Abstract
Neurotrophin 3 (Ntf3) is expressed in Sertoli cells and acts as a chemo-attractant for cell migration from the mesonephros into the developing testis, a process critical to the early morphological events of testis cord formation. The male sex-determining gene Sry initiates the process of testicular development. Sox9 is a key regulator of male sex determination and is directly regulated by SRY. Information on other downstream target genes of SRY is limited. The current study demonstrates an interaction of SRY with the Ntf3 promoter both in vitro and in vivo. The Ntf3 promoter in both rat and mouse contains at least one putative SRY binding site in the −0.6 kb promoter region. In a luciferase reporter assay system, both SRY and SOX9 stimulated the Ntf3 promoter in vitro through an interaction with this SRY-binding motif. In an immunoprecipitation-based pull-down assay, recombinant SRY protein bound the Ntf3 promoter fragment containing an intact SRY binding site, whereas the same protein did not interact with the fragment containing a mutated SRY motif. Specific antibodies against SRY were used in a chromatin immunoprecipitation (ChIP) assay of embryonic testis and were found to precipitate the Ntf3 promoter region. The SRY ChIP assay confirmed the direct interaction between SRY and the Ntf3 promoter in vivo during male sex determination. Observations suggest that SRY physically interacts with the Ntf3 promoter during male sex determination to coordinate cell migration in the testis to form testis cords.
Keywords: Ntf3, rat, sex determination, Sry, testis development
SRY directly regulates the Ntf3 promoter in vitro and in vivo during male sex determination.
INTRODUCTION
In most mammals, male sex is determined after expression of Sry (sex-determining region Y) in precursor Sertoli cells [1, 2]. SRY induces the differentiation of Sertoli cells, which are thought to orchestrate differentiation of other cell types, ensuring testis development [3, 4]. In Sertoli cells SRY synergistically interacts with NR5A1 (nuclear receptor subfamily 5, group A, member 1; previously called steroidogenic factor 1 [SF1]), an orphan nuclear receptor, at a testis-specific enhancer, Tesco, located in the upstream region of the Sox9 promoter to regulate Sox9 expression [5]. The Sox9 gene is expressed at the time of sex determination in Sertoli cells, with increased expression beginning at the peak of Sry expression [6–9]. SOX9 binds to Tesco to establish an autoregulatory positive feedback loop to maintain its expression in the Sertoli cells [5]. Interestingly, Sry expression declines once the Sox9 expression reaches its maximum. SOX9 thereafter takes over the function of SRY and acts independently on downstream genes that are associated with testicular differentiation [10]. An important molecular interaction in the positive feedback loop that ensures testicular development is interaction of SOX9 with fibroblast growth factor 9 (Fgf9) and prostaglandin D2 synthase (brain) (Ptgds) genes [11–13].
Another major event in testis development is the orchestration of somatic and germ cells into cord-like structures that occur at the critical time of male gonadal sex determination. Migrating somatic cells from the mesonephros and vascular endothelial cells from the coelomic epithelium contribute to testis cord formation [4, 14–19]. Studies have shown that SRY is necessary for mesonephric cell migration and the formation of cords in the testis [3, 4]. The growth factors produced by fetal Sertoli cells, such as neurotrophin 3 (NTF3) and fibroblast growth factor 9 (FGF9), have been shown to act on mesonephric cells and to control their migration into the gonad [4, 17]. The mRNAs for these two genes remain elevated in Sertoli cells following sex determination [17, 20].
Since the discovery of SRY as a master initiator of male gonadal sex determination in almost all eutherian and metatherian mammals, studies have focused on the relationship between SRY and Sox9. It is not clear whether Sox9 gene is the only functional target of SRY or it is one of many targets. Studies conducted by Bradford et al. [21] have confirmed cerebellin 4 precursor protein (Cbln4) as a direct target of both SRY and SOX9; however, the exact function of CBLN4 product in testicular development is unknown. Given that SRY-induced pre-Sertoli cells direct mesonephric cell migration in the testis during male sex determination and neurotrophin 3 present in Sertoli cells acts as a chemo-attractant for this migration [4, 17], the hypothesis was developed that SRY may directly or indirectly interact with Ntf3 gene to complete this cell migration process. In order for SRY to directly interact with its targets, there should be SRY response elements in the promoter of the target genes. Analysis of promoters of the Ntf3 genes of mouse, rat, and human (NTF3) revealed at least one SRY binding motif on all the promoters within 600 bp upstream of the transcription start site. The current study demonstrates that SRY and SOX9 stimulate and bind to the Ntf3 promoter in vitro in both a luciferase promoter assay system and in vitro pull-down assays. Observations with a unique in vivo chromatin immunoprecipitation (ChIP) assay demonstrate SRY physically interacts with the Ntf3 promoter at the time of male gonadal sex determination, indicating Ntf3 is one of the downstream and direct targets of SRY.
MATERIALS AND METHODS
Animals and Histology
Sprague-Dawley rats were kept in a temperature-controlled environment and given food and water ad libitum. Estrous cycles of female rats were monitored by cellular morphology from vaginal smears [22]. Rats in early estrus were bred overnight, and matings were confirmed by sperm-positive smears, denoted Day 0 of pregnancy. Pregnant rats were euthanized at Embryonic Day (E) 13, E14, and E16 of pregnancy, and embryonic gonads were collected for histological analysis. Sex was determined by PCR using primers specific for Sry on genomic DNA isolated from embryo tails as previously described [19]. All procedures were approved by the Washington State University Animal Care and Use Committee (IACUC approval 02568-018). Tissue specimens were fixed in Bouin solution for 1 h and embedded in paraffin using standard procedures. Serial sections of 4 μm were stained with hematoxylin and eosin using standard procedures. Sections were visualized by light microscopy.
