Abstract
To determine the interaction site(s) of ATP-sensitive K+ (KATP) channels for G-proteins, sulfonylurea receptor (SUR2A or SUR1) and pore-forming (Kir6.2) subunits were reconstituted in the mammalian cell line, COS-7. Intracellular application of the G-protein βγ2-subunits (Gβγ2) caused a reduction of ATP-induced inhibition of Kir6.2/SUR channel activities by lessening the ATP sensitivity of the channels. Gβγ2 bound in vitro to both intracellular (loop-NBD) and C-terminal segments of SUR2A, each containing a nucleotide-binding domain (NBD). Furthermore, a single amino acid substitution in the loop-NBD of SUR (Arg656Ala in SUR2A or Arg665Ala in SUR1) abolished the Gβγ2-dependent alteration of the channel activities. These findings provide evidence that Gβγ modulates KATP channels through a direct interaction with the loop-NBD of SUR.
Keywords: ATP-sensitive K+ channel/ATP sensitivity/channel modulation/G-protein βγ-subunits/sulfonylurea receptor
Introduction
ATP-sensitive K+ (KATP) channels (Noma, 1983), which are composed of the inwardly rectifying K+ channel (Kir6.0) and the sulfonylurea receptor (SUR) subunit (Inagaki et al., 1995), play an important role in various cellular responses by linking the metabolic status of the cell to its membrane potential (Ashcroft, 1988). KATP channels modulate action potential duration and excitability in heart, and control insulin secretion in pancreatic β cells. Furthermore, KATP channels mediate hypoxic vasodilatation in coronary and cerebral vascular smooth muscles. A direct effect of hypoxia on Ca2+ channels may also be critical for the decrease in smooth muscle force (Taggart and Wray, 1998). Whereas neuronal Ca2+ channels are regulated negatively by guanine nucleotide binding regulatory proteins (G-proteins) (Clapham and Neer, 1993; Hille, 1994), KATP channels are regulated positively by G-protein-coupled receptors such as galanin (Dunne et al., 1989), adenosine A1 (Kirsch et al., 1990) and somatostatin (Ribalet and Eddlestone, 1995) receptors. This regulation of KATP channels is thought to be mediated by the α-subunits (Gαs) of Gi family G-proteins (Kirsch et al., 1990; Terzic et al., 1994; Ribalet and Eddlestone, 1995; Sánchez et al., 1998). It remains to be elucidated, however, which subunit(s) of the G-protein complex (Gα or the βγ-subunits, Gβγ) preferentially mediates this modulation of KATP, which subunit of the KATP channel (SUR and/or Kir) interacts with G-proteins, and whether the interaction between KATP and G-protein is direct or indirect.
To address these issues at the molecular level, KATP channels were functionally expressed in mammalian COS-7 cells by introducing Kir6.2 and SUR (SUR2A or SUR1) cDNAs, and the effects of Gβγ2 proteins on the channel activities were studied using the inside-out configuration of the patch–clamp technique. In addition, a direct association of Gβγ with KATP channels was determined by in vitro binding using glutathione S-transferase (GST) proteins fused with intracellular loop and C-terminal segments of SUR2A, and N- and C-terminal segments of Kir6.2. Finally, an amino acid residue critical for the Gβγ-dependent modulation of KATP channels was identified within the Gβγ-binding site of SUR by site-directed mutagenesis.
Results
Cloning of rat Kir6.2 and SUR1
Determination of the nucleotide and predicted amino acid sequences of the inserts of clones pVGK1 and pBGK1 (see Materials and methods) revealed that GK15 is a rat version of Kir6.2 (BIR; Inagaki et al., 1995). Three nucleotide changes in the sequences were shown not to alter the coding amino acid residues: A (15), T (40) and T (327) of rat BIR (Isomoto et al., 1996) were G, C and C in our clones, respectively.
In addition, determination of the nucleotide and predicted amino acid sequences of the inserts of clone pRSUR1 revealed 15 nucleotide changes from the sequence of rat SUR1 (Aguilar-Bryan et al., 1995), which resulted in nine amino acid substitutions and one amino acid insertion: Ala116, Thr487, Pro626, Ala630, Ile699, Pro835, Gly836, Gly1313 and Pro1562 were determined as Gly, Ser, Ser, Thr, Thr, Gln, Arg, Arg and Ser, encoded by GGT, AGC, TCC, ACA, ACC, CAG, CGA, CGT and TCG, respectively. Ser encoded by AGC was inserted at position 741. Twenty-one nucleotide changes in the sequences were shown not to alter the coding amino acid residues: C (21), C (24), G (27), G (1971), C (1977), T (1978), A (1986), G (1992), G (1995), C (2109), T (2310), T (2610), T (2634), C (3762), A (3765), A (3777), C (3780), T (4723), C (4724), C (4725) and T (4728) were G, G, A, T, G, C, G, A, A, T, C, A, C, T, T, G, G, A, G, T and C in our clones, respectively. Thus, the insert of this clone contained a cDNA sequence encoding SUR1.
Functional expression of Kir/SUR channels in COS cells
To study the modulation of KATP channels by G-proteins at the molecular level, KATP channels were expressed in mammalian COS-7 cells by introducing SUR (SUR2A or SUR1) and Kir6.2 cDNAs.
