reg Integr Comp Physiol. 2005;288:678–684. doi: 10.1152/ajpregu.00590.2004. [DOI] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- Escobar C, Díaz-Muñoz M, Encinas F, Aguilar-Roblero R. Persistence of metabolic rhythmicity during fasting and its entrainment by restricted feeding schedules in rats. Am J Physiol. 1998;274:R1309–1316. doi: 10.1152/ajpregu.1998.274.5.R1309. [DOI] [PubMed] [Google Scholar]
- Díaz-Muñoz M, Vázquez-Martínez O, Aguilar-Roblero R, Escobar C. Anticipatory changes of liver metabolism and entrainment of insulin, glucagon and corticosterone in food-restricted rats. Am J Physiol Regul Integr Comp Physiol. 2000;279:R48–R56. doi: 10.1152/ajpregu.2000.279.6.R2048. [DOI] [PubMed] [Google Scholar]
- Báez-Ruiz GA, Vázquez-Martínez O, Escobar C, Aguilar-Roblero R, Díaz-Muñoz M. Metabolic adaptation or liver mitochondria during restricted feeding schedules. Am J Physiol Gastrointest Liver Physiol. 2005;289:G1015–G1023. doi: 10.1152/ajpgi.00488.2004. [DOI] [PubMed] [Google Scholar]
- Martínez-Merlos MT, Angeles-Castellanos M, Díaz-Muñoz M, Aguilar-Roblero R, Mendoza J, Escobar C. Dissociation between adipose tissue signals, behavior and the food-entrained oscillator. J Endocrinol. 2004;181:53–63. doi: 10.1677/joe.0.1810053. [DOI] [PubMed] [Google Scholar]
- Díaz-Muñoz M, Vázquez-Martínez O, Báez-Ruiz A, Martínez-Cabrera G, Soto-Abraham MV, Avila-Casado MC, Larriva-Sahd J. Daytime food restrictions alters liver glycogen, triacylglycerols, and cell size. A histochemical, morphometric, and ultrastructural study. Comp Hepatol. 2010;9:5. doi: 10.1186/1476-5926-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Zavala JB, Báez-Ruiz A, Díaz-Muñoz M. Changes in the 24 h rhythmicity of liver PPARs and peroxisomal markers when feeding is restricted to two daytime hours. PPAR Res. 2011. [DOI] [PMC free article] [PubMed]
- Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froy O. The circadian clock and metabolism. Clin Sci. 2011;120:65–72. doi: 10.1042/CS20100327. [DOI] [PubMed] [Google Scholar]
- Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–37. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan K, Sassone-Corsi P. Metabolism control by the circadian clock and vice versa. Nature Struct Mol Biol. 2009;16:462–467. doi: 10.1038/nsmb.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ, Bootman MD, Roderick HL. Calcium signalings: dynamics, homeostasis and remodeling. Nature Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- Uhlén P, Fritz N. Biochemistry of calcium oscillations. Biochem Biophys Res Commun. 2010;396:28–32. doi: 10.1016/j.bbrc.2010.02.117. [DOI] [PubMed] [Google Scholar]
- Barrit GJ, Chen J, Rychkov GY. Ca2+-permeable channels in the hepatocyte plasma membrane and their roles in hepatocyte physiology. Biochem Biophys Acta. 2008;1783:651–672. doi: 10.1016/j.bbamcr.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;28:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Sugiyama T, Wallace CS, Gompf HS, Yoshioka T, Miyawaki A, Allen C. Circadian dynamics of cytosolic and nuclear Ca2+ in single suprachiasmatic nucleus neurons. Neuron. 2003;38:253–263. doi: 10.1016/S0896-6273(03)00164-8. [DOI] [PubMed] [Google Scholar]
- Hamada T, Liou SY, Fukushima T, Maruyama T, Watanabe S, Mikoshiba K, Ishida N. The role of Inositol trisphosphate-induced Ca2+ release from IP3 receptor in the rat suprachiasmatic nucleus on circadian entrainment mechanism. Neurosci Lett. 1999;263:125–128. doi: 10.1016/S0304-3940(99)00111-1. [DOI] [PubMed] [Google Scholar]
- Lundkvist GB, Kwak Y, Hajime T, Block GD. A calcium flux is required for circadian rhythm generation in mammalian pacemaker neurons. J Neurosci. 2005;25:7682–7686. doi: 10.1523/JNEUROSCI.2211-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K, Takeuchi Y, Yamaguchi S, Okamura H, Fukunaga K. Involvement of calcium/calmodulin-dependent protein kinase II in the induction of mPer1. J Neurosci Res. 2003;72:384–392. doi: 10.1002/jnr.10581. [DOI] [PubMed] [Google Scholar]
- Refinetti R, Cornélissen G, Halberg F. Procedures for numerical analysis of circadian rhythms. Biol Rhythm Res. 2007;38:275–325. doi: 10.1080/09291010600903692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AJ, Poole AS, Yamazaki S, Menaker M. Is the food-entrainable oscillator in the digestive system? Genes Brain Behav. 2003;2:32–39. doi: 10.1034/j.1601-183X.2003.00005.x. [DOI] [PubMed] [Google Scholar]
- Treiman M, Caspersen C, Christensen SB. A tool coming of age: thapsigargin as an inhibitor of sarco-endoplasmic reticulum Ca2+-ATPase. Trends Pharmacol Sci. 1998;19:131–135. doi: 10.1016/S0165-6147(98)01184-5. [DOI] [PubMed] [Google Scholar]
- Chen J, Tao R, Sun H, Tse H, Lau C, Li G. Multiple Ca2+ signaling pathways regulate intracellular Ca2+ activity in human cardiac fibroblasts. J Cell Physiol. 2010;223:68–75. doi: 10.1002/jcp.22010. [DOI] [PubMed] [Google Scholar]
