Abstract
Continual spermatogenesis at a quantitatively normal level is required to sustain male fertility. The foundation of this process relies on maintenance of an undifferentiated spermatogonial population consisting of spermatogonial stem cells (SSCs) that self-renew as well as transient amplifying progenitors produced by differentiation. In mammals, type Asingle spermatogonia form the SSC population, but molecular markers distinguishing these from differentiating progenitors are undefined and knowledge of mechanisms regulating their functions is limited. We show that in the mouse male germline the transcriptional repressor ID4 is expressed by a subpopulation of undifferentiated spermatogonia and selectively marks Asingle spermatogonia. In addition, we found that ID4 expression is up-regulated in isolated SSC-enriched fractions by stimulation from GDNF, a key growth factor driving self-renewal. In mice lacking ID4 expression, quantitatively normal spermatogenesis was found to be impaired due to progressive loss of the undifferentiated spermatogonial population during adulthood. Moreover, reduction of ID4 expression by small interfering RNA treatment abolished the ability of wild-type SSCs to expand in vitro during long-term culture without affecting their survival. Collectively, these results indicate that ID4 is a distinguishing marker of SSCs in the mammalian germline and plays an important role in the regulation of self-renewal.
Keywords: ID4, self-renewal, spermatogonial stem cell, undifferentiated spermatogonia
The expression of ID4 localizes to single spermatogonia in the germline of adult mice and is regulated by GDNF signaling, influencing self-renewal capacity of spermatogonial stem cells to maintain quantitatively normal spermatogenesis.
INTRODUCTION
Spermatogenesis is a tissue-specific stem cell-dependent process, relying on self-renewal and differentiation of spermatogonial stem cells (SSCs) to support continual production of millions of spermatozoa daily [1]. In rodents, SSCs represent a subfraction of the undifferentiated spermatogonial population that also consists of Apaired (Apr, cohorts of 2 cells) and Aaligned (Aal, cohorts of 4, 8, or 16 cells) spermatogonia [1, 2]. The traditional model states that SSCs are single spermatogonia, referred to as type Asingle or As, capable of self-renewal and production of progenitors that remain connected by an intercellular bridge [1–4]. Thus, generation of Apr spermatogonia represents initial SSC differentiation, and this cohort of germ cells has the potential to develop further as a syncytium into Aal spermatogonia, eventually giving rise to spermatozoa. Recent studies suggest that in the mouse germline some chained spermatogonia (Apr and short Aal cohorts) can fragment to produce new single spermatogonia that may become As [5], but evidence that these cells contribute to the SSC pool has not been described. Therefore, it has been postulated that some chained undifferentiated spermatogonia are not irreversibly committed to differentiation and can contribute to replenishment of the SSC pool in certain instances when As spermatogonia are lost [5]. Regardless, it is currently widely accepted that As spermatogonia represent the actual SSC population in mammalian testes.
Similar to other tissue-specific stem cells, SSC fate decisions are influenced by a niche microenvironment that is dictated by a main support cell population, which are the Sertoli cells in mammalian testes [6]. In rodents, glial cell line-derived neurotrophic factor (GDNF) is expressed by Sertoli cells to drive self-renewal and survival of SSCs [7–9]. While key pieces of information regarding the testis stem cell niche have been discovered, knowledge of molecular mechanisms regulating SSC fate decisions is limited. Studying SSC self-renewal and differentiation is challenging due to lack of specific markers to distinguish As from Apr and Aal progenitor spermatogonia. While a pure population of SSCs cannot currently be isolated from mammalian testes, specific populations enriched for SSCs can be fractionated based on cell surface phenotypes, particularly the thymus cell antigen 1 (THY1)-expressing cell population, which is enriched 30-fold for SSCs and contains nearly the entire SSC pool in testes of adult mice [8]. Culture of THY1(+) spermatogonia in defined media conditions with GDNF supplementation supports maintenance of a germ cell population consisting of SSCs and Apr/Aal-like undifferentiated spermatogonia [9, 10]. Also, self-renewal and initial differentiation of SSCs is supported for long periods in these cultures, providing an in vitro model that can be used to intensively study molecular mechanisms controlling SSC functions [8, 11].
The effects of niche factors such as GDNF on SSC fate decisions are mediated by activation or suppression of specific molecular networks. Of particular interest are transcription factors up-regulated by GDNF signaling that subsequently affect expression of other genes to influence self-renewal or differentiation. The inhibitor of DNA binding (ID) proteins are a class of helix-loop-helix molecules consisting of four different isoforms (ID1–ID4) that are expressed preferentially in undifferentiated cell populations where they play diverse roles in fate determination by functioning as transcriptional repressors [12–16]. Examination of ID isoform expression in the mouse testis has revealed that ID1 is expressed exclusively in spermatocytes, with ID2 and ID3 expression being localized to Sertoli cells [17]. Interestingly, only ID4 expression was detected in the type-A spermatogonial population [17]. However, this population is heterogeneous, and whether expression is localized specifically to certain subpopulations of type-A spermatogonia or plays a role in germ cell development is unknown. In previous studies, we identified genes with enriched expression in the THY1(+) germ cell fraction from mouse testes and genes influenced by GDNF signaling in cultured THY1(+) germ cells [11, 18]. Examination of those databases for expression of ID proteins revealed that ID4 is highly enriched in the THY1(+) fraction and up-regulated by GDNF signaling. Thus, in the current study, we tested the hypothesis that ID4 expression is restricted to the SSC (i.e., As spermatogonia) population in the mouse germline and plays an important role in the regulation of SSC functions.
MATERIALS AND METHODS
Animals
Cultures of wild-type THY1(+) germ cells were generated from B6;129S-Gt(ROSA)26Sor/J (mice designated ROSA; The Jackson Laboratory). All the germ cell stages in these mice express a LacZ transgene and are easily identifiable in recipient seminiferous tubules following transplantation. Recipient mice for SSC transplantations were F1 offspring from mating 129ScvP × C57BL/6, which are immunologically compatible with ROSA donor mice. Id4−/− mice generated by knock-in of a GFP transgene have been described previously [19]. All the animal procedures were approved by the Pennsylvania State University Institutional Animal Care and Use Committee.
Antibodies
Staining for ID4 expression was achieved using a mouse anti-human ID4 polyclonal antibody (Abnova) at 1:100–1:200 dilutions. Detection of PLZF expression was achieved by staining with a rabbit anti-human PLZF polyclonal antibody (Santa Cruz Biotechnology) at 1:100–1:200 dilutions. GCNA1 expression was identified by staining with a rat anti-mouse GCNA1 IgM monoclonal antibody (a generous gift from Dr. George Enders) used at 1:40 dilution. Detection of GFP expression was conducted with a fluorescein isothiocyanate (FITC)-conjugated goat anti-GFP antibody (Abcam, Inc.) at 1:300 dilution. Secondary antibodies included donkey anti-mouse IgG conjugated to Alexa 546 (Invitrogen, Inc.), biotinylated goat anti-mouse IgG (Santa Cruz Biotechnology), goat anti-rat IgM conjugated to Alexa 488 (Invitrogen, Inc.), and biotinylated rabbit anti-goat IgG (Santa Cruz Biotechnology). All the secondary antibodies were used at dilutions of 1:1000–1:2000.