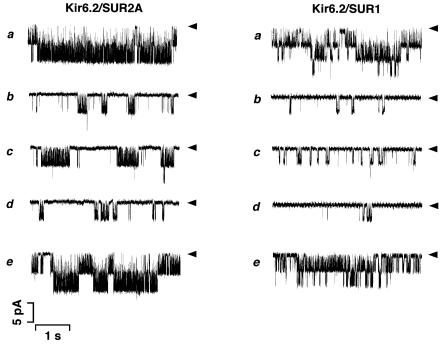
Figure 1 illustrates unitary K+ currents recorded from excised inside-out membrane patches of COS-7 cells that have been transfected with Kir6.2/SUR2A (left) or Kir6.2/SUR1 (right) cDNAs. Although unitary K+ currents through Kir6.2/SUR2A and Kir6.2/SUR1 channels were scarcely observed in cell-attached patches (n = 30), the channels were spontaneously activated upon formation of inside-out patches (Figure 1a). The open probability without ATP, NPoi (see Materials and methods), was 2.39 ± 0.84 (n = 9) (Kir6.2/SUR2A) or 0.96 ± 0.37 (n = 9) (Kir6.2/SUR1). These channel activities were dramatically inhibited by application of 100 µM ATP to the cytoplasmic side of the excised patches (Figure 1b) and recovered after wash-out of ATP (Figure 1e). The channels reconstituted with Kir6.2/SUR2A or Kir6.2/SUR1 had a unitary conductance of 81.1 ± 3.4 (n = 6) or 80.1 ± 2.9 (n = 3) pS, respectively, at a membrane potential of –50 mV, the values that were similar to those reported (Inagaki et al., 1995, 1996). These findings indicate that KATP channels are functionally reconstituted in COS-7 cells by transfection with a combination of Kir6.2/SUR2A or Kir6.2/SUR1 cDNAs.
Fig. 1. Effects of Gβγ2 on Kir6.2/SUR2A and Kir6.2/SUR1 channels expressed in COS-7 cells. Single channel currents were recorded continuously from COS-7 cells expressing Kir6.2/SUR2A (left) or Kir6.2/SUR1 (right) channels, using the inside-out patch configuration. (a) Control (in the absence of ATP); (b) in the presence of 100 µM ATP; (c) in the presence of both 100 µM ATP and 50 pM Gβγ2; (d) after wash-out of Gβγ2; (e) after wash-out of both Gβγ2 and ATP. The membrane potential was –50 mV. Arrowheads, the zero current levels. Down indicates inward in the current traces.
Modulation of Kir/SUR channels by Gβγ
As shown in Figure 1c (left), bath (i.e. intracellular) application of 50 pM Gβγ2 (purified from bovine brain) caused an ‘enhancement’ of Kir6.2/SUR2A channel activities in the presence of 100 µM ATP (or a ‘reduction’ of ATP-induced inhibition of channel activities). The unitary conductance remained unchanged, but the relative NPo increased ∼2.5-fold (Figure 2). This enhancement of channel activities by Gβγ2 was abolished after wash-out of Gβγ2 (Figure 1d, left). Gβγ2 exerted similar effects on Kir6.2/SUR1 channels (Figure 1c and d, right). In contrast, Gβγ2 could not influence either Kir6.2/SUR2A or Kir6.2/SUR1 channels unless ATP was present (n = 5), indicating that Gβγ2 increases NPo only when the channel activities are suppressed by ATP.
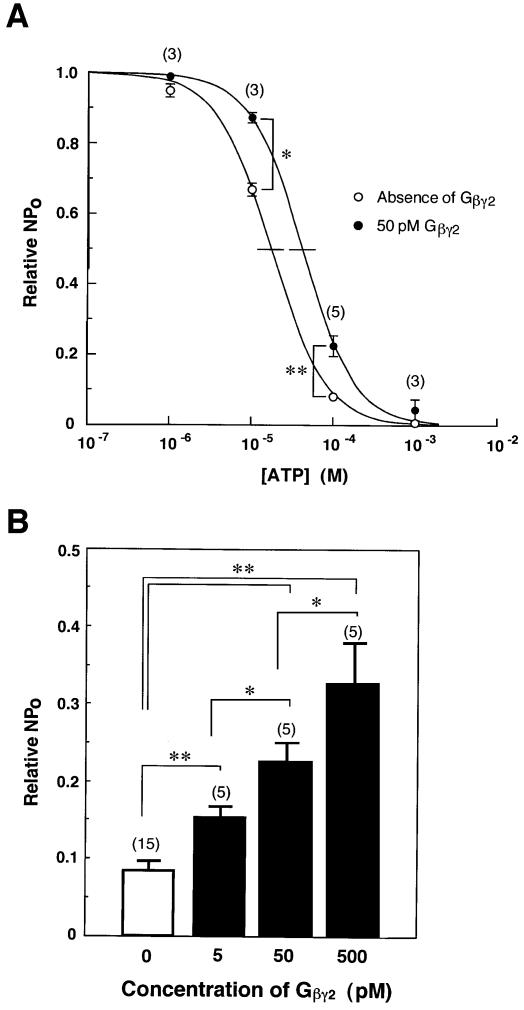
Fig. 2. ‘Antagonistic’ actions of Gβγ2 against ATP-induced inhibition of Kir6.2/SUR2A channels. (A) Concentration–inhibition relationships for ATP in the absence (open circles) or presence (filled circles) of 50 pM Gβγ2 in Kir6.2/SUR2A channels. (B) Dose-dependent response relationships for Gβγ2 observed in Kir6.2/SUR2A currents in the presence of 100 µM ATP. Current responses (NPo) were normalized to those in the absence of ATP (Relative NPo, see Materials and methods). The number of experiments is indicated in parentheses. *P<0.05. **P<0.001.
The pattern of channel activities induced by Gβγ2 in the presence of ATP was different for Kir6.2/SUR2A and Kir6.2/SUR1 channels (Figure 1b and c). Gβγ2 increased the burst length rather than the opening frequency of Kir6.2/SUR2A channels, whereas Gβγ2 increased the opening frequency of Kir6.2/SUR1 channels without obvious changes in the burst duration. These suggest that there is a different mechanism for the Gβγ2-induced modulation of SUR2A- and SUR1-linked Kir6.2 channels.
To characterize further the modulation of Kir6.2/SUR2A channels by Gβγ2, their responses to Gβγ2 were studied with various concentrations of ATP (Figure 2A). The activities of Kir6.2/SUR2A channels were inhibited by ATP in a concentration-dependent manner (Figure 2A, open circles). The inhibition constant (Ki) of ATP was 16.8 µM, and the Hill coefficient (nH) was 1.35.