- Chu A, Díaz-Muñoz M, Hawkes MJ, Brush K, Hamilton SL. Ryanodine as a probe for the functional state of the skeletal muscle sarcoplasmic reticulum calcium release channel. Mol Pharmacol. 1990;37:735–741. [PubMed] [Google Scholar]
- Ding JM, Buchanan GF, Chen D, Kuriashkina L, Faiman LE, Alster JM, McPherson PS, Campbell KP, Gillette MU. A neural ryanodine receptor mediates light-induced phase delays of the circadian clock. Nature. 1998;394:381–384. doi: 10.1038/28639. [DOI] [PubMed] [Google Scholar]
- Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, Thompson CB, Evans RM. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh-hashi K, Naruse Y, Tanaka M. Intracellular calcium mobilization induces period genes via MAP kinase pathways in NIH3T3 cells. FEBS Lett. 2002;516:101–105. doi: 10.1016/S0014-5793(02)02510-3. [DOI] [PubMed] [Google Scholar]
- Aguilar-Roblero R, Mercado C, Alamilla J, Laville A, Díaz-Muñoz M. Ryanodine receptor Ca2+ release channels are an output pathway for the circadian clock in the rat suprachiasmatic nuclei. Eur J Neurosci. 2007;26:575–582. doi: 10.1111/j.1460-9568.2007.05679.x. [DOI] [PubMed] [Google Scholar]
- Colwell CS. Circadian modulation of calcium levels in cells in the suprachiasmatic nucleus. Eur J Neurosci. 2000;12:571–576. doi: 10.1046/j.1460-9568.2000.00939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Muñoz M, Dent M, Granados-Fuentes D, Hall AC, Hernández-Cruz A, Harrington M, Aguilar-Roblero R. Circadian modulation of the Ryanodine receptor type 2 in the SCN of rodents. Neuroreport. 1999;10:481–486. doi: 10.1097/00001756-199902250-00007. [DOI] [PubMed] [Google Scholar]
- Hamada T, Niki T, Ziging P, Sugiyama T, Watanabe S, Mikoshiba K, Ishida N. Differential expression patterns of Inositol trisphosphate receptor types 1 and 3 in the rat suprachiasmatic nucleus. Brain Res. 1999;838:131–135. doi: 10.1016/S0006-8993(99)01719-9. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Maecacci L, Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured rat-1 fibroblast. Curr Biol. 2000;10:1291–1294. doi: 10.1016/S0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- Goto K, Laval-Martin DL, Edmunds LN. Biochemical modeling of an autonomously oscillatory circadian clock in Euglena. Science. 1985;228:1284–1288. doi: 10.1126/science.2988128. [DOI] [PubMed] [Google Scholar]
- Khalsa SB, Ralph MR, Block GD. The role of extracellular calcium in generating and in phase-shifting the Bulla ocular circadian rhythm. J Biol Rhythms. 1993;8:125–139. doi: 10.1177/074873049300800203. [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Kikuchi H, Ishizaki S, Tamura A, Tsukahara Y, Nakaoka Y, Iwai E, Sato T. Simple fluctuation of Ca2+ elicits the complex circadian dynamics of cyclic AMP and cyclic GMP in Paramecium. J Cell Sci. 1999;112:201–207. doi: 10.1242/jcs.112.2.201. [DOI] [PubMed] [Google Scholar]
- Xu X, Hotta CT, Dodd AN, Love J, Sharrock R, Lee YW, Xie Q, Johnson CH, Webb AA. Distincts light and clock modulation of cytosolic free Ca2+ and rhythmic Clorophyll A/B binding protein 2 promoter activity in Arabidopsis. Plant Cell. 2007;19:3474–3490. doi: 10.1105/tpc.106.046011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrisingh MC, Wu Y, Lnenicka GA, Nitabach MN. Intracellular Ca2+ regulates free-running circadian clock oscillations In Vivo. J Neurosci. 2007;27:12489–12499. doi: 10.1523/JNEUROSCI.3680-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergner A, Huber RM. Regulation of the endoplasmic reticulum Ca2+-store in cancer. Anticancer Agents Med Chem. 2008;8:705–709. doi: 10.2174/187152008785914734. [DOI] [PubMed] [Google Scholar]
- Pittendrigh C, Daan S. A functional analysis of circadian pacemakers in nocturnal rodents. J Comp Physiol. 1976;106:223–252. doi: 10.1007/BF01417856. [DOI] [Google Scholar]
- Harms E, Young MW, Saez L. Posttranscriptional and posttranslational regulation of clock genes. J Biol Rhythms. 2004;19:361–373. doi: 10.1177/0748730404268111. [DOI] [PubMed] [Google Scholar]
- Um JH, Yang S, Yamazaki S, Kang H, Viollet B, Foretz M, Chung JH. Activation of 5'-AMP-activated kinase with diabetes drug metformin induces casein kinase I epsilon (CKIepsilon)-dependent degradation of clock protein mPer2. J Biol Chem. 2007;282:20794–20798. doi: 10.1074/jbc.C700070200. [DOI] [PubMed] [Google Scholar]
- Harrisingh MC, Nitabach MN. Integrating circadian timekeeping with cellular physiology. Cell. 2008;320:879–880. doi: 10.1126/science.1158619. [DOI] [PubMed] [Google Scholar]
- Weber F, Hung H, Maurer C, Kay SA. Second messenger and Ras/MAPK signaling pathways regulate CLOCK/CYCLE-dependent transcription. J Neurochem. 2006;98:248–257. doi: 10.1111/j.1471-4159.2006.03865.x. [DOI] [PubMed] [Google Scholar]