Immunohistochemistry
Embryonic testis sections were deparaffinized, rehydrated through a graded ethanol series, boiled 10 min in 10 mM sodium citrate buffer to expose the antigens, washed with 0.1% Triton-X solution, and then blocked with 10% goat serum (Vector Laboratories Inc., Burlingame, CA) for 30 min prior to incubation with 0.5 μg/ml primary anti-NTF3 antibody for 18 h (Santa Cruz Biotechnology, Santa Cruz, CA). The sections were then washed with PBS and incubated with Alexa Fluor 488-labeled secondary antibody (diluted 1:3000) for 45 min (goat anti-rabbit IgG; Invitrogen, Carlsbad, CA). Slides were mounted with Vecta-Shield plus 4′,6′-diamidino-2-phenylindol (DAPI; Vector Laboratories Inc.), sealed with cover slips, and analyzed using fluorescence confocal microscopy. Negative control experiments were performed using an affinity-purified nonimmune primary antibody at 1 μg/ml (rabbit IgG; Sigma, St. Louis, MO). NTRK3 (the receptor for NTF3) and AMH localizations were performed using 5 μg/ml primary anti-NTRK3 antibody (Santa Cruz Biotechnology) and anti-AMH antibody (R&D Systems, Minneapolis, MN), respectively, followed by an Alexa Fluor 488-labeled secondary antibody (donkey anti-goat IgG; 1:3000; Invitrogen) using the protocol above. Diaminobenzidine staining was performed with the NTRK3 localization.
Plasmid Construction
Mouse and rat Ntf3 promoter/reporter vectors
A 636-bp fragment of each of the mouse and rat Ntf3 promoters was amplified from respective genomic DNA and cloned into the pGL3-basic luciferase reporter vector (Promega, Madison, WI) using the NheI and KpnI sites of the multiple cloning region. To generate mutant promoter/reporter constructs, the SOX binding consensus sites in the mouse and rat Ntf3 promoters were mutated using complementary oligos, with a total of four point mutations to the consensus sequence A/TAACAAA/T. Oligos were purified and mutagenesis performed using QuikChange Site-Directed Mutagenesis (Stratagene, La Jolla, CA) according to the manufactur's directions. All constructed promoter vectors were sequence verified.
Sry and Sox9 expression vectors
A full-length rat Sry expression plasmid with an MYC tag was produced by amplifying the single exon from rat genomic DNA (forward: AGAGCTTTGGGAGCAGTGACAGTT, reverse: TCTTTGCTGAGGTGCTCCTGCTAT) and cloned into pCMV-Myc expression vectors (Clontech, Mountain View, CA) using the BglII and NotI restriction sites. HA-tagged mouse Sry and Sox9 expression vectors in a pcDNA 3.1 vector were kindly donated by Dr. Peter Koopman (Queensland University, Brisbane, Australia) [12].
Cell Preparation and Culture
Sertoli cell culture
Sertoli cells were isolated from 20-day-old rat testes by sequential enzymatic digestion [23–25]. All animal procedures and protocols were approved by the Washington State University Animal Care and Use Committee. Decapsulated testes were minced with razor blades. Fragments were then digested with trypsin (2.5%; Invitrogen) to remove the interstitial cells, followed by collagenase (type 1; 12.5 mg/ml; Sigma) for removal of peritubular cells and hyaluronidase (25 mg/ml; Sigma) for removal of germ cells. Sertoli cells were plated under serum-free conditions in 24-well plates at 1 × 106 cells per well. Cells were maintained in 5% CO2 atmosphere in Ham F-12 medium (Thermo Fisher Scientific, Waltham, MA) at 32°C.
E13 testis cell culture
E13 embryos were collected from timed-pregnant females as described above. Gonads from E13 animals were dissected, and each pair of gonads from individual animals was placed into one well of a 24-well plate with 300 μl Ham F-12 medium until embryos could be sexed [19]. The male gonads were then pooled and digested with trypsin (2.5%) and collagenase (type I; 12.5 mg/ml) to disassociate the cells. All the cells from the digested testes were then cultured on 100-mm plates in Ham F-12 with 10% bovine calf serum (Thermo Fisher). Cells initially multiplied well in culture and were split as they reached confluence 1:2 twice, at which point cell division slowed down considerably. Cells were maintained in culture, with medium changed every three days, until growth plaques were observed at approximately 1 mo. These growth plaques were then collected for further propagation, and frozen stocks were prepared for subsequent cell splitting such that cells were maintained with less than 12 subcultures for use.
Transfection Procedures
Sertoli cell transfection
Sertoli cells cultured for 48 h were transfected by the calcium phosphate method coupled with hyperosmotic shock (10% glycerol) as previously described [23]. Briefly, 2 μg of promoter reporter plasmid with or without 1 μg expression plasmid in 150 μl transfection buffer (250 mM CaCl2 mixed 1:1 (v:v) with 2× HEBES [28 mM NaCl, 50 mM HEPES, and 1.47 mM Na2HPO4; pH 7.05]) was added to each well of a 24-well plate containing 106 Sertoli cells in 1 ml Ham F-12 medium and incubated at 32°C for 3.5 h. Following incubation, the cells were subjected to hyperosmotic shock. The medium was aspirated, and 1 ml of 10% glycerol in Hanks Balanced Salt Solution (HBSS; Invitrogen) was added for 3 min. Wells were washed twice in HBSS before fresh Ham F-12 with 0.01% bovine serum albumin (BSA) and 1% serum were added. Following a 72-h incubation at 32°C, cells were harvested for luciferase assays. Medium was aspirated and 100 μl of 1× cell lysis solution (Promega) was added per well. Plates were frozen and thawed before cell lysate was collected. For detection of luciferase reporter activity, 20 μl of Sertoli cell lysate was mixed with 50 μl of luciferase substrate (Promega), and luciferase activity was detected on a Wallac Victor II 1420 instrument (PerkinElmer, Waltham, MA).