When Gβγ2 complex was added to the intracellular solution at a concentration of 50 pM (Figure 2A, filled circles), the Kir6.2/SUR2A channel activities were enhanced by 2.8- and 1.3-fold in the presence of 100 and 10 µM ATP, respectively. Moreover, an increase in the Ki value of ATP from 16.8 to 40.7 µM was observed after addition of Gβγ2. In contrast, an nH of 1.30 was comparable to that (1.35) before addition of Gβγ2. Thus, Gβγ2 produced a rightward shift of the concentration–inhibition curves for ATP without affecting the Hill coefficient. Similar effects of Gβγ2 on the concentration–inhibition curves for ATP were observed in Kir6.2/SUR1 channels (n = 3), in which the Ki value of ATP was markedly increased from 3.43 to 18.9 µM by application of Gβγ2, while nH was not significantly influenced (1.33 and 1.28) in the absence and presence of Gβγ2, respectively.
The Ki values of ATP for Kir6.2/SUR1 (3.43 µM) and for Kir6.2/SUR2A (16.8 µM) indicate that these channels are 3- or 5-fold more sensitive to ATP than those reported, respectively (Inagaki et al., 1995, 1996). This may be accounted for, at least in part, by different levels of membrane phospholipids (Baukrowitz et al., 1998; Shyng and Nichols, 1998) in various cell lines used. However, a higher sensitivity of Kir6.2/SUR1 channels to ATP (Inagaki et al., 1996) was conserved in ATP-induced inhibition as compared with Kir6.2/SUR2A channels.
On the other hand, the ATP-induced inhibition of Kir6.2/SUR2A channels was reduced by Gβγ2 in a concentration-dependent manner (Figure 2B). These results indicate that Gβγ2 ‘antagonizes’ ATP-induced inhibition of Kir6.2/SUR channels, as observed in Giα-mediated modulation (Kirsch et al., 1990; Terzic et al., 1994; Sánchez et al., 1998). On the other hand, Goα at 10 pM failed to facilitate Kir6.2/SUR2A channels in the presence of ATP (n = 3), whereas Gβγ2 at a lower concentration of 5 pM appreciably facilitated the channels (Figure 2B), indicating that Goα is not involved in the G-protein-mediated modulation of KATP channels (Kirsch et al., 1990). The activities of Goα were verified by examining its ability to couple to adenosine A1 receptors as reported previously (Asano et al., 1995). Recently, Gβγ–i1 and Gβγ–i2 have been reported not to affect Kir6.2/SUR1 channels (Sánchez et al., 1998). The discrepancy between our results and earlier reports may reflect different isoforms of Gβγ that were purified from bovine brain by using different procedures (Asano et al., 1993).
Moreover, bath application of 100 µM 1-(5-isoquinolinylsulfonyl)-2-methyl-piperazine (H7, n = 5), an inhibitor of cyclic nucleotide-dependent protein kinase and protein kinase C (PKC), bath application of 50 µM wortmannin (n = 4), an inhibitor of phosphatidylinositol (PI) 3-kinase, or pre-treatment with 2 mM neomycin sulfate (n = 4), an inhibitor of phospholipase C (PLC), exerted no effect on the Gβγ2-induced facilitation of Kir6.2/SUR2A channels in the presence of 100 µM ATP. These results are consistent with the view that protein kinase A, PKC, PI 3-kinase and PLC are not primarily involved in this channel modulation by Gβγ, although adenylyl cyclase, PI 3-kinase and PLC are known to be stimulated by Gβγ.
Effects of mutant Kir6.2 subunits on Gβγ-dependent modulation of KATP channels
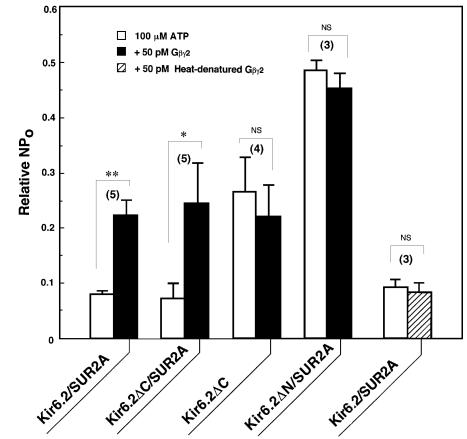
To determine which subunit of KATP channels (Kir6.2 and/or SUR) interacts with Gβγ, we studied the effect of Gβγ2 on deletion mutant Kir6.2 channels when co-expressed with wild-type SUR2A (Figure 3). The mutant Kir6.2ΔC, where the C-terminal region (amino acid residues 366–390) of Kir6.2 had been removed, showed channel activities without the SUR2A subunit (Kir6.2ΔC, open box; Tucker et al., 1997), in which the sensitivity to ATP-induced inhibition was lower than that of wild-type Kir6.2/SUR2A channels (Kir6.2/SUR2A, open box). Moreover, Gβγ2 did not further enhance the channel activities of Kir6.2ΔC in the presence of ATP (Kir6.2ΔC, filled box). In contrast, co-expression of SUR2A with Kir6.2ΔC gained a high sensitivity to ATP (Kir6.2ΔC/SUR2A, open box; Tucker et al., 1997) and also a Gβγ2-mediated reduction of ATP-induced inhibition (Kir6.2ΔC/SUR2A, filled box), as observed in the wild-type Kir6.2/SUR2A channels. Thus, Gβγ reversed the high sensitivity of Kir6.2ΔC to ATP that was induced by SUR2A.
Fig. 3. Mutational effects of the Kir6.2 subunit on the Gβγ2-dependent modulation of KATP channels. The mutant Kir6.2 (Kir6.2ΔC or Kir6.2ΔN) was co-expressed with the wild-type SUR2A in COS-7 cells (Kir6.2ΔC/SUR2A or Kir6.2ΔN/SUR2A) or expressed alone (Kir6.2ΔC). For comparison, co-expression of the wild-type Kir6.2 and the SUR2A was also carried out (Kir6.2/SUR2A). Current responses (NPo) to 100 µM ATP were measured in the absence (open boxes) or presence (filled boxes) of 50 pM Gβγ2. Some Gβγ2 was heat treated at 90°C for 3 min (hatched box). NPo are expressed as a ratio to those in the absence of ATP. The number of experiments is indicated in parentheses. *P<0.05. **P<0.001. NS, not significant.