Assessment of Fertility and Spermatogenesis
Fertility and spermatogenesis of Id4−/− and control Id4+/− male mice were examined at the following postpubertal stages: young adult (2–3 mo of age), mature adult (4–5 mo of age), and aged (7–8 mo of age). For assessing fertility status at each age point, males of both genotypes were housed with two pubertal (2–3 mo of age) wild-type C57BL/6 female mice for 3–4 wk and the number of litters sired for each was recorded. Males were considered fertile if one litter of pups was born from the mating. Spermatogenesis was assessed by determining spermatozoa concentration in the epididymis, measuring testis weight, and examining cross-sections of seminiferous tubules. For spermatozoa concentration, the epididymis of each testis was removed and homogenized in a standard volume of ice-cold PBS, pH 7.4. The number of spermatozoa was then quantified using a hemocytometer. Testis weight was measured with intact tunica albuginea prior to fixation. Cross-sections of testes were examined by light microscopy at 10–100× magnification. The percentage of seminiferous tubules with disrupted spermatogenesis was determined by counting the number of round tubules with a Sertoli cell-only phenotype or remnants of differentiating germ cells only in five random fields of view for each cross-section and dividing by the total number of round tubules within the same field of view. Two cross-sections of each testis were examined. The number of ID4(+) and PLZF(+) spermatogonia per seminiferous tubule cross-section in testes of adult wild-type mice was determined by counting the number of stained cells after immunohistochemistry analysis in five round tubules of five random fields of view for each cross-section of three different animals. Quantification of PLZF(+) spermatogonia in cross-sections of seminiferous tubules from Id4−/− and Id4+/− mice involved evaluation of 75 round tubules from each genotype at all the age points. Serum testosterone concentration in adult Id4−/− and Id4+/− mice was determined by commercial ELISA (parameter testosterone immunoassay; R&D Systems, Inc.).
Digital Imaging
Tissue cross-sections, cells, and whole-mount seminiferous tubules were viewed with an IX51 inverted fluorescent microscope equipped with FITC, DAPI (4′,6-diamidino-2-phenylindole), and TRITC (tetramnethylrhodamine-5/6-isothiocyanate) filters. Digital images were captured with a DP71 digital microscope camera and Cell Sense software (Olympus, Inc.).
Immunohistochemistry
Testes were fixed in Bouin solution according to procedures described previously [20], dehydrated, embedded in paraffin, and 5-μm thick cross-sections adhered to glass slides. Following deparaffinization, antigen retrieval was achieved by boiling in citrate buffer (10 mM citric acid, 0.05% Tween-20, pH 6.0). Sections were then blocked for endogenous activity of both peroxidases by incubation in 0.03% H2O2 and biotin using a commercial kit (avidin/biotin blocking kit; Invitrogen). Next, nonspecific antibody binding was blocked by incubation with 10% normal serum for 30 min. Sections were then incubated with primary antibodies overnight at 4°C, followed by washing in PBS and incubation with secondary antibody for 1 h at room temperature. For colorimetric staining, samples were again washed and developed with a horseradish peroxidase (HRP)-conjugated streptavidin kit (Vector Labs) and hematoxylin counterstaining. For immunofluorescence, sections were washed, and glass coverslips were mounted with aqueous medium containing DAPI (Invitrogen, Inc.).
Immunocytochemistry
For examination of freshly isolated THY1(+) germ cells, ∼5 × 104 cells were adhered to poly-L-lysine-coated coverslips prior to fixation. Examination of cultured THY1(+) germ cell clumps was achieved by fixation within culture wells on STO feeder cell monolayers. For examination of cultured THY1(+) germ cell clumps as single cells, the clumps were gently removed from STO feeders by gentle pipetting and disassociated by trypsin/EDTA (ethylenediaminetetraacetic acid) digestion. Approximately 5 × 104 cells were then adhered to poly-L-lysine-coated coverslips. All the cells were fixed in 4% paraformaldehyde for 10 min at room temperature followed by incubation with ice-cold methanol for 2 min to permeablize the membranes. Nonspecific antibody binding was blocked by incubation with 10% normal serum for 5 min at room temperature. Cells were then washed with PBS and incubated with primary antibodies diluted in PBS containing 0.5% BSA at 4°C overnight. On the next day, cells were washed in PBS and incubated with secondary antibody for 1 h at room temperature. Cells were again washed and incubated with DAPI in PBS to label the cell nuclei and examined by fluorescent microscopy. The percentage of isolated THY1(+) cells expressing ID4 was determined by counting the number of red fluorescent cells within five random fields of view and dividing by the total number of DAPI-labeled nuclei within the same fields.
Isolation and Culture of THY1(+) Germ Cells
The THY1(+) germ cell fraction was isolated from both pup and adult mice using magnetic-activated cell sorting (MACS) as described previously [21]. For cultures, THY1(+) germ cells were maintained on mitotically inactivated STO feeder cell monolayers in mouse serum-free medium (mSFM) [8] supplemented with 20 ng/ml recombinant human GDNF (PeproTech) and 1 ng/ml recombinant human FGF2 (BD Biosciences).
Quantitative Real-Time PCR
RNA was extracted using Trizol reagent (Invitrogen, Inc.), treated with DNase I, and reverse transcribed using oligo(d)T priming and Moloney murine leukemia virus reverse transcriptase. Relative transcript abundance in each sample for Id4 and the constitutively expressed gene ribosomal protein S2 (Rps2) was determined using validated TaqMan probe assays (Applied Biosystems). Differences in Id4 expression between samples were determined by normalization to Rps2 as described previously [18, 22].
Small Interfering RNA Treatments
Cultured THY1(+) germ cells (105 cells) were treated with 75 pmol of pooled nontargeting control small interfering RNAs (siRNAs) that lack homology to any known protein-encoding sequence or Id4-specific siRNAs (both purchased from Dharmacon, Inc.) using lipofectamine 2000 reagent (Invitrogen, Inc.) as described previously [18, 22]. On the next day, mSFM was changed with supplementation of GDNF and FGF2 growth factor.
SSC Transplantation
To assay for stem cell content of cultured ROSA THY1(+) germ cell populations, single cell suspensions of germ cell clumps were generated by trypsin/EDTA digestion and washed in mSFM, and total germ cell numbers were determined by counting with a hemacytometer. Cells were then suspended at a concentration of 2 × 106 cells/ml in mSFM and microinjected into the seminiferous tubules of recipient mice that were treated with busulfan (60 mg/kg of body weight) at least 6 wk prior to eliminate endogenous spermatogenesis as described previously [21]. For each recipient testis, ∼7 μl of cell suspension was infused filling 80%–90% of the surface tubules. Testes were then examined for colonies of donor-derived spermatogenesis by X-Gal (G-Biosciences) staining 2 mo after transplantation. The number of colonies was determined by manual counting using a SZX51 stereo zoom microscope (Olympus, Inc.). SSC number in transplanted cultures was determined using the equation: SSC number = (number of donor-derived colonies of spermatogenesis/105 THY1(+) cells injected) × (105 total cells harvested from the culture well). To determine expansion of SSCs over time in vitro, SSC numbers were multiplied by subculture dilution ratios.