E13 cultured testis cell transfection
Cells between sub 8 and 12 were transfected using Lipofectamine 2000 (Invitrogen). Cells were serum starved for 24 h in Ham F-12 medium without antibiotics. Promoter reporter plasmid (2 μg) with or without 0.5 μg expression plasmid was mixed with 2 μl Lipofectamine 2000 in 100 μl Opti-MEM medium (Invitrogen) for each well of a 24-well plate. This 100-μl mix was incubated for 20 min at room termperature and then added to each well containing ∼90% confluent-cultured E13 testis cells in 1 ml Ham F-12 medium without antibiotics and incubated at 32°C for 24 h. After 24 h, medium was aspirated from cells and replaced with 1 ml Ham F-12 with 10% serum. Cells were incubated 72 h and collected for luciferase assays as described above.
Immunoprecipitation Pull-Down Assay
SRY and SOX9 recombinant proteins with HA tag were synthesized in vitro by using mouse expression constructs and an in vitro transcription and translation kit according to the manufacturer's instructions (Promega). Oligonucleotides corresponding to rat Ntf3 promoter with or without the SRY/SOX9 binding site were used. DNA-protein binding was performed in 30 ul reaction mixture containing 100 mM KCl, 1 mM MgCl2, 10 μM ZnSO4, 10 mM Tris (pH 7.5), 4% glycerol, 0.1% Triton X-100, 1 mg/ml BSA, 1 μg of poly(dIdC)/poly(dAdT), 0.5 mM dithiothreitol, and protease inhibitor cocktail. Binding reaction continued for 1 h at room temperature. Protein-A Sepharose beads (Sigma) were preswollen in the incubation buffer overnight. Before use, beads were incubated with anti-HA antibody or with nonimmune IgG as a negative control. Following gentle centrifugation (600 × g) for 5 min, beads were treated with protein-oligo mixture and incubated with gentle rotation overnight at 4°Celsius. Beads were washed three times with wash buffer containing 50 mM, 100 mM, and 150 mM NaCl. Pulled-down oligos were eluted in a buffer containing 0.5% SDS and purified in PCR purification columns according to the manufacturer's instruction (Promega). Purified DNA was subjected to PCR with primers designed to flank the rat Ntf3 promoter (primer sequences were AGAAGGGTCTGAGGTTTGGA [forward] and ACTTTGGAGTTTGCTCTTGAGA [reverse]).
In Vivo ChIP Assay
A carrier ChIP (cChIP) assay method was established [26], and conditions were modified to meet the condition for immunoprecipitating rat SRY. The mouse SRY antibody was kindly provided by Dr. Dagmar Wilhelm and Dr. Peter Koopman (Queensland University, Brisbane, Australia). A second commercially available SRY antibody was obtained from Santa Cruz Biotechnology. To run the ChIP assay, male and female gonads (gonad plus mesonephros) were dissected from approximately twenty rat embryos at 13 days postcoitum (dpc; 16–18 tail somite stage) and snap-frozen in liquid nitrogen. E13 rat embryos contain 12–18 tail somites, which is equivalent to E11.5 dpc mouse embryos with 15–17 tail somites. Drosophila SL2 cells were used as a carrier. Densely grown cells (approximately 5 × 107 cells) were pelleted and washed three times in ice-cold PBS and 5 mM sodium butyrate and resuspended in 0.5 ml NB buffer (15 mM Tris-HCL [pH 7.4], 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 0.1 mM ethylene glycol bis(trichloroacetate), 0.5 mM 2-mercaptoethanol, and 0.1 mM PMSF). Frozen testis samples were thawed, mixed with SL2 cells, and homogenized to make a single-cell suspension. Nuclei were pelleted, resuspended in 20 ml NB buffer and 5% (vol/vol) sucrose, pelleted, and resuspended again in 5 ml digestion buffer (50 mM Tris-HCl [pH 7.4], 0.32 M sucrose, 4 mM MgCl2, 1 mM CaCl2, and 0.1 mM PMSF). Following micrococcal nuclease digestion (New England BioLabs, Ipswich, MA) for 10 min at 37°C, the digested samples were gently centrifuged (800 × g) for 10 min. A fraction (50 ul) of the supernatant was put aside as an input sample. The extract was split into two equal aliquots and incubated with either anti-SRY or nonimmune IgG at 4°C overnight. After incubation with 100 μl of preswollen Protein-A Sepharose for 2 h at 4°C, the immunoprecipitates were washed five times with wash buffer (50 mM Tris-HCl [pH 7.5], 10 mM ethylenediaminetetraacetic acid, 5 mM Na butyrate, and 50–150 mM NaCl). The protein-DNA complexes were eluted with elution buffer (1% SDS in incubation buffer). Co-immunoprecipitated DNA was purified by proteinase K digestion, phenol/chloroform extraction, and ethanol precipitation. Final concentration of immunoprecipitated DNA varied from 100 to 300 ng per assay. Two rounds of PCR reactions were run (for 25 cycles each) before gel electrophoresis. The identity of the amplified fragments was verified by sequencing. Primers used to investigate the enrichment of Ntf3 promoter fragments by PCR were the same as above for the SRY/SOX response element and for the nonrelated Ntf3 site were: 5′-TGCTTCCTGTCCCATATGCTGGT-3′ (forward) and 5′-ACCTGTGGAGAATGCTCGTCCT −3′ (reverse). Nested PCR primers for the SRY/SOX response element in the Ntf3 promoter site were: 5′CAAATCAAAGATATTAGCGTGTA-3′ (forward) and 5′GCCCGCAGTGACTTG-3′ (reverse).