On the other hand, another mutant Kir6.2 (Kir6.2ΔN) channels with a deletion of the N-terminal region (amino acid residues 2–14 of Kir6.2) were less sensitive to ATP when compared with wild-type or Kir6.2ΔC channels, even though wild-type SUR2A was co-expressed (Kir6.2ΔN/SUR2A, open box; Koster et al., 1999; Reimann et al., 1999). The Kir6.2ΔN/SUR2A channels showed no reduction of the ATP-induced inhibition in the presence of Gβγ2 (Kir6.2ΔN/SUR2A, filled box), standing in contrast to Kir6.2/SUR2A and Kir6.2ΔC/SUR2A channels. This ‘antagonistic’ action of Gβγ2 against ATP-induced inhibition as typically observed in the wild-type Kir6.2/SUR2A channels was abolished by heat denaturation of Gβγ2 proteins added (Kir6.2/SUR2A, hatched box). These findings indicate that SUR is necessary for KATP channels to allow Gβγ to ‘antagonize’ the ATP-induced channel inhibition. It is further suggested that Gβγ might interfere with the interaction between SUR and Kir6.2 by acting directly on SUR, but not through diffusible second messengers, and that Gβγ-mediated modulation of KATP channels might take place preferentially in a high ATP-sensitive channel state that is closely associated with SUR.
Gβγ2 binds to the intracellular loop and C-terminus of SUR
SUR, a member of the ATP-binding cassette (ABC) superfamily, has two domains (NBD-1 and NBD-2) for binding and hydrolysis of nucleotides (Aguilar-Bryan et al., 1995). ATP/ADP binds to NBDs of SUR (Ueda et al., 1997), and NBDs in turn function as ATP/ADP sensors (Nichols et al., 1996; Gribble et al., 1997), and thus, SUR regulates the ATP inhibition of KATP channels (Babenko et al., 1999). On the other hand, Kir6.2, a pore-forming subunit of KATP channels, is also defined as the primary site of the ATP-induced channel inhibition (Tucker et al., 1997). The C- and N-termini of Kir6.2 are both involved in this ATP inhibition (Drain et al., 1998; Tucker et al., 1998). Therefore, Gβγ might interact with these ATP-binding sites, thereby reducing the inhibitory action of ATP.
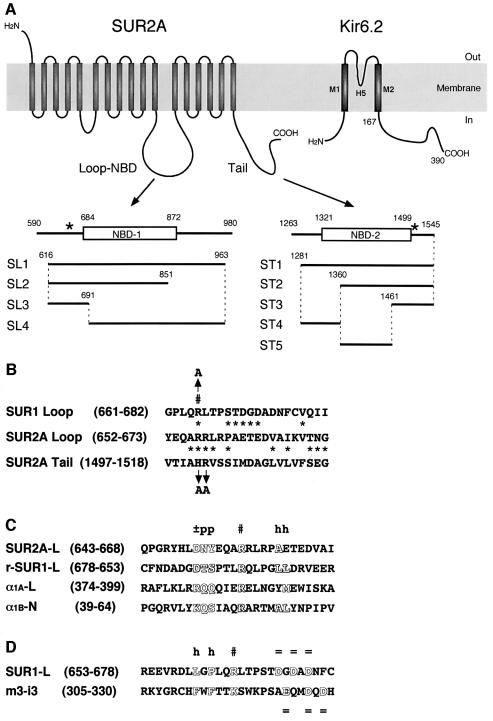
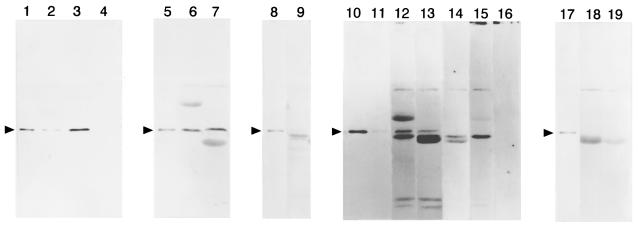
To examine this hypothesis, two fragments (SL1 and ST1) from the intracellular loop containing NBD-1 (referred to as ‘loop-NBD’ in the present study) and the C-terminus containing NBD-2 of SUR2A, were expressed in Escherichia coli as GST fusion proteins, respectively (Figure 7A, SL1 and ST1). In addition, the N-terminus (corresponding to amino acid residues 2–73) and the C-terminus (corresponding to amino acid residues 169–390) of the Kir6.2 subunit (KN1 and KT1) were also fused with GST, respectively. Then, as shown in Figure 4, their ability to bind Gβγ was tested by immunoblotting analysis using the antibody M-14 against Gβ1. As described previously (Furukawa et al., 1998), Gβγ2 did not bind to GST proteins alone.
Fig. 7. Schematic representation of SUR2A fragments produced as fusion proteins of GST. (A) The loop-NBD (SL1 to SL4) and C-terminal (ST1 to ST5) fragments of SUR2A, which were fused with GST. Positions of the fragments, loop-NBD (NBD-1, open box) and C-termini (NBD-2, open box) are indicated by the number of the amino acid residues for SUR2A and Kir6.2. Asterisks, the sites mutagenized. (B) Comparison of amino acid sequences from the loop-NBD of SUR1 (SUR1 Loop) and of SUR2A (SUR2A Loop), and from the C-terminus of SUR2A (SUR2A Tail). Residue numbers are indicated in parentheses. Asterisks, similar amino acid residues between SUR2A Loop and either SUR1 Loop or SUR2A Tail; #, arginine residues critical for the interaction with Gβγ complex (see text); arrows, amino acid substitutions. (C and D) Comparison of amino acid sequences from the loop-NBD of SUR1 (SUR1-L) and of SUR2A (SUR2A-L) with those from the I–II loop of Ca2+ channel α1A subunit (α1A-L) (De Waard et al., 1997), the N-terminus of Ca2+ channel α1B subunit (α1B-N) (Cantí et al., 1999), and the third intracellular loop of the m3 subtype of muscarinic receptor (m3-i3) (Wu et al., 2000), which are determined as interaction sites with Gβγ. Arginine residues critical for the interaction with Gβγ and lysine residue affecting the Gβγ binding are aligned (#). A reversed amino acid sequence for SUR1-L (r-SUR1-L) was compared as well. Residue numbers are indicated in parentheses. Outlined letters, consensus amino acids residues; ±, p, h and =, charged, polar, hydrophobic and negatively charged amino acids, respectively.