Western Blot Analysis
For determination of ID4 protein levels following siRNA transfections, cultured THY1(+) germ cells were lysed in RIPA (Santa Cruz Biotechnology) 48 h after transfection. Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes followed by blocking in PBS containing nonfat milk powder. Membranes were then incubated with rabbit anti-mouse ID4 polyclonal antibody (Santa Cruz Biotechnology) at 1:1000 dilution for 2 h at room temperature followed by washing in TBS-T (10 mM Tris, pH 7.4, 100 mM NaCl, and 0.1% Tween-20). Blots were then incubated with an HRP-conjugated goat anti-rabbit IgG polyclonal antibody (Santa Cruz Biotechnology) at 1:5000 dilution for 1 h at room temperature followed by washing in TBS-T. Detection of proteins was achieved by developing with a chemiluminescent substrate and viewed with a ChemiDoc imager (Bio-Rad). Digital images were captured for further analyses, and blots were striped for incubation with rabbit anti-human tubulin-beta antibody (Novus Biologicals) at 1:10 000 dilution to detect total tubulin-beta within each sample. Expression of ID4 was compared between cells transfected with Id4-specific or control siRNA by normalization of the ID4 band density with that of tubulin-beta.
Whole Mount Immunofluorescence
Seminiferous tubules were separated by digestion with 1 mg/ml collagenase in Hanks Balanced Salt Solution (HBSS; Invitrogen) at 37°C for 10 min and gentle agitation. Tubules were then washed three times in HBSS by gravity sedimentation on ice to remove interstitial cells and fixed in 4% paraformaldehyde for 3 h at 4°C. Samples were then dehydrated by incubation in increasing concentrations (25%, 50%, 75%, and 100%) of TBS-T (10 mM Tris, pH 7.4, 100 mM NaCl, and 0.01% Triton X-100) for 30-min increments. Nonspecific antibody binding was blocked by incubation in 10% normal serum, and tubules were incubated with primary antibodies diluted in PBS containing 0.5% BSA and 0.1% Triton X-100 overnight at 4°C. On the next day, tubules were washed in PBS and incubated with secondary antibody for 1 h at room temperature and then washed extensively in PBS. For imaging, the tubules were spread on glass slides. Stained cells within 25 μm of neighboring cells were considered clones of spermatogonia, whereas individual cells separated by a distance of 25 μm or greater were considered single spermatogonia.
Statistical Analyses
Differences between means were determined using the general linear model one-way ANOVA function of SPSS statistical software (IBM Corporation). Multiple comparisons were conducted using the Tukey post hoc test for significance.
RESULTS
ID4 Is Expressed Selectively by Undifferentiated Spermatogonia in the Male Mouse Germline
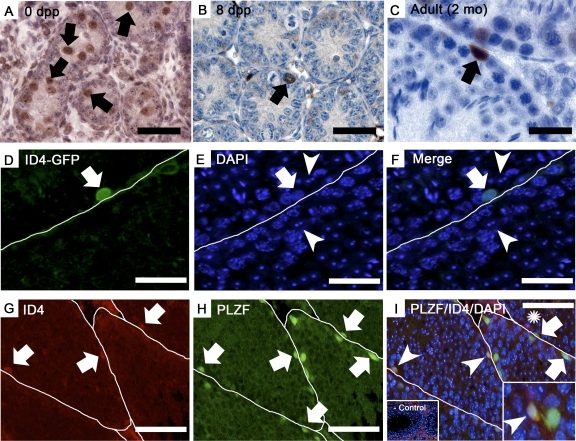
During postnatal development, prospermatogonia, also referred to as gonocytes, migrate from the center of seminiferous tubules to the basement membrane during the period of 0–6 days postpartum (dpp) [23, 24]. After this translocation, prospermatogonia transition into SSCs that serve to sustain spermatogenesis from puberty until old age by continually generating Apr/Aal progenitor spermatogonia or differentiating spermatogonia that continue their development to provide the first round of spermatogenesis [2, 25]. Here we used immunohistochemistry to characterize further the expression pattern of ID4 in the male mouse germline. Several commercially available antibodies were examined for their specificity of staining the germline in cross-sections of testes from adult mice, and we found one that stained spermatogonia only while others stained Sertoli cells and preleptotene spermatocytes in addition to spermatogonia (see Supplemental Discussion and Supplemental Fig. S1; all the supplemental data are available online at www.biolreprod.org). Previous studies revealed that the N-terminal region of ID isoforms is conserved [15] and that ID1–ID3 are expressed by Sertoli cells and spermatocytes [17]. Thus, we reasoned that antibodies staining more than spermatogonia were not specific for ID4 and likely cross-reacted with the other ID isoforms. Indeed, these antibodies were found to stain Sertoli cells and germ cells in cross-sections of testes from Id4−/− mice, further indicating cross-reactivity with the other ID isoforms (see Supplemental Discussion and Supplemental Fig. S2). In contrast, the one antibody that stained spermatogonia only in testes of wild-type mice did not stain any cells in the Id4−/− mice, confirming its specificity for the detection of ID4. Therefore, we used this antibody to further characterize expression of ID4 in the male germline. At 0 dpp, expression of ID4 was restricted to prospermatogonia in seminiferous cords (Fig. 1A). In testes of mice at 8 dpp and adults at 2–3 mo of age, which contain the entire array of undifferentiated spermatogonia, expression of ID4 was observed by individual spermatogonia residing along the basement membrane of seminiferous tubules (Fig. 1, B and C, and Supplemental Fig. S1). In adults, all the ID4(+) spermatogonia observed contained oval nuclei and were flattened along the basement membrane, which are defining features of undifferentiated spermatogonia in mice [26]. A bias of ID4-stained cells in seminiferous tubules of adult mice at certain stages of the seminiferous cycle was not evident (n = 3 testes cross-sections examined and 15-staged round seminiferous tubules), consistent with the notion that the number of As and Apr spermatogonia is constant during the cycle of the seminiferous epithelium [27]. To confirm the specificity of ID4 expression in type-A spermatogonia, we used immunofluorescence to localize the expression of GFP in cross-sections of testes from homozygous Id4(−)/Gfp(+) knock-in mice. In this model, ID4-expressing cells are marked by the presence of GFP [19], and these analyses revealed that GFP expression is localized to spermatogonia only (Fig. 1, D–F), thereby validating findings from immunohistochemistry analyses of wild-type mice. Collectively, these observations demonstrate that expression of ID4 in the germline of male mice is restricted to prospermatogonia (i.e., gonocytes) during neonatal development and retained in the type-A spermatogonial population of adult testes.
FIG. 1.
Expression of ID4 in the undifferentiated spermatogonial population. A–C) Immunohistochemistry staining for ID4 expression in cross-sections of testes from mice at the neonatal age of 0 dpp (A), prepubertal age of 8 dpp (B), and adulthood at 2 mo of age (C). D–F) Immunofluorescent staining for GFP expression in a testis cross-section from an adult (2 mo of age) homozygous Id4(−)/Gfp(+) knock-in mouse. Green fluorescence indicates GFP(+) expression, which was observed by individual spermatogonia only (arrow) and not by Sertoli cells (arrowhead). DAPI was used to stain all the cell nuclei; white lines mark the basement membrane of adjacent seminiferous tubules. G–I) Immunofluorescent staining for ID4 and PLZF expression in cross-sections of testes from adult mice (4 mo of age). ID4 staining (G, arrows) is observed in only a few spermatogonia. In contrast, PLZF staining (H, arrows) is observed in many spermatogonia within seminiferous tubules. In the merged image (I), three different types of spermatogonia are observed, including those that stain for expression of the undifferentiated spermatogonial marker PLZF (arrows), germ cells expressing both PLZF and ID4 (arrowheads), and germ cells expressing ID4 only (asterisks). The left inset is the negative control with normal IgG in place of primary antibodies; note the nonspecific red fluorescence of interstitial cells that can also be observed in cross-sections stained for expression of ID4. The right inset is a magnified view of spermatogonia stained for expression of both PLZF and ID4 (arrowhead). White lines mark the boundaries of seminiferous tubules. Bars are 50 μm for A–F and 200 μm for G–I.