RESULTS
NTF3 Protein Is Present in Sertoli Cells
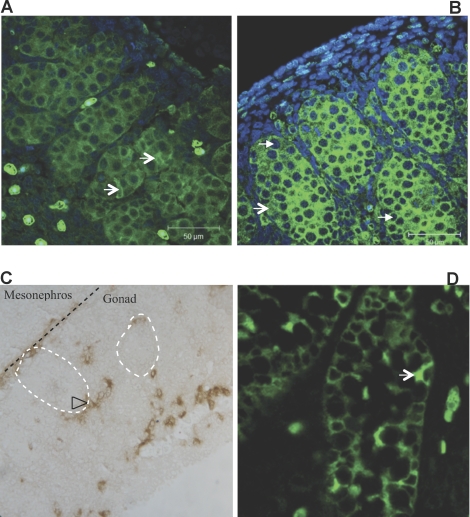
To identify the expression pattern of NTF3 in differentiating testis, immunohistochemical localization was performed. NTF3 was localized on E14 and E16 embryonic rat testis sections using an NTF3-specific antibody. To assist in identifying the specific cell types in which NTF3 is expressed, comparison with the Sertoli cell-specific marker genes SOX9 and anti-Müllerian hormone (AMH) was also performed. NTF3 was expressed by Sertoli cells at E14 and E16 (Fig. 1, A and B). At E14, NTF3 stained Sertoli cells with characteristic irregular shapes, in contrast to germ cells with a lack of NTF3 staining and that contain large, round DAPI-stained nuclei. The receptor for NTF3, neurotrophic tyrosine kinase receptor type 3 (NTRK3), was localized in interstitial cells and also layered between mesonephros and the testis (Fig. 1C). Localization of NTF3 at E16 also shows expression in germ cells and Leydig cells (Fig. 1B). As a control, AMH showed the expected expression in Sertoli cells at E16, when it was initially expressed (Fig. 1D).
FIG. 1.
Immunohistochemical localization of NTF3 in E14 (A) and E16 (B) testis. Green color denotes NTF3, blue denotes DAPI. C) Multiple cell types showed immunolocalization for NTRK3, the receptor for NTF3, in the E14 testis (original magnification ×400). D) Immunolocalization for the Sertoli cell marker AMH in the E16 testis (original magnification ×400). V-shaped arrow, Sertoli cells; triangle-shaped arrow, germ cells; open arrowhead, interstitial cells (likely to be Leydig cells). Representative of micrographs from three different experiments.
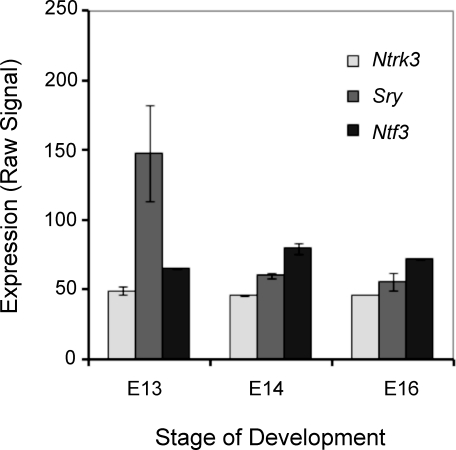
The expression of Ntf3 and Sry during embryonic testis development was also determined using microarray analysis [27]. The expression of Sry was highest at rat Embryonic Day 13, with Ntf3 following at E14 (Fig. 2). Rat Sox9 expression could not be determined due to the absence of a probe set for this gene on the microarray chip used. The isolation of rat Embryonic Day 13 embryos was found to have approximately 18 tail somites, which corresponds to E11.5 in the mouse embryo [28, 29], as has been previously used in male gonadal sex determination [12]. Therefore, the rat E13 is comparable to the mouse E11.5 during development.
FIG. 2.
Ntf3 and Sry expression profiles during embryonic testis development in the rat from a microarray analysis data set [27]. Relative expression versus embryonic day of development is presented. Original data were reanalyzed with Partek software (Affymetrix, Palo Alto, CA) and RMA (R) analysis with the mean ± SEM presented. All data points have error bars; if an error bar is not apparent, then it is small and covered by the data point symbol.
Ntf3 Promoter Contains SRY-Binding Motif
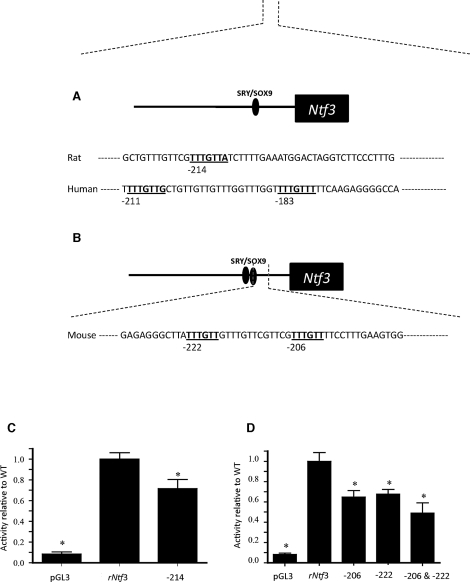
The expression of Ntf3 in the Sertoli cell at the onset of testis differentiation suggested that SRY and/or SOX9 might directly regulate Ntf3 transcription. To test this hypothesis, the genomic regions upstream of the mouse, human, and rat Ntf3 transcription start sites were compared using a ClustalW alignment (available on the EMBL-EBI web server). Over 500 bp upstream of the transcription start site are highly conserved between human, mouse, and rat. Motif Search (available through GenomeNet, http://www.genome.jp) was used to identify the consensus SRY/SOX binding sites to which the conserved HMG box DNA binding region of SOX proteins such as SOX9 and SRY can bind. A single site was found in the rat promoter 214 bp upstream of the putative transcriptional start site (Fig. 3A), and two of these SRY/SOX binding sites, separated by 9 bp, were found in the mouse promoter at 222 and 206 bp upstream of the transcriptional start site (Fig. 3B).
FIG. 3.
A) Rat Ntf3 promoter with one SRY/SOX site 214 bp upstream of the transcription start site. B) Mouse Ntf3 promoter with two SRY/SOX sites at 206 and 222 bp upstream of the transcription start site. Ntf3 promoter mutational assays. C) Rat Ntf3 promoter activity is compared to the activity of a promoter with the 214 SRY/SOX site mutated. D) Mouse Ntf3 promoter activity is compared to the activity of promoters with the 206, 222, or both 206 and 222 SRY/SOX sites mutated. Activity within assays was normalized to wild-type promoter activity, and empty pGL3 vector activity shown represents background activity. Data are shown as the mean of a minimum of three different experiments, each with triplicate samples for each condition. The means ± SEM are presented. *Indicates statistical difference from wild-type promoter activity, P ≤ 0.05.