Fig. 4. Direct binding of Gβγ2 complex to the SUR2A subunit of KATP channels. Bovine brain Gβγ2 released from glutathione beads was detected by the anti-Gβ1 antibody M-14. Gβγ2 was incubated with the beads that had been immobilized with GST proteins fused with one of the following SUR2A fragments: SL1 (lane 2), ST1 (lane 3), KT1 (lane 4), SL2 (lane 6), SL3 (lane 7), SL4 (lane 9), ST2 (lanes 12 and 15), ST3 (lanes 13 and 14), ST4 (lane 18) and ST5 (lane 19) (see Figure 7A). For immunoblot analysis, proteins bound to the beads were separated by 12% (lanes 1–7 and 10–16) or 11% (lanes 8, 9 and 17–19) SDS–PAGE, together with 25 ng (lanes 10 and 16) or 12.5 ng (lanes 1, 5, 8, 11 and 17) of purified Gβγ2. The antibody was pre-incubated with the peptide antigen sc-261P (lanes 14–16). Arrowheads, positions of purified Gβ (Furukawa et al., 1998).
The antibody M-14 reacted with a 36 kDa Gβ purified from brain (Figure 4, lanes 1, 5, 8, 10, 11 and 17), as well as a 36 kDa polypeptide released from both SL1- and ST1-bound beads (Figure 4, lanes 2 and 3) that had been incubated with the purified Gβγ2. These reactivities of M-14 with the 36 kDa polypeptides were inhibited by pre-incubation of M-14 with the peptide antigen sc-261P for M-14 (lane 16; n = 3), indicating that the 36 kDa immunoreactive polypeptide was recognized specifically by the antibody against Gβ when released from the loop-NBD and C-terminus of SUR2A. In contrast, this antibody scarcely detected a 36 kDa polypeptide released from KT1- (lane 4) or KN1-bound (n = 3) beads, the beads that had been incubated with the purified Gβγ2. These results support the idea obtained from the electrophysiological experiments that the Kir6.2/SUR channel is under the regulation of Gβγ interacting directly with the SUR subunit.
To confirm further the binding sites of Gβγ on SUR, various fragments derived from the loop-NBD (Figure 7A, SL2 to SL4) and the C-terminus (Figure 7A, ST2 to ST5) of SUR2A were also fused with GST and tested for Gβγ binding in vitro. The antibody M-14 reacted with a 36 kDa polypeptide released from both SL2- and SL3-bound beads (Figure 4, lanes 6 and 7), but not from SL4-bound beads (lane 9), when these beads had been incubated with the purified Gβγ2. Moreover, M-14 reacted with a 36 kDa polypeptide released from both ST2- and ST3-bound beads (lanes 12 and 13), but not from both ST4- and ST5-bound beads (lanes 18 and 19). These reactivities of M-14 with the 36 kDa polypeptide were inhibited by pre-incubation with the peptide antigen sc-261P (lanes 14 and 15; n = 3). Thus, the results indicate that Gβγ interacts with both intracellular (loop-NBD) and C-terminal segments of SUR2A corresponding to SL3 (amino acid residues 616–691) and ST3 (amino acid residues 1461–1545), respectively (see Figure 7A).
Molecular determination of the critical amino acid residue(s) for Gβγ-dependent modulation of Kir/SUR channels
As shown in Figure 7B (SUR2A Loop, SUR2A Tail), a homology search has revealed that the regions SL3 and ST3 of SUR2A contain similar amino acid sequences. Aiming at identifying the critical amino acid residues on the SUR subunit interacting with Gβγ2, conserved basic amino acid residues within these amino acid sequences, Arg656 and Arg657 in SL3, and His1501 and Arg1502 in ST3, were substituted by Ala to generate four mutant SUR2As: SLM1 (Arg656,657Ala), SLM2 (Arg657Ala), SLM3 (Arg656Ala) and STM1 (His1501Ala and Arg1502Ala).
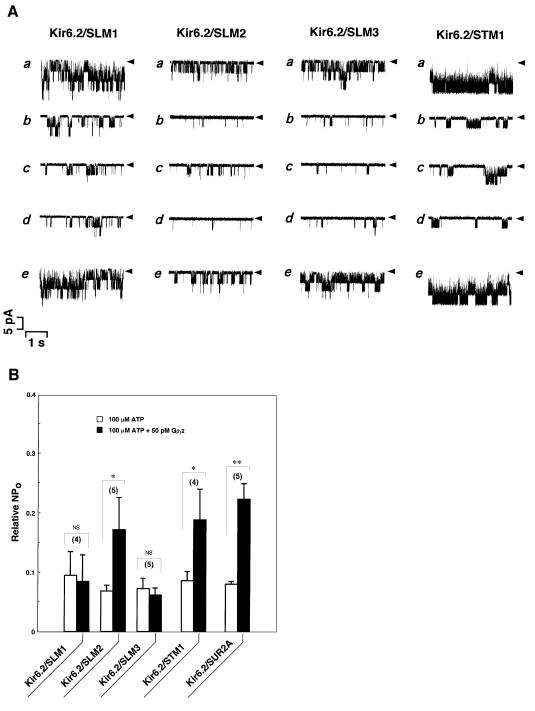
These three kinds of mutant SUR2As with substitution of amino acids in loop-NBD (SLM1, SLM2 or SLM3) were co-expressed for each with wild-type Kir6.2, and ATP-dependent channel activities were observed (Figure 5, Kir6.2/SLM1, Kir6.2/SLM2 and Kir6.2/SLM3; A, a, b, e; B, open boxes), as shown previously for wild-type SUR2A (Figures 1 and 5B, Kir6.2/SUR2A, open box). Again, Gβγ2 reduced ATP-induced inhibition of Kir6.2/SLM2 channels (Figure 5, Kir6.2/SLM2; A, c, d; B, filled box), as observed in Kir6.2/SUR2A channels (Figure 5B, Kir6.2/SUR2A, filled box). In contrast, Gβγ2 failed to enhance the channel activities that were inhibited by ATP in either Kir6.2/SLM1 (Figure 5, Kir6.2/SLM1; A, c, d; B, filled box) or Kir6.2/SLM3 channels (Figure 5, Kir6.2/SLM3; A, c, d; B, filled box). When three kinds of segment in SL3 of SUR2A, namely amino acid residues 616–689 (SLΔ1, n = 10), 616–658 (SLΔ2, n = 5) or 616–640 (SLΔ3, n = 5), were deleted, no channel activities were detectable in spite of the co-expression with wild-type Kir6.2.