Expression of ID4 Marks a Subtype of the Undifferentiated Spermatogonial Population
Next, we further examined which type-A spermatogonia express ID4. Previous studies showed that PLZF is expressed by As/Aal spermatogonia [28, 29]; thus, we used this marker to examine ID4 expression in the context of the undifferentiated spermatogonial population in vivo. Based on examination of tissue cross-sections from testes of adult mice, only 1.2 ± 0.6 ID4(+) spermatogonia (n = 3 cross-sections examined from different testes) were observed per seminiferous tubule cross-section that contained ID4(+) cells (Fig. 1G), whereas 3.2 ± 1.5 PLZF(+) spermatogonia (n = 3 cross-sections examined from different testes) were observed per tubule (Fig. 1H). Coimmunofluorescent staining of cross-sections from testes of adult mice revealed three distinct spermatogonial populations, including 1) those expressing both ID4 and PLZF, 2) germ cells expressing PLZF only, and 3) germ cells expressing ID4 only (Fig. 1I). The PLZF(+) population was mostly ID4(−) and only ∼50% of the ID4(+) cells also expressed PLZF. These observations indicate that germ cells coexpressing ID4 and PLZF are a subfraction of the undifferentiated spermatogonial population, whereas ID4(+) germ cells that do not coexpress PLZF may represent another subtype.
Expression of ID4 Is Restricted to Asingle Spermatogonia in Seminiferous Tubules of Adult Mice
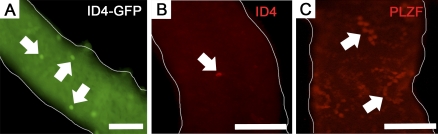
The SSC designation has been applied to As spermatogonia [1–4], whereas Apr and Aal spermatogonia do not function as stem cells during steady-state spermatogenesis, being referred to as transient amplifying progenitors [30]. Currently, molecular markers distinguishing As, Apr, and Aal spermatogonia are undefined. Here we used whole-mount imaging of seminiferous tubules to examine whether ID4 expression is localized to single and/or cohorts (i.e., Apr and Aal) of spermatogonia. Examination of seminiferous tubules from adult (2–3 mo of age) homozygous Id4(−)/Gfp(+) knock-in mice revealed expression by single cells (Fig. 2A), and cohorts of two or greater GFP(+) cells were not observed in any sample (n = 3 different mice and 6 testes examined). To confirm this finding, whole-mount immunofluorescent staining was performed with seminiferous tubules from adult (2–3 mo of age) wild-type C57BL/6 mice (n = 3 different mice and 6 testes examined). Similarly, only single cells were found to stain for expression of ID4 (Fig. 2B), which is in stark contrast to the cohorts of spermatogonia that stain for expression of the undifferentiated spermatogonial marker PLZF (Fig. 2C). Collectively, these observations indicate that expression of ID4 selectively marks at least some of the As spermatogonia in seminiferous tubules of adult mice.
FIG. 2.
Whole-mount imaging of ID4 expressing cells in mouse seminiferous tubules. A) Representative image of seminiferous tubules from adult homozygous Id4(−)/Gfp(+) knock-in mice. Only single GFP(+) cells were observed (arrows). B) Representative image of immunofluorescent staining (red fluorescence) for expression of ID4 in seminiferous tubules from adult wild-type mice. Only single ID4(+) cells were observed (arrow). C) Representative image of immunofluorescent staining (red fluorescence) for expression of the undifferentiated spermatogonial marker PLZF in seminiferous tubules from adult wild-type mice. In contrast to the staining of ID4 expression in single cells only, PLZF staining is observed in cohorts of spermatogonia (arrows). Bars are 100 μm for A and 50 μm for B and C.
Expression of ID4 Is Enriched in the THY1(+) Germ Cell Fraction of Mouse Testes and Regulated GDNF Signaling
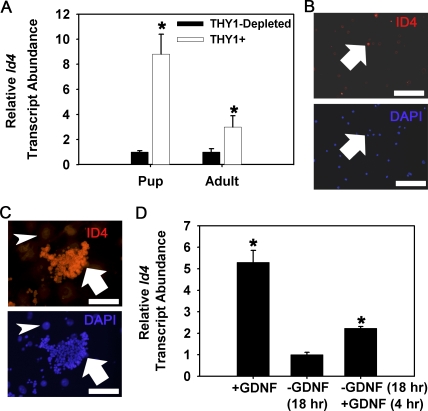
The THY1(+) germ cell fraction from mouse testes is enriched for SSCs and contains nearly the entire pool of SSCs in the total testis cell population [8, 31]. When maintained in serum-free conditions and exposed to GDNF, THY1(+) germ cells form clumps consisting of both SSCs and Apr/Aal-like spermatogonia [9, 10]. Also, SSC self-renewal is supported in these clumps in a GDNF-dependent manner for extended periods of time [9]. In a previous microarray study, we found that Id4 gene expression is enriched in the THY1(+) germ cell fraction of prepubertal pups [11]. Confirming and expanding on that finding, Id4 transcript abundance was determined to be 9-fold and 3-fold greater in the THY1(+) germ cell fraction compared to the THY1-depleted cell population from testes of prepubertal pups and adult mice, respectively (Fig. 3A). Using immunocytochemical staining of the MACS-isolated THY1(+) cell fraction from prepubertal pups at 8 dpp, a developmental stage in which all undifferentiated spermatogonial subtypes are present, we found that ID4(+) cells represent a subpopulation comprising ∼11% (n = 2 different isolated cell populations) of the total cells (Fig. 3B). Examination of gene expression in cultures of THY1(+) germ cells derived from prepubertal pups, validated to contain SSCs by transplantation following withdrawal and replacement of GDNF, revealed that Id4 gene expression is significantly up-regulated by exposure to GDNF (Fig. 3D). In addition, an uneven staining pattern for ID4 expression was observed within cultured THY1(+) germ cell clumps following immunocytochemical staining (Fig. 3C). Further examination of disassociated clumps as single cell suspensions revealed that ∼31% (n = 3 different cultures) of the cells express ID4, indicating greater enrichment compared to the freshly isolated cell fraction. Together, these results demonstrate that ID4 is expressed by a subpopulation of the SSC-containing THY1(+) germ cell fraction in testes of mice and suggests an important role in regulation of SSC self-renewal.
FIG. 3.
Expression of ID4 in the SSC-containing THY1(+) germ cell fraction. A) Quantitative real-time PCR analyses for Id4 gene expression in the THY1(+) germ cell fraction compared to the THY1-depleted testis cell population of testes from prepubertal pup (8 dpp) and adult (3 mo of age) mice. Relative Id4 transcript abundance was calculated by normalization to the constitutively expressed gene Rps2. Data are mean ± SEM for three different MACS-isolated cell populations. *Denotes significant difference at P < 0.05. B) Representative images of immunocytochemical staining for ID4-expressing cells in the MACS-isolated THY1(+) germ cell fraction from prepubertal mice at 8 dpp that contain the entire array of spermatogonia found in the male germline. ID4(+) cells (red fluorescence, arrow) were found to comprise only ∼11% of the cell population. DAPI (blue fluorescence) was used to stain all the cell nuclei. Bars = 100 μm. C) Immunocytochemical staining for ID4 expression in cultured THY1(+) germ cell clumps. Nuclear staining was observed in germ cell clumps (red fluorescence, arrow) but not feeder cell monolayers (arrowhead). DAPI (blue fluorescence) was used to stain all the cell nuclei. Bars are 100 μm. D) Quantitative real-time PCR analyses for GDNF-regulation of Id4 gene expression in cultured THY1(+) germ cells. Germ cell clumps continually cultured with GDNF supplementation (+GDNF) were subjected to 18 h withdrawal of GDNF (−GDNF 18 h) followed by replacement of GDNF for 4 h (+GDNF 4 hr). Relative Id4 transcript abundance was determined by normalization to the constitutively expressed gene Rps2. Data are mean ± SEM for three different THY1(+) germ cell cultures and expressed as fold-difference from the −GDNF treatment. *Denotes significant difference (P < 0.05) from −GDNF treatment.