SRY/SOX9 Regulation of Ntf3 Promoter
To determine whether the SRY/SOX binding sites are important for Ntf3 promoter activity, a mutational analysis was performed. Introduction of four point mutations into the rat 214 site resulted in a statistically significant decrease in promoter activity compared to that of the wild-type promoter, when transfected on Postnatal Day 20 cultured Sertoli cells (Fig. 3C). In mouse, mutation of either the 222 or 206 site, or both sites, also resulted in a significant decrease (P ≥ 0.05) in promoter activity (Fig. 3D). Similar trends in Ntf3 promoter activity were seen when this experiment was repeated in E13 cultured testis cells. These observations suggest that the SRY/SOX sites are important for full Ntf3 promoter activity in mouse and rat.
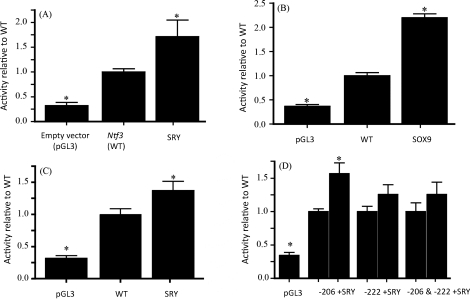
To test if SRY or SOX9 expression has an effect on Ntf3 promoter activity, co-transfection of Ntf3 promoter reporter plasmids with Sry or Sox9 expression plasmids or an empty expression plasmid as a negative control was performed. When the rat Ntf3 promoter was co-transfected with a rat Sry expression vector, the promoter was stimulated significantly over the Ntf3 promoter in the absence of SRY (Fig. 4A). A similar effect was seen when mouse Ntf3 promoter was co-transfected with mouse SRY (Fig. 4C). A mouse SOX9 expression vector was also co-transfected with mouse and rat Ntf3 promoters. The results show that SOX9 is also able to stimulate the Ntf3 promoter of rat (Fig. 4B) and mouse in vitro. These results show that SRY and SOX9 have the ability to stimulate Ntf3 promoter activity.
FIG. 4.
Effects of SRY and SOX9 expression on Ntf3 promoter activity. A) Rat Ntf3 promoter activity with or without SRY co-transfection. B) Rat Ntf3 promoter activity stimulation with SOX9. C) Mouse Ntf3 promoter activity stimulation with SRY. D) Mutant mouse Ntf3 promoter activity stimulation with SRY. Activity within assays was normalized to wild-type promoter activity. Empty pGL3 vector activity represents background activity. Data represent mean ± SEM from a minimum of three experiments. *Indicates statistical difference from wild-type promoter activity, P ≤ 0.05.
SRY/SOX9 Bind to the SOX Motifs in Ntf3 Promoter
To determine if SRY and SOX9 are acting specifically at the SRY/SOX binding sites, co-transfections of protein expression vectors with mutated Ntf3 promoters were also analyzed. Results showed an inability of SRY or SOX9 to stimulate Ntf3 promoter when SRY/SOX binding sites were mutated, suggesting SRY and SOX9 act specifically at the SRY/SOX binding-response elements for promoter activation in these assays. Interestingly, mutating only the 206 site in mouse still allowed activation by SRY and SOX9, while mutating only the 222 site did not (Fig. 4D). Together these data suggest that the SRY/SOX sites are required for Ntf3 promoter stimulation by SRY and SOX9 and that the 222 site may be more important for mouse Ntf3 activation.
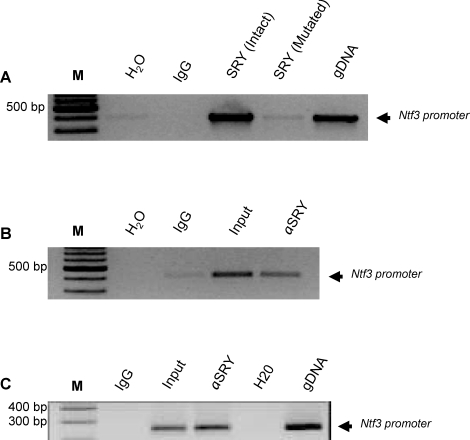
Since SRY acted in a site-specific manner in the transfection assays, it was hypothesized that it may bind directly to the SRY/SOX motifs. To test if binding is direct and site- specific, in vitro pull down assays were performed using both intact and mutated rat promoters. The recombinant protein for mouse SRY contained an HA tag. Therefore, the antibody against HA was used to pull down the protein-DNA complex. Anti-HA antibody recognized and precipitated the oligo-protein complex with an intact SRY/SOX binding motif and did not immunoprecipitate those with mutated sites (Fig. 5A).
FIG. 5.
SRY binding to Ntf3 promoter. A) In vitro transcribed and translated SRY protein with HA tag was subjected to binding with a 300-bp Ntf3 promoter fragment containing either intact or mutated SRY/SOX binding sites. An anti-HA antibody immunoprecipitated the Ntf3 promoter fragment containing the SRY binding site, but not the mutated one. Nonimmune IgG was used as a negative control. Bands indicate PCR products amplified by rat Ntf3 promoter-specific primer flanking SRY binding site. Lanes are: Water (H2O); non-immune IgG (negative control; IgG); Ntf3 promoter with intact SRY binding site (SRY (Intact)); Ntf3 promoter with mutated SRY binding site (SRY (Mutated)); positive control for PCR (genomic DNA). Representative of a minimum of three experiments. B and C) In vivo SRY binding used a cChIP assay. Chromatin from 20 pairs of testes were used for immunoprecipitation with anti-SRY antibodies, Koopman antibody (B) and commercial Santa Cruz antibody (C), followed by centrifugation. Immunoprecipitated DNA was purified with a phenol chloroform method and used for PCR. Two rounds of PCR reaction were used to get the expected band, which was later sequence verified. Nonimmune IgG was used as a negative control. Bands indicate PCR products amplified by rat Ntf3 promoter-specific primer flanking SRY binding site for water control (H2O); nonimmune IgG (IgG); input chromatin DNA (Input); anti-SRY antibody ChIP (αSRY). Representative of a minimum of three different experiments.