Fig. 5. Mutational effects of the SUR2A subunit on the Gβγ2-dependent modulation of KATP channels. (A) Single channel currents were recorded continuously from COS-7 cells co-expressed with the wild-type Kir6.2 subunit together with four kinds of mutant SUR2A subunit (SLM1, SLM2, SLM3 or STM1). (a) Control (in the absence of ATP); (b) in the presence of 100 µM ATP or (c) 100 µM ATP plus 50 pM Gβγ2; and (d) after wash-out of Gβγ2 or (e) Gβγ2 plus ATP. Arrowheads, the zero current levels. (B) Current responses (NPo) to 100 µM ATP were measured in the absence (open boxes) or presence (filled boxes) of 50 pM Gβγ2 in the wild-type or mutant channels as indicated, and expressed as a ratio to those in the absence of ATP. The number of experiments is indicated in parentheses. *P<0.05. **P<0.001. NS, not significant.
On the other hand, the mutant STM1, with substitution of two amino acid residues in the C-terminus, also produced ATP-sensitive channels when co-expressed with wild-type Kir6.2 (Figure 5, Kir6.2/STM1; A, a, b, e; B, open box). Moreover, Gβγ2 reversibly ‘antagonized’ the ATP-induced inhibition of Kir6.2/STM1 channels (Figure 5, Kir6.2/STM1; A, c, d; B, filled box), as observed in wild-type channels (Figures 1 and 5B, Kir6.2/SUR2A, filled box). These results clearly indicate that a single amino acid residue in the loop-NBD of SUR2A, i.e. Arg656, plays a critical role in the modulation of Kir6.2/SUR2A channels by Gβγ.
Similarly, Kir6.2/SUR1 channels were investigated with respect to the importance of loop-NBD in the regulation of Gβγ-dependent modulation. Arg665 in SUR1, which is a basic amino acid residue located at a position homologous to Arg656 in SUR2A (Inagaki et al., 1996), was replaced with Ala by site-directed mutagenesis to generate mutant S1LM1 (Figure 7B, SUR1 Loop).
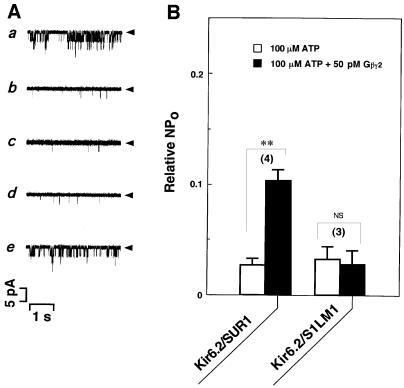
When the mutant S1LM1 was co-expressed with wild-type Kir6.2, ATP-sensitive channel activities were observed (Figure 6A, a, b, e; B, Kir6.2/S1LM1, open box), as shown in wild-type SUR1 (Figures 1 and 6B, Kir6.2/SUR1, open box). In contrast to wild-type SUR1 (Figure 6B, Kir6.2/SUR1, filled box), Gβγ2 did not reduce the inhibition of channel activities in the presence of ATP in Kir6.2/S1LM1 channels (Figure 6A, c, d; B, Kir6.2/S1LM1, filled box).
Fig. 6. Mutational effects of the SUR1 subunit on the Gβγ2-dependent modulation of KATP channels. (A) Single channel currents were recorded continuously from COS-7 cells co-expressed with wild-type Kir6.2 subunit together with the mutant SUR1 subunit, S1LM1. (a) Control (in the absence of ATP); (b) in the presence of 100 µM ATP or (c) 100 µM ATP plus 50 pM Gβγ2; and (d) after wash-out of Gβγ2 or (e) Gβγ2 plus ATP. Arrowheads, the zero current levels. (B) Current responses (NPo) to 100 µM ATP were measured in the absence (open boxes) or presence (filled boxes) of 50 pM Gβγ2 in the wild-type or mutant channels as indicated, and expressed as a ratio to those in the absence of ATP. The number of experiments is indicated in parentheses. **P<0.001. NS, not significant.
None of the mutants examined exerted a significant effect on the amplitude of single-channel currents nor the level of ATP sensitivities (n = 3). All these findings provide conclusive evidence that the loop-NBD of SUR plays a critical role in the modulation of KATP channels by Gβγ.
Discussion
In the present study, we found that an interaction of Gβγ with KATP produced a channel facilitation by ‘antagonizing’ the ATP-induced inhibition of Kir6.2/SUR channels in a membrane-delimited manner. The loop-NBD and the C-terminus of SUR2A were both defined as the interaction sites of Gβγ, based on the direct binding of Gβγ2 in vitro to SUR and Kir6.2 segments. Finally, a single arginine residue in the loop-NBD of SUR (656 in SUR2A or 665 in SUR1) was determined to be essential for this interaction between Gβγ and KATP channel, by using site-directed mutagenesis. This mutation produced a channel phenotype that lacked Gβγ-induced reduction of channel inhibition in the presence of ATP, while keeping the ATP sensitivity intact as the wild-type did. Taken together, these findings provide evidence that Gβγ interacts directly with the SUR subunit of KATP and thereby facilitates its activity by lessening the ATP sensitivity. Because these arginine residues lie close to the Walker motif in NBD-1 (Walker et al., 1982), Gβγ might alter ATP binding, either by competing for the overlapping site or by exerting an allosteric effect on the ATP-binding site. Moreover, two interaction sites for Gβγ on the loop-NBD and the C-terminus may be not distant from each other in its folded structure because of physical association between NBD-1 and NBD-2 (Mikhailov and Ashcroft, 2000).