ID4 Loss-of-Function Results in Impaired Male Fertility
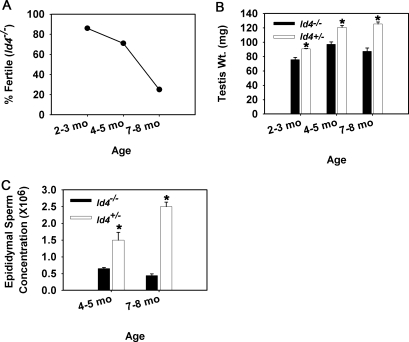
To investigate the importance of ID4 in regulating SSC fate, we examined the fertility and spermatogenesis of Id4-null mice generated by replacing exons 2 and 3 of the Id4 locus with a Gfp-neo transgene [19], referred to hereafter as Id4−/−. A hallmark of impaired SSC function is aging-related loss of the undifferentiated spermatogonial population resulting in disrupted spermatogenesis and fertility defects [28, 29]. Thus, using young adult (2–3 mo of age), mature adult (4–5 mo of age), and aged (7–8 mo of age) mice, we examined the fertility of the Id4−/− male mice by mating them with wild-type C57BL/6 females as well as their spermatozoal production. Aging-related defects in fertility and spermatogenesis were determined by comparison to age-matched control Id4+/− male littermates. Previous studies found no measurable deficiencies of Id4+/− mice compared to wild-type Id4+/+ mice, and Id4+/− mice are thus a suitable control for comparison to Id4−/− littermates [19, 32]. Most (∼86%) of the Id4−/− males (n = 7) and all the control males (n = 7) examined were fertile at 2–3 mo of age, siring at least one litter. The single Id4−/− male that did not sire a litter at this age did produce offspring at 4 mo of age; thus, it is assumed that all Id4−/− males were fertile at 2–3 mo of age. At the mature adult stage of 4–5 mo of age, the percentage of Id4−/− males (n = 7) that were fertile was reduced to 71%, and fertility continued to decline to only 25% for older males at 7–8 mo of age (n = 7; Fig. 4A). In comparison, all the Id4+/− control males examined were fertile at both the mature adult (n = 7) and aged (n = 7) points of development. Testis weight, a measure of normal spermatogenesis, was significantly (P < 0.05) reduced for Id4−/− males to 84%, 81%, and 66% of Id4+/− controls at 2–3, 4–5, and 7–8 mo of age, respectively (Fig. 4B). Importantly, spermatozoa concentration within the epididymis of Id4−/− male mice was also significantly (P < 0.05) reduced to only 56% and 18% of that in Id4+/− control males at 4–5 and 7–8 mo of age, respectively, when major fertility defects were prominent (Fig. 4C).
FIG. 4.
Reproductive phenotype of adult ID4-deficient male mice. A) Fertility assessment of ID4-deficient male mice (Id4−/−) at the developmental stages of young adult (2–3 mo of age), mature adult (4–5 mo of age), and aged (7–8 mo of age). Fertility was assessed based on whether a male sired pups when exposed to two pubertal wild-type female mice. Data are derived from seven Id4−/− male mice at each age point. B) Comparison of testes weights, a measure of normal spermatogenesis, between ID4-deficient mice (Id4−/−) and control mice with sufficient expression of ID4 (Id4+/−) at developmental stages of young adult (2–3 mo of age), mature adult (4–5 mo of age), and aged (7–8 mo of age). Data are mean ± SEM for three different mice of both genotypes at each age point. *Denotes significant difference at P < 0.05. C) Comparison of epididymal sperm concentration between Id4−/− and control Id4+/− male mice at the developmental stages of mature adult (4–5 mo of age) and aged (7–8 mo of age), periods when fertility defects are prominent. Data are mean ± SEM for three different mice of both genotypes at each age point. *Denotes significant difference at P < 0.05.
Previous studies revealed reduced brain size of Id4−/− mice [19, 32], which could disrupt the function of the hypothalamic-pituitary-gonadal (HPG) axis and contribute to impaired spermatogenesis and fertility. To explore this possibility, we measured serum testosterone concentration in adult mice (3–4 mo of age) and found it to be no different (P > 0.05) in Id4−/− males (2.0 ± 0.6 ng/ml, n = 3 different mice) compared to control Id4+/− males (2.2 ± 1.1 ng/ml, n = 3 different mice), indicating that the lack of ID4 does not impair normal HPG function. Overall, these results demonstrate that the lack of ID4 expression results in a subfertile condition in male mice and that ID4 is needed for quantitatively normal spermatogenesis. Moreover, the fertility defects are due to deficiencies in the germ cell population and not as a result of altered function of the endocrine system.
Disrupted Spermatogenesis and Loss of the Undifferentiated Spermatogonial Population in Mice with Impaired Expression of ID4
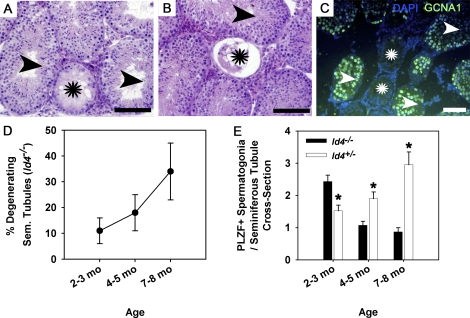
To investigate further the cause of subfertility for Id4−/− mice, we examined cross-sections of testes for qualitatively normal spermatogenesis. At all the ages evaluated, seminiferous tubules with disrupted spermatogenesis were observed, including complete degeneration of the seminiferous epithelium with a Sertoli cell-only phenotype (Fig. 5A) and tubules containing remnants of differentiated germ cells but no spermatogonia (Fig. 5B). Tubules with an apparent Sertoli cell-only phenotype were confirmed by lack of staining for the general germ cell marker GCNA1 (Fig. 5C). These phenotypes are indicative of SSC failure in mouse testes due to lack of successive generations of spermatogonia to provide the next cohort of developing germ cells. Interestingly, the percentage of disrupted tubules increased with age (Fig. 5D). At 2–3 mo of age, 11% of tubules had impaired spermatogenesis, increasing to 18% and 34% at 4–5 and 7–8 mo of age (n = 3 different mice examined and 6 total testes at each age), respectively. Degenerative seminiferous tubules were not seen in testes of control Id4+/− males at any age point examined (n = 5 testes examined at each age).
FIG. 5.