SRY Directly Interacts with the Ntf3 Promoter In Vivo at the Time of Sex Determination
The in vitro promoter mutation analysis does not necessarily reflect regulation of Ntf3 in vivo. To determine if SRY associates with the Ntf3 promoter in vivo, a ChIP assay was developed. Since the amount of embryonic gonadal tissue is limited, the large amount of starting material required to run traditional ChIP assays is problematic. Therefore, a procedure was used that involved Drosophila SL2 cells as carrier cells to support gonad samples during the immunoprecipitation process. SL2 cells do not contain Ntf3 promoter consensus sequences in their genome. An anti-SRY antibody from Dr. Peter Koopman successfully immunoprecipitated the Ntf3 promoter (Fig. 5B), whereas primers flanking the promoter region 1 kb from the SRY/SOX binding site were not immunoprecipitated. A commercially available anti-SRY antibody from Santa Cruz Biotechnology also successfully immunoprecipitated the Ntf3 promoter (Fig. 5C). In order to confirm whether the immunoprecipitated fragments belonged to Ntf3, the PCR product was sequence verified. The observation that the SRY antibodies immunoprecipitated the Ntf3 promoter out of the freshly isolated E13 testicular chromatin suggests that Ntf3 is a direct target of SRY. Previously the SRY antibody has been shown to be specific for SRY, but the SOX9 antibody has been shown to cross react with both SRY and SOX9 [12]. Therefore, the ability of SOX9 to bind directly the Ntf3 promoter regions requires further investigation. However, SRY does directly bind the Ntf3 promoter in vivo at the time of gonadal sex determination.
DISCUSSION
Previously NTF3 was shown to act as a chemo-attractant for cell migration from the mesonephros to the developing testis [17, 19], a process critical to early morphological events of testis cord formation. The expression of Ntf3 as determined in previous studies suggested a potentially direct regulation of this gene by the key regulators of sex determination, SRY and SOX9 [30–32]. In this study the molecular mechanism of male-specific activation of Ntf3 during gonadal sex differentiation was investigated. Specifically, Ntf3 promoter regulation by SRY and SOX9 was studied to determine if Ntf3 is a direct target of SRY action in the testis. This investigation is particularly important as only a few direct targets of SRY action have been identified since its discovery in 1990 [1].
Localization of NTF3 in the developing rat testis revealed initial expression at E13 in Sertoli cells only, whereas its receptor NTRK3 was found in both mesonephros and gonadal cells. Cells that stained for NTRK3 were found in the border between mesonephros and testis [17, 19]. Both peritubular cells and interstitial cells also showed positive staining for NTRK3, suggesting that mesonephric cells migrated to the testis. The initial expression of Ntf3 in Sertoli cells at the onset of testis morphogenesis is consistent with the induction of Ntf3 expression by SRY or SOX9, which are initially expressed in rat Sertoli cells at E13 and E13.5, respectively. Interestingly, by E16, NTF3 is also expressed in germ cells and interstitial cells. It has been shown that expression of SRY in pre-Sertoli cells can act in a cell-autonomous manner to increase Sox9 expression and through an intercellular signaling mechanism to increase Sox9 in neighboring cells to promote Sertoli cell differentiation [33, 34]. Similar mechanisms may be involved in the initiation of Ntf3 expression, first in Sertoli cells, then in other testicular cell types. Specifically, Ntf3 appears to be initially expressed after SRY in Sertoli cells, followed by its expression later in development in other testis cell types.
Analysis of the Ntf3 promoter revealed consensus SRY/SOX family protein binding sites in the promoters for human, mouse, and rat. Interestingly, there are two SRY/SOX sites in the proximal 600 bp of the promoters in human and mouse and only a single site in the rat. For SOX9 target genes identified previously, paired SRY/SOX sites oriented in opposing directions with 3–4 bp between them appear to be important for recognition and activation [12, 35, 36]. In the testis, however, it has been speculated that paired SRY/SOX sites may not be required for transcriptional activity. This suggestion arose after the identification of a patient with a Sox9 mutation disrupting the ability of SOX9 to dimerize. Several tissues in which SOX9 dimerization is required were adversely affected; however, the patient was not sex reversed despite the fact that Sox9 is required for male sex determination [37]. The SRY/SOX sites identified in the mouse Ntf3 and human NTF3 promoters are oriented in the same rather than opposing directions, and in the rat there is only one site. This means that the potential mechanism of action of SOX9 or SRY at these sites may differ from that of other SOX9 targets and may be different between mouse and rat. Furthermore, the DNA-dependent dimerization domains found in SOX9 are not present in SRY [37–39]. Therefore, SRY action does not likely depend on dimerization or reverse-oriented, paired SRY/SOX binding sites. This further supports a hypothesis that SRY may directly regulate Ntf3.
To determine if the consensus SRY/SOX binding sites in the Ntf3 promoter are important for gene expression, mutational analysis of these sites using mouse and rat Ntf3 promoter reporter plasmids was performed. Results show that the SRY/SOX site in rat and both sites in mouse are important for full promoter activity in their respective species. Coexpression of these reporter constructs with SRY or SOX9 expression plasmids showed that both SRY and SOX9 were able to stimulate the promoters in a site-specific manner. This is in contrast to a previous study of prostaglandin D synthase (Ptgds) regulation in which only SOX9 was found to stimulate SRY/SOX sites in the Ptgds promoter and only as a dimer [12]. In the mouse Ntf3 promoter there are two SRY/SOX sites 9 bp apart in the same orientation. Both sites are important for full promoter activity, but the two sites do not play equal roles. The mouse Ntf3 promoter can still be stimulated by SRY and SOX9 in the absence of a consensus SRY/SOX 206 site, but not when the 222 site is mutated. This suggests the 222 site is more important for activation of the mouse promoter then the 206 site. Investigation of a protein complex with SRY or SOX9 may be useful in the future to determine the difference in the importance of the mouse SRY/SOX sites and the difference in number of sites between mouse and rat.