Gβγ interaction motif in the loop-NBD of SUR
When predicted amino acid sequences were compared between SUR2A and SUR1 (Inagaki et al., 1996), a large diversity of amino acid sequences was observed in segments flanking the critical arginine residue (Arg656 in SUR2A and Arg665 in SUR1) for the interaction of SUR with Gβγ (Figure 7B), which were positioned between the conserved sequences corresponding to the 11th putative transmembrane and the NBD-1 (Figure 7A). Their lack of sequence homology predicts that the mode of interaction of Gβγ with SUR will be different among the subtypes of SUR (or between SUR2A and SUR1). This prediction is supported by the fact that the flanking segments from SUR2A (Figure 7C, SUR2A-L) and SUR1 (Figure 7D, SUR1-L) have sequence homologies to two different sequence motifs contributed to the interaction with Gβγ. The segment SUR2A-L showed a similarity to Gβγ-interaction sites determined in the I–II loop (De Waard et al., 1997; Zamponi et al., 1997) and the N-terminus (Cantí et al., 1999) of Ca2+ channel α1B and/or α1A subunits (Figure 7C, α1A-L and α1B-N, respectively). On the other hand, the segment SUR1-L showed homology to a Gβγ-interaction site recently demonstrated in the third intracellular loop of the m3 subtype of muscarinic acetylcholine receptor (Figure 7D, m3-i3) (Wu et al., 2000).
We can define consensus motif for Gβγ interaction in the segment SUR2A-L as ‘c-p-p-x-x-x-R-x4–5-h’, where c, p, h and x represent charged, polar, hydrophobic and any amino acids, respectively. On the other hand, consensus sequence for Gβγ interaction in the segment SUR1-L can be defined as ‘h-x-h-x-x-R/K-x5–6-nc’, in which nc represents a negatively charged region. Interestingly, a backward reading amino acid sequence corresponding to the SUR1-L segment, r-SUR1-L (Figure 7C, r-SUR1-L), shares the motif of ‘c-p-p-x-x-x-R-x4–5-h’. Since a suitable foldability and functionality are conserved in retro-proteins such as the B-domain of protein A (Olszewski et al., 1996), metallothionein-2 (Pan et al., 1999), cell-adhesive peptide (Jois et al., 1999) and leucine zipper transcription activator (Mittl et al., 2000), there is another possibility that the Gβγ-binding motif is not a linear amino acid sequence, but more likely involves general characteristics or conformations of a larger protein domain. None of these two motifs was shared by the C-terminal fragment ST3 of SUR2A or the corresponding fragment of SUR1.
Physiological significance of KATP channel modulation by Gβγ
Since the intracellular ATP concentration in most cells lies between 3 and 7 mM, it was initially argued that KATP channels should never be open under ATP-rich, physiological conditions. As observed in the Gβγ-mediated modulation of KATP channels, however, MgADP (Dunne and Petersen, 1986; Kakei et al., 1986) and phosphatidylinositol phosphates (PIPs) (Baukrowitz et al., 1998; Shyng and Nichols, 1998) also facilitate the channel activities in the presence of ATP by ‘antagonizing’ the ATP-induced channel inhibition. Gβγ modulated KATP channels through interaction with the loop-NBD of SUR, while MgADP (Nichols et al., 1996; Ueda et al., 1997) or PIPs (Baukrowitz et al., 1998; Shyng and Nichols, 1998) regulate the channels by binding primarily to the NBD-2 of SUR or by interacting with the C-terminus of Kir6.2, respectively. In addition, the concentration of Gβγ sufficient to modulate channel activities was three orders of magnitude lower than those of MgADP and PIPs. Moreover, neither wortmannin nor neomycin affected the Gβγ-mediated modulation. These findings indicate that the channel modulation via Gβγ is distinguished from that by MgADP or PIPs, and that KATP channels are regulated by multiple molecules under physiological conditions.
In conclusion, our results provide a new insight into the mechanism of KATP channel regulation by G-proteins. It is becoming clear that, as with SUR, many other ABC transporters regulate ion channels and that this kind of channel regulation will be of clinical and pharmacological relevance in understanding the molecular mechanism of diseases and in the development of a new type of drug. Some of the mode of regulation by G-proteins may be generally applicable to the regulation of other ABC transporters.
Materials and methods
cDNA cloning
PCR was performed using the G2 sense primer (Takao et al., 1994) and an antisense primer [CCGGATCCTACGGAGGG(G/T)T(A/G/C/T)GG(A/G/C/T)AC(C/T)TC] corresponding to amino acid residues 269–275 and 350–356 of GIRK1 according to the procedures described previously (Takao et al., 1994). One clone (pGK15) carrying an ∼260 bp PCR product was found to encode Kir6.2 (BIR; Inagaki et al., 1995). Using this cDNA fragment as a probe, cDNA clones (pVGK1 and pBGK1) encoding a rat version of Kir6.2, GK15 (DDBJ/EMBL/GenBank accession No. D61687), were isolated from rat ventricular and brain λgt10 cDNA libraries according to the methods described previously (Nakamura et al., 1991).