Effects of ID4 loss-of-function on spermatogenesis in testes of adult mice. A and B) Representative images of a cross-section from testes of Id4−/− mice. Seminiferous tubules with depletion of the germ cell population, a Sertoli cell-only phenotype (stars), and qualitatively normal spermatogenesis (arrowheads) were observed. C) Representative image of immunofluorescent staining of a testis cross-section from an adult Id4−/− mouse for expression of the general germ cell marker GCNA1 (green fluorescence). Seminiferous tubules completely lacking germ cells (stars), confirming a Sertoli cell-only phenotype, are observed along with tubules containing germ cells (arrowheads). DAPI (blue fluorescence) was used to stain all the cell nuclei. D) Assessment of the percentage of seminiferous tubules containing a degenerating phenotype and lacking active spermatogenesis in cross-sections of testes from Id4−/− mice at the developmental stages of young adult (2–3 mo of age), mature adult (4–5 mo of age), and aged (7–8 mo of age). Data are mean ± SEM for three different mice at each age point. E) Comparison of the number of undifferentiated spermatogonia in cross-sections of seminiferous tubules from testes of Id4−/− and control Id4+/− mice at young adult (2–3 mo of age), mature adult (4–5 mo of age), and aged (7–8 mo of age) developmental stages. Undifferentiated spermatogonia were identified based on immunohistochemical staining for expression of the marker PLZF. Data are mean ± SEM for three different mice and 75 round seminiferous tubules of both genotypes at each age point. *Denotes significant difference at P < 0.05.
While degenerative phenotypes were observed in testes of Id4−/− mice, the majority of tubules contained qualitatively normal spermatogenesis, but reduced epididymal spermatozoa concentration suggested impairment of quantitatively normal spermatogenesis. Previous studies indicate that ID proteins influence differentiation of the neural progenitor population, and loss of ID4 causes premature differentiation [19, 32]. Thus, we decided to examine whether expression of ID4 is important for sustaining a quantitatively normal pool of undifferentiated spermatogonia. To address this, the number of cells expressing the undifferentiated spermatogonial marker PLZF in seminiferous tubules of Id4−/− males was examined and compared to testes of control Id4+/− mice (Fig. 5E). Surprisingly, testes of 2–3 mo old Id4−/− mice were found to contain significantly (P < 0.05) more PLZF(+) spermatogonia per seminiferous tubule by 62% compared to control Id4+/− males (Fig. 4F). Thereafter, the number of PLZF(+) spermatogonia per seminiferous tubule was significantly (P < 0.05) decreased in Id4−/− mice to only 54% and 34% of that in Id4+/− controls at 4–5 and 7–8 mo of age, respectively. Overall, these results indicate that the lack of ID4 expression results in premature differentiation of SSCs followed by aging-related decline of the undifferentiated spermatogonial population, suggesting an important role for ID4 in regulating SSC self-renewal.
Reduction of ID4 Expression Impairs Self-Renewal of SSCs In Vitro
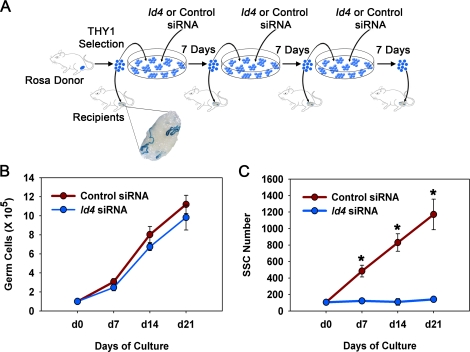
Examining SSC self-renewal in vivo is challenging because the phenotype of testes in which SSC survival and differentiation is impaired would be similar to those with disrupted SSC self-renewal, each resulting in loss of the germ cell population. Culture of THY1(+) germ cells in serum-free conditions with GDNF supplementation supports the formation of germ cell clumps consisting of SSCs and Apr/Aal-like spermatogonia. In this system, SSC self-renewal and differentiation is supported for extended periods, providing an in vitro model to study the effects of experimental manipulations on SSC fate decisions [9]. Therefore, we decided to examine whether expression of ID4 is important for GDNF-regulation of SSC self-renewal in these cultures. Because the cultured cell population is heterogeneous, consisting of SSCs and non-stem-cell spermatogonia, transplantation analyses were utilized to determine the effects of impairing ID4 expression by siRNA treatment on maintenance of SSCs over a 21-day period (Fig. 6A). SSC doubling time is ∼6 days in vitro [9]; thus, these analyses examined stem cell expansion over greater than three self-renewal cycles.
FIG. 6.
Effects of impairing ID4 expression on GDNF-induced self-renewal of SSCs from wild-type mice. A) Experimental strategy for examining the effects of impairing ID4 expression by siRNA treatment on expansion of SSCs in cultures of THY1(+) germ cells supplemented with GDNF over a 21-day period, which constitutes greater than three self-renewal cycles in vitro. THY1(+) germ cells from ROSA donor mice (6–8 dpp) that express a LacZ marker transgene in all the germ cells were treated with nontargeting control or Id4-specific siRNA and maintained in serum-free conditions with GDNF and FGF2 supplementation for 7 days. Single cell suspensions were then created, and a portion of the cells were transplanted into testes of recipient mice to determine SSC content based on the ability to reestablish colonies of spermatogenesis. The remaining cells were again treated with nontargeting control or Id4-specific siRNA and cultured another 7 days before collection and transplantation or retreatment with siRNA. This protocol was conducted three times over the 21-day culture period to track SSC expansion. B) Assessment of overall germ cell expansion in cultures of THY1(+) germ cells treated with nontargeting control or Id4-specific siRNA over a 21-day period. Data are mean ± SEM for three different THY1(+) germ cell cultures. *Denotes significant difference at P < 0.05. C) Assessment of SSC expansion in cultures of THY1(+) germ cells treated with nontargeting control or Id4-specific siRNA over a 21-day period. SSC numbers were determined based on donor-derived colonies of spermatogenesis following transplantation into recipient testes. Data are mean ± SEM for three different THY1(+) germ cell cultures. *Denotes significant difference at P < 0.01. Note that the error bars do not extend beyond the circles for Id4 siRNA treatment.
Transient reduction of ID4 expression in THY1+ germ cell cultures generated from wild-type ROSA donors was achieved by treatment with a pool of targeting siRNAs, and transplantation assays were conducted to determine the effects on SSC content after three successive self-renewal cycles. After 24 h, Id4 siRNA treatment reduced transcript abundance by ∼59% compared to cells treated with nontargeting control siRNA; protein abundance was found to be reduced by ∼75% 48 h after transfection (see Supplemental Fig. S3). Importantly, this effect was transient, and Id4 transcript abundance was not different compared to control siRNA-treated cells by 7 days after transfection; thus, ID4 expression was not different at the time of transplantation. During the 21-day treatment period, total germ cell expansion (SSCs and non-stem-cell spermatogonia) was 9.6-fold (n = 3 different cultures) in Id4 siRNA-treated cultures, which was not different (P > 0.05) compared to the 11.2-fold (n = 3 different cultures) increase in control siRNA-treated cultures (Fig. 6B). Transplantation of both control and Id4 siRNA-treated cells produced colonies of spermatogenesis within recipient seminiferous tubules, confirming the presence of SSCs (Fig. 6A). Quantification of colony numbers, a measure of SSC content of the injected cell suspension, revealed an 11.1-fold (n = 3 different cultures) expansion of SSCs in control siRNA-treated cultures over the 21-day period. In contrast, in Id4 siRNA-treated cultures, expansion of SSCs was essentially abolished, increasing by only 1.3-fold, which was significantly (P < 0.01) reduced compared to control siRNA-treated cultures (Fig. 6C). Interestingly, the number of SSCs did not significantly (P > 0.05) decline over the 21-day period in Id4 siRNA-treated cultures, indicating no major effect on SSC survival. In addition, the effects of Id4 siRNA treatment were greater for expansion of SSCs, being reduced by 8.5-fold compared to control siRNA (11.1-fold increase in control siRNA/1.3-fold increase in Id4 siRNA), than for total germ cell numbers in Id4 siRNA-treated cultures, which was only reduced by 1.2-fold compared to controls (11.2-fold increase in control siRNA/9.6-fold increase in Id4 siRNA). Collectively, these results indicate that ID4 plays an important role in regulating the rate of SSC self-renewal, and the effects of impairing ID4 are greater in SSCs compared to the other non-stem-cell spermatogonia also present in the cultured THY1(+) germ cell population.