To determine if SRY has the potential to bind the Ntf3 promoter directly, a hybrid electrophoretic mobility shift assay (EMSA)/IP assay using protein DNA binding reaction set up as in the EMSA was used, followed by immunoprecipitation with an antibody specific for the HA tag on the SRY protein and PCR for DNA of interest. This allowed the detection of bound DNA without the interference of nonspecific shifting seen with the standard EMSA protocol. SRY recombinant protein bound to oligo containing intact SRY/SOX9 binding sites, but not those containing mutations, indicating an interaction of SRY with the Ntf3 promoter in that specific site.
To further confirm these results in vivo, a sensitive cChIP assay was adopted [26] and modified to meet parameters for the present study. Two different anti-SRY antibodies were used in this ChIP assay on a chromatin mixture of testis from E13 (14–18 tail somite stage) embryos and pulled down a promoter fragment of the Ntf3 promoter containing an SRY binding site. NTF3 has previously been shown to direct mesonephric cell migration into the testis and facilitate testis cord formation [17, 19, 22]. The current results provide evidence that the SRY interacts with the Ntf3 promoter during this critical time of male sex determination. The NTF3 produced then acts as a chemo-attractant to promote mesonephros migration and cord formation [17].
Combined observations demonstrate that both SRY and SOX9 can activate mouse and rat Ntf3 in vitro in a site-specific manner and that both proteins can act at a single SRY/SOX response element binding site, suggesting that dimerization is not required. In vitro binding assays and in vivo ChIP assays further validate the results obtained with promoter assays, suggesting that SRY directly interacts with Ntf3 during male gonadal sex determination. Therefore, Ntf3 appears to be one of the direct targets of SRY.
ACKNOWLEDGMENTS
We thank Ms. Ellyn Schinke and Dr. Marina Savenkova for technical assistance and Ms. Heather Johnson for assistance in preparation of the manuscript.
Footnotes
Supported by a National Institutes of Health grant to M.K.S. This research was performed in part when the laboratory was associated with the School of Molecular Biosciences at Washington State University.
These authors contributed equally to this work.
REFERENCES
- Koopman P, Munsterberg A, Capel B, Vivian N, Lovell-Badge R. Expression of a candidate sex-determining gene during mouse testis differentiation. Nature 1990; 348: 450 452. [DOI] [PubMed] [Google Scholar]
- Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature 1991; 351: 117 121. [DOI] [PubMed] [Google Scholar]
- Brennan J, Capel B. One tissue, two fates: molecular genetic events that underlie testis versus ovary development. Nat Rev Genet 2004; 5: 509 521. [DOI] [PubMed] [Google Scholar]
- Capel B, Albrecht KH, Washburn LL, Eicher EM. Migration of mesonephric cells into the mammalian gonad depends on Sry. Mech Dev 1999; 84: 127 131. [DOI] [PubMed] [Google Scholar]
- Sekido R, Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 2008; 453: 930 934. [DOI] [PubMed] [Google Scholar]
- Koopman P, Bullejos M, Bowles J. Regulation of male sexual development by Sry and Sox9. J Exp Zool 2001; 290: 463 474. [DOI] [PubMed] [Google Scholar]
- Bergstrom DE, Young M, Albrecht KH, Eicher EM. Related function of mouse SOX3, SOX9, and SRY HMG domains assayed by male sex determination. Genesis 2000; 28: 111 124. [PMC free article] [PubMed] [Google Scholar]
- Moreno-Mendoza N, Harley V, Merchant-Larios H. Cell aggregation precedes the onset of Sox9-expressing preSertoli cells in the genital ridge of mouse. Cytogenet Genome Res 2003; 101: 219 223. [DOI] [PubMed] [Google Scholar]
- Sekido R, Bar I, Narvaez V, Penny G, Lovell-Badge R. SOX9 is up-regulated by the transient expression of SRY specifically in Sertoli cell precursors. Dev Biol 2004; 274: 271 279. [DOI] [PubMed] [Google Scholar]
- Wilhelm D, Palmer S, Koopman P. Sex determination and gonadal development in mammals. Physiol Rev 2007; 87: 1 28. [DOI] [PubMed] [Google Scholar]
- Kim Y, Kobayashi A, Sekido R, DiNapoli L, Brennan J, Chaboissier MC, Poulat F, Behringer RR, Lovell-Badge R, Capel B. Fgf9 and Wnt4 act as antagonistic signals to regulate mammalian sex determination. PLoS Biol 2006; 4: e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm D, Hiramatsu R, Mizusaki H, Widjaja L, Combes AN, Kanai Y, Koopman P. SOX9 regulates prostaglandin D synthase gene transcription in vivo to ensure testis development. J Biol Chem 2007; 282: 10553 10560. [DOI] [PubMed] [Google Scholar]
- Kashimada K, Koopman P. Sry: the master switch in mammalian sex determination. Development 2010; 137: 3921 3930. [DOI] [PubMed] [Google Scholar]
- Cool J, Carmona FD, Szucsik JC, Capel B. Peritubular myoid cells are not the migrating population required for testis cord formation in the XY gonad. Sex Dev 2008; 2: 128 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magre S, Jost A. Dissociation between testicular organogenesis and endocrine cytodifferentiation of Sertoli cells. Proc Natl Acad Sci U S A 1984; 81: 7831 7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Tung PS, Fritz IB. Cooperativity between Sertoli cells and testicular peritubular cells in the production and deposition of extracellular matrix components. J Cell Biol 1985; 100: 1941 1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupp AS, Uzumcu M, Skinner MK. Chemotactic role of neurotropin 3 in the embryonic testis that facilitates male sex determination. Biol Reprod 2003; 68: 2033 2037. [DOI] [PubMed] [Google Scholar]