DISCUSSION
Tissue-specific stem cells comprise subfractions of heterogeneous undifferentiated cell populations that are few in number and defined based on functional abilities to maintain and reestablish tissue homeostasis. In the mammalian testis, SSCs represent a rare subfraction of the heterogeneous undifferentiated spermatogonial population that also consists of Apr and Aal progenitor germ cells. The traditional stem cell model of mammalian spermatogenesis states that As spermatogonia form the SSC population and that initial differentiation generates Apr and Aal progenitors [1–4]. Until now, molecular markers of As spermatogonia that are not expressed by Apr and Aal germ cells have not been described. In this study, we show that ID4 is expressed by a subpopulation of the undifferentiated spermatogonial population. Importantly, expression was observed exclusively by single cells within seminiferous tubules, indicative of As spermatogonia in the germline. While the expression of other markers has been described, including PLZF [28, 29], GDNF family receptor α 1 [33], nanos homologue 2 [34], nanos homologue 3 [35, 36], neurogenin 3 [37], and LIN28 [38], none of these is expressed exclusively by As spermatogonia. Thus, ID4 represents the first described molecular marker to distinguish As from other progenitor spermatogonia. However, further experimentation is required to determine if all As spermatogonia are ID4(+) or only a portion of the population. Recent studies examining expression of other undifferentiated spermatogonial markers indicate that the As spermatogonial population in testes of mice is heterogeneous [38–40]. Thus, it is possible that ID4 expression marks only some As spermatogonia, and because ID proteins are known to control cell cycle progression, expression may be temporal, occurring during cell division. Also, we found that ID4 expression does not appear to overlap entirely with the more widespread expression of PLZF by undifferentiated spermatogonia, suggesting that ID4(+)/PLZF(−) germ cells are another subpopulation of undifferentiated spermatogonia, further strengthening the notion of heterogeneity in the As spermatogonial population.
While we observed expression of ID4 by As spermatogonia only, the possibility that some Apr cells are also ID4(+) cannot be ruled out. During the process of self-renewal, SSCs must transition through a state of being paired cells prior to the completion of mitosis and migration away from each other to become As spermatogonia. Thus, it is possible that some SSCs could be visualized as false Apr spermatogonia, a phenomenon that has been described previously [41, 42]. In this regard, if all SSCs express ID4, then rare paired spermatogonia could be seen as ID4(+). However, the analyses performed in the current study did not observe any paired ID4(+) spermatogonia, possibly because complete quantification of all As and Apr germ cells within seminiferous tubules was not conducted.
Impairment of SSC functions manifests as infertility as a result of progressive loss of the undifferentiated spermatogonial population, causing formation of seminiferous tubules with a Sertoli cell-only phenotype. In addition, reduced activity by the SSC population in providing insufficient numbers of undifferentiated spermatogonia could cause subfertility from lowered number of sperm in the ejaculate. Impairment of either self-renewal or differentiation of SSCs could cause an identical subfertile phenotype; thus, distinguishing between them from disrupted expression of certain molecules is challenging. In this study, we found that lack of ID4 expression results in formation of some seminiferous tubules with a Sertoli cell-only phenotype, indicating complete loss of the SSC population. This finding implies an important role for ID4 in the process of self-renewal that is needed to maintain the SSC population, thereby preventing loss of the germline. In addition, the majority of tubules in ID4-null mice were found to contain qualitatively normal spermatogenesis but with reduced numbers of undifferentiated spermatogonia that became progressively more severe with increasing age. Interestingly, at 2 mo of age, the number of PLZF(+) spermatogonia in testes of ID4-deficient mice was found to be significantly greater compared to mice with sufficient expression of ID4 and then dramatically declined as the animals aged. These findings indicate premature differentiation of SSCs that generated an overabundance of progenitor spermatogonia following completion of the first round of spermatogenesis and impaired rate of self-renewal, leading to a deficiency in the SSC pool from which the next round of progenitor spermatogonia could be derived. This conclusion was supported by results of experiments showing complete inhibition of SSC self-renewing expansion in cultures of wild-type SSCs when ID4 expression was transiently impaired. Overall, these combined findings from in vivo and in vitro studies indicate that ID4 plays an important role in regulating both the rate and process of SSC self-renewal that is needed to maintain the stem cell pool.
Most, if not all, tissues within the mammalian body function based on stem and progenitor cell populations, and it is likely that many of the mechanisms regulating self-renewal and differentiation are conserved. The mammalian testis with its associated SSC population is a valuable model system to study these mechanisms because of the availability of a robust functional transplantation assay and method for long-term culture of primary stem cells. For several progenitor cell populations, ID proteins play a role in maintaining an undifferentiated state by functioning as transcriptional repressors [12–16, 19, 32, 43]. Expression of ID4 appears to be a conserved regulator of stem and progenitor cell populations in several tissues, including the germline. Previous studies showed that mice lacking ID4 expression have reduced brain size due to premature differentiation of neural progenitors [19, 32]. Here we show that ID4 is a regulator of SSC self-renewal to support quantitatively normal spermatogenesis and male fertility. Collectively, findings from the current study and that of others with ID4-null mice indicate that ID4 is an important regulator of many undifferentiated progenitor cell populations. With mice, loss of ID4 expression manifests as an age-related decline in tissue function that is highlighted in the current study by increased fertility defects of male mice with advancing age after puberty. Thus, in humans, dysfunction of ID proteins may be underlying causes of several disease states induced by progressive loss of tissue homeostasis.
ACKNOWLEDGMENTS
We thank Dr. Mark A. Israel for the generous donation of Id4 mutant mice and Dr. Fred Sablitzky for helpful discussions.
Footnotes
Supported by grants HD061665 and HD058137 awarded to J.M.O. from the National Institutes of Health.
REFERENCES
- Oatley JM, Brinster RL. Regulation of spermatogonial stem cell self-renewal in mammals. Ann Rev Cell Dev Biol 2008; 24: 263 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij DG, Russell LD. All you wanted to know about spermatogonia but were afraid to ask. J Androl 2000; 21: 776 798. [PubMed] [Google Scholar]
- Huckins C. The spermatogonial stem cell population in adult rats. I. Their morphology, proliferation and maturation. Anat Rec 1971; 169: 533 557. [DOI] [PubMed] [Google Scholar]
- Oakberg EF. Spermatogonial stem-cell renewal in the mouse. Anat Rec 1971; 169: 515 531. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Sharma M, Nabeshima Y, Braun RE, Yoshida S. Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science 2010; 328: 62 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley MJ, Racicot KE, Oatley JM. Sertoli cells dictate spermatogonial stem cell niches in the mouse testis. Biol Reprod 2011; 84: 639 645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Lindahl M, Hyvönen ME, Parvinen M, de Rooij DG, Hess MW, Raatikainen-Ahokas A, Sainio K, Rauvala H, Lakso M, Pichel JG, Westphal H, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 2000; 287: 1489 1493. [DOI] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL. Culture conditions and single growth factors affect fate determination of mouse spermatogonial stem cells. Biol Reprod 2004; 71: 722 731. [DOI] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc Natl Acad Sci U S A 2004; 101: 16489 16494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley JM, Kaucher AV, Avarbock MR, Brinster RL. Regulation of mouse spermatogonial stem cell differentiation by STAT3 signaling. Biol Reprod 2010; 83: 427 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley JM, Oatley MJ, Avarbock MR, Tobias JW, Brinster RL. Colony stimulating factor 1 is an extrinsic stimulator of mouse spermatogonial stem cell self-renewal. Development 2009; 136: 1191 1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 1990; 61: 49 59. [DOI] [PubMed] [Google Scholar]
- Sun XH, Copeland NG, Jenkins NA, Baltimore D. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol 1991; 11: 5603 5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen Y, Weintraub H, Benezra R. Overexpression of Id protein inhibits the muscle differentiation program: in vivo association of Id with E2A proteins. Genes Dev 1992; 6: 1466 1479. [DOI] [PubMed] [Google Scholar]
- Riechmann V, van Crüchten I, Sablitzky F. The expression pattern of Id4, a novel dominant negative helix-loop-helix protein, is distinct from Id1, Id2 and Id3. Nucleic Acids Res 1994; 22: 749 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deed RW, Jasiok M, Norton JD. Lymphoid-specific expression of the Id3 gene in hematopoietic cells. Selective antagonism of E2A basic helix-loop-helix protein associated with Id3-induced differentiation of erythroleukemia cells. J Biol Chem 1998; 273: 8278 8286. [DOI] [PubMed] [Google Scholar]
- Sablitzky F, Moore A, Bromley M, Deed RW, Newton JS, Norton JD. Stage- and subcellular-specific expression of Id proteins in male germ and Sertoli cells implicates distinctive regulatory roles for Id proteins during meiosis, spermatogenesis, and Sertoli cell function. Cell Growth Differ 1998; 9: 1015 1024. [PubMed] [Google Scholar]
- Oatley JM, Avarbock MR, Telaranta AI, Fearon DT, Brinster RL. Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc Natl Acad Sci U S A 2006; 103: 9524 9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun K, Mantani A, Garel S, Rubenstein J, Israel MA. Id4 regulates neural progenitor proliferation and differentiation in vivo. Development 2004; 131: 5441 5448. [DOI] [PubMed] [Google Scholar]
- Russell LD, Ettlin RA, Hikim AP, Clegg ED. Mammalian spermatogenesis. : Histological and Histopathological Evaluation of the Testis. St. Louis: Cache River Press; 1990: 1 40. [Google Scholar]
- Oatley JM, Brinster RL. Spermatogonial stem cells. Methods Enzymol 2006; 419: 259 282. [DOI] [PubMed] [Google Scholar]
- Oatley JM, Avarbock MR, Brinster RL. Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. J Biol Chem 2007; 282: 25842 25851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckins C, Clermont Y. Evolution of gonocytes in the rat testis during late embryonic and early post-natal life. Arch Anat Histol Embryol 1968; 51: 341 354. [PubMed] [Google Scholar]
- Bellve AR, Cavicchia JC, Millette CF, O'Brien DA, Bhatnagar YM, Dym M. Spermatogenic cells of the prepubertal mouse: isolation and morphological characterization. J Cell Biol 1977; 74: 68 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Sukeno M, Nakagawa T, Ohbo K, Nagamatsu G, Suda T, Nabeshima Y. The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development 2006; 133: 1495 1505. [DOI] [PubMed] [Google Scholar]
- Chiarini-Garcia H, Russell LD. High-resolution light microscopic characterization of mouse spermatogonia. Biol Reprod 2001; 65: 1170 1178. [DOI] [PubMed] [Google Scholar]
- Chiarini-Garcia H, Hornick JR, Griswold MD, Russell LD. Distribution of type A spermatogonia in the mouse is not random. Biol Reprod 2001; 65: 1179 1185. [DOI] [PubMed] [Google Scholar]
- Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, Braun RE. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet 2004; 36: 647 652. [DOI] [PubMed] [Google Scholar]
- Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet 2004. 36: 653 659. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Nabeshima Y, Yoshida S. Functional identification of the actual and potential stem cell compartments in mouse spermatogenesis. Dev Cell 2007; 12: 195 206. [DOI] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL. Spermatogonial stem cells share some, but not all, phenotypic and functional characteristics with other stem cells. Proc Natl Acad Sci U S A 2003; 100: 6487 6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford L, Walker R, Kondo T, van Crüchten I, King ER, Sablitzky F. Id4 is required for the correct timing of neural differentiation. Dev Biol 2005; 280: 386 395. [DOI] [PubMed] [Google Scholar]
- Grisanti L, Falciatori I, Grasso M, Dovere L, Fera S, Muciaccia B, Fuso A, Berno V, Boitani C, Stefanini M, Vicini E. Identification of spermatogonial stem cell subsets by morphological analysis and prospective isolation. Stem Cells 2009; 27: 3043 3052. [DOI] [PubMed] [Google Scholar]
- Sada A, Suzuki A, Suzuki H, Saga Y. The RNA-binding protein nanos2 is required to maintain murine spermatogonial stem cells. Science 2009; 325: 1394 1398. [DOI] [PubMed] [Google Scholar]
- Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y. Conserved role of nanos proteins in germ cell development. Science 2004; 301: 1239 1241. [DOI] [PubMed] [Google Scholar]
- Lolicato F, Marino R, Paronetto MP, Pellegrini M, Dolci S, Geremia R, Grimaldi P. Potential role of nanos3 in maintaining the undifferentiated spermatogonia population. Dev Biol 2008; 313: 725 738. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Takakura A, Ohbo K, Abe K, Wakabayashi J, Yamamoto M, Suda T, Nabeshima Y. Neurogenin3 delineates the earliest stages of spermatogenesis in the mouse testis. Dev Biol 2004; 269: 447 458. [DOI] [PubMed] [Google Scholar]
- Zheng K, Wu X, Kaestner KH, Wang PJ. The pluripotency factor LIN28 marks undifferentiated spermatogonia in mouse. BMC Dev Biol 2009; 9: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Sada A, Yoshida S, Saga Y. The heterogeneity of spermatogonia is revealed by their topology and expression of marker proteins including the germ cell-specific proteins Nanos2 and Nanos3. Dev Biol 2009; 336: 222 231. [DOI] [PubMed] [Google Scholar]
- Tokuda M, Kadokawa Y, Kurahashi H, Marunouchi T. CDH1 is a specific marker for undifferentiated spermatogonia in mouse testes. Biol Reprod 2007; 76: 130 141. [DOI] [PubMed] [Google Scholar]
- Huckins C. The spermatogonial stem cell population in adult rats. I. Their morphology, proliferation and maturation. Anat Rec 1971; 169: 533 557. [DOI] [PubMed] [Google Scholar]
- de Rooij DG. Morphometric description of spermatogonial stem cells and expansion of their clonal derivatives. In: Orwig KE, Hermann BP (eds), Male Germline Stem Cells: Developmental and Regenerative Potential. New York: Springer Science/Business Media; 2010: 89 105 [Google Scholar]
- Kreider BL, Benezra R, Rovera G, Kadesch T. Inhibition of myeloid differentiation by the helix-loop-helix protein Id. Science 1992; 27: 1700 1702. [DOI] [PubMed] [Google Scholar]