- Combes AN, Wilhelm D, Davidson T, Dejana E, Harley V, Sinclair A, Koopman P. Endothelial cell migration directs testis cord formation. Dev Biol 2009; 326: 112 120. [DOI] [PubMed] [Google Scholar]
- Levine E, Cupp AS, Skinner MK. Role of neurotropins in rat embryonic testis morphogenesis (cord formation). Biol Reprod 2000; 62: 132 142. [DOI] [PubMed] [Google Scholar]
- Colvin JS, Green RP, Schmahl J, Capel B, Ornitz DM. Male-to-female sex reversal in mice lacking fibroblast growth factor 9. Cell 2001; 104: 875 889. [DOI] [PubMed] [Google Scholar]
- Bradford ST, Hiramatsu R, Maddugoda MP, Bernard P, Chaboissier MC, Sinclair A, Schedl A, Harley V, Kanai Y, Koopman P, Wilhelm D. The cerebellin 4 precursor gene is a direct target of SRY and SOX9 in mice. Biol Reprod 2009; 80: 1178 1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzumcu M, Dirks KA, Skinner MK. Inhibition of platelet-derived growth factor actions in the embryonic testis influences normal cord development and morphology. Biol Reprod 2002; 66: 745 753. [DOI] [PubMed] [Google Scholar]
- Chaudhary J, Skinner MK. E-box and cyclic adenosine monophosphate response elements are both required for follicle-stimulating hormone-induced transferrin promoter activation in Sertoli cells. Endocrinology 1999; 140: 1262 1271. [DOI] [PubMed] [Google Scholar]
- Chaudhary J, Skinner MK. Role of the transcriptional coactivator CBP/p300 in linking basic helix-loop-helix and CREB responses for follicle-stimulating hormone-mediated activation of the transferrin promoter in Sertoli cells. Biol Reprod 2001; 65: 568 574. [DOI] [PubMed] [Google Scholar]
- Saxlund MA, Sadler-Riggleman I, Skinner MK. Role of basic helix-loop-helix (bHLH) and CREB transcription factors in the regulation of Sertoli cell androgen-binding protein expression. Mol Reprod Dev 2004; 68: 269 278. [DOI] [PubMed] [Google Scholar]
- O'Neill LP, VerMilyea MD, Turner BM. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nat Genet 2006; 38: 835 841. [DOI] [PubMed] [Google Scholar]
- Clement TM, Anway MD, Uzumcu M, Skinner MK. Regulation of the gonadal transcriptome during sex determination and testis morphogenesis: comparative candidate genes. Reproduction 2007; 134: 455 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witschi E. Development: Rat. : Altman PL, Dittmer DS. (eds.), Growth Including Reproduction and Morphological Development. Washington DC: Federation of American Societies for Experimental Biology; 1962: 304 314. [Google Scholar]
- O'Rahilly R. Early human development and the chief sources of information on staged human embryos. Eur J Obstet Gynecol Reprod Biol 1979; 9: 273 280. [DOI] [PubMed] [Google Scholar]
- Cupp AS, Kim GH, Skinner MK. Expression and action of neurotropin-3 and nerve growth factor in embryonic and early postnatal rat testis development. Biol Reprod 2000; 63: 1617 1628. [DOI] [PubMed] [Google Scholar]
- Russo MA, Giustizieri ML, Favale A, Fantini MC, Campagnolo L, Konda D, Germano F, Farini D, Manna C, Siracusa G. Spatiotemporal patterns of expression of neurotrophins and neurotrophin receptors in mice suggest functional roles in testicular and epididymal morphogenesis. Biol Reprod 1999; 61: 1123 1132. [DOI] [PubMed] [Google Scholar]
- Robinson LL, Townsend J, Anderson RA. The human fetal testis is a site of expression of neurotrophins and their receptors: regulation of the germ cell and peritubular cell population. J Clin Endocrinol Metab 2003; 88: 3943 3951. [DOI] [PubMed] [Google Scholar]
- Wilhelm D, Martinson F, Bradford S, Wilson MJ, Combes AN, Beverdam A, Bowles J, Mizusaki H, Koopman P. Sertoli cell differentiation is induced both cell-autonomously and through prostaglandin signaling during mammalian sex determination. Dev Biol 2005; 287: 111 124. [DOI] [PubMed] [Google Scholar]
- Adams IR, McLaren A. Sexually dimorphic development of mouse primordial germ cells: switching from oogenesis to spermatogenesis. Development 2002; 129: 1155 1164. [DOI] [PubMed] [Google Scholar]
- Jenkins E, Moss JB, Pace JM, Bridgewater LC. The new collagen gene COL27A1 contains SOX9-responsive enhancer elements. Matrix Biol 2005; 24: 177 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers G, Wilson M, Wilhelm D, Koopman P. SOX8 is expressed during testis differentiation in mice and synergizes with SF1 to activate the Amh promoter in vitro. J Biol Chem 2003; 278: 28101 28108. [DOI] [PubMed] [Google Scholar]
- Bernard P, Tang P, Liu S, Dewing P, Harley VR, Vilain E. Dimerization of SOX9 is required for chondrogenesis, but not for sex determination. Hum Mol Genet 2003; 12: 1755 1765. [DOI] [PubMed] [Google Scholar]
- Peirano RI, Wegner M. The glial transcription factor Sox10 binds to DNA both as monomer and dimer with different functional consequences. Nucleic Acids Res 2000; 28: 3047 3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sock E, Pagon RA, Keymolen K, Lissens W, Wegner M, Scherer G. Loss of DNA-dependent dimerization of the transcription factor SOX9 as a cause for campomelic dysplasia. Hum Mol Genet 2003; 12: 1439 1447. [DOI] [PubMed] [Google Scholar]