Based on the cDNA sequence for the rat SUR (DDBJ/EMBL/GenBank accession No. L40624), 10 oligodeoxyribonucleotide primers were synthesized. The sense primers corresponded to amino acid residues 1–10 (primer SS1), 415–422 (SS2), 659–666 (SS3), 872–880 (SS4) and 1256–1263 (SS5): SS1, CGGTCGACACCATGCCTTTGGCCTT(C/T)TG (C/T)GG(A/G/C/T)AC(A/G/C/T)GA(A/G)AA; SS2, GGCAGATCTG (C/T)AA(C/T)CT(A/G/C/T)GT(A/G/C/T)GC; SS3, CTCTTGGGCCC (A/G/C/T)CT(A/G/C/T)CA(A/G)AG(A/G)CT; SS4, CATCCTCGAGC TGCT(A/G/C/T)CG(A/G/C/T)GA(C/T)GA; SS5, GGAGCATGCGTG GT(A/G/C/T)CT(A/G/C/T)AT(A/C/T)GC. The antisense primers corresponded to amino acid residues 411–419 (primer AS1), 657–663 (AS2), 867–875 (AS3), 1251–1259 (AS4) and 1573–1582 (AS5): AS1, TGCAG ATCTGCCC(A/G/C/T)GC(A/G/C/T)GTCAT(C/T)TC; AS2, CAGTGG GCCCA(A/G)(A/G/C/T)AG(A/G)TC(A/G/C/T)CG; AS3, CAGCTCGA-GGAT(A/G/C/T)CC(A/G/C/T)GC(C/T)TGCAT(C/T)A; AS4, CACGC ATGC(A/G/C/T)CC(A/G/T)AT(A/G)TA(C/T)TCCATGC; AS5, GGA CTAGTCATTTGTCCGCGCGGAC(A/G)AA(A/G)(G/C) (A/T) (A/G/C/T)GC(A/G)AA(A/G/C/T)AC. The first cDNA strands were synthesized by the AS1-, AS2-, AS3-, AS4- or AS5-primed reverse transcription of rat pancreas poly(A)+RNA (Clontech) using an RNA LA PCR kit (Takara, Japan). PCRs were carried out for 0.5 min at 94°C, 0.5 min at 48–65°C, and 1.5 min at 72°C for 50 cycles, by adding the sense primer into the first strand reaction mixture containing the antisense primer in combination with SS1/AS1, SS2/AS2, SS3/AS3, SS4/AS4 and SS5/AS5. In PCR, the five sets of primers were utilized to produce 1270, 750, 650, 1160 and 990 bp products, which were then purified, digested with SalI and BglII (for 1270 bp product), BglII and ApaI (for 750 bp product), ApaI and XhoI (for 650 bp product), XhoI and SphI (for 1160 bp product), and SphI and SpeI (for 990 bp product), and reconstructed into the SalI–SpeI site of pBluescript SK(+) to yield pRSUR1 containing the entire coding sequence of rat SUR1. Nucleotide sequence analysis was performed using a DNA sequencer (ABI Prism 377, PE Applied Biosystems).
Dual expression of Kir6.2 and SUR subunits in COS-7 cells
The sense (SK1, GGCCCGCGGAGCCATGCTGTCCCGAAA) and antisense (AK1, ATGGTGAAAATGAGCAGCGTGT) primers corresponding to amino acid residues 1–5 and 70–77 were synthesized, and PCR was carried out for 0.5 min at 94°C, 0.5 min at 60°C, and 1 min at 72°C for 25 cycles, using pVGK1 (see above) as a template. The 80 bp SacII–KpnI fragment from the 240 bp product amplified by the PCR was ligated with the 4.1 kb SacII–KpnI fragment from pVGK1 to yield pVGK1-N, in which the 5′ non-coding sequence of Kir6.2 was shortened.
In the same orientation with respect to cytomegalovirus gene transcription, the 1.2 kb SacII(blunted)–EcoRI(blunted) fragment containing the entire coding sequence for Kir6.2 from pVGK1-N was inserted into the KpnI(blunted)–XbaI(blunted) site of pTracer-CMV (Invitrogen), an expression vector having a green fluorescent protein (GFP)–zeocin fusion gene as selection markers, to produce pTKir1. Then, the 1.3 kb BglII(blunted)–PvuII fragment containing a cytomegalovirus gene transcription unit from pTracer-CMV was inserted into the BglII(blunted) site of pTKir1 to produce a plasmid pTKir1-dCMV, which possessed two sets of cytomegalovirus gene transcription units. For dual expression of both Kir6.2 and SUR1 cDNAs under the control of cytomegalovirus promoters in COS-7 cells, the 4.7 kb SalI(blunted)–SpeI(blunted) fragment from pRSUR1 (see above) was inserted into the EcoRV site of pTKir1-dCMV in the same orientation with respect to the gene transcription, to yield pTSUR1-Kir1. The 4.7 kb NotI–SalI fragment containing the entire coding sequence for rat SUR2A (Inagaki et al., 1996) was recombined into the NotI–SalI site of pBluescript SK(+) to yield pRSUR2A. The rat SUR2A cDNA was kindly provided by Dr Susumu Seino. For dual expression of both Kir6.2 and SUR2A cDNAs, the 4.7 kb NotI(blunted)–XhoI(blunted) fragment from pRSUR2A was similarly inserted into the EcoRV site of pTKir1-dCMV to produce pTSUR2A-Kir1.
Construction of mutant and chimeric SUR2A and Kir6.2 subunits
Kir6.2ΔC. The plasmid pVGK1-N (see above) carrying rat Kir6.2 cDNA was digested with NruI, ligated with PacI linker (dCCTTAATTAAGG; New England Biolabs), digested with PacI, and ligated. In this recombinant plasmid pKir6.2ΔC, the codon CGA for Arg365 and GGG for Gly366 were replaced with the codon CCT for Pro and TAA for termination, respectively.
Kir6.2ΔN. The 4.1 kb SacII–KpnI fragment excised from pVGK1-N was ligated with the 33 bp HpaII–KpnI fragment from pVGK1-N and the annealed oligonucleotides GGCCAACGGAGCCATGACC and CGGGTCATGGCTCCGTTGGCCGC. In this plasmid, the segment encoding amino acid residues 2–14 of the Kir6.2 subunit was deleted.
SLΔ1, SLΔ2, SLΔ3. The plasmid pRSUR2A (see above) was digested with PvuII, ligated with EcoRI linker (dCCGAATTCGG; Takara, Japan), and digested with AflII and EcoRI. The resulting 340 bp fragment was ligated with the 7.1 kb AflII–EcoRI fragment excised from pRSUR2A to yield pSLΔ1. In this plasmid, the segment encoding amino acid residues 616–689 of SUR2A was deleted. Next, genetic mutations of the intracellular loop containing NBD-1 (loop-NBD) of SUR2A were performed with PCR (see above) using two kinds of sense primers and an antisense primer. The sense primers were S2LΔ2 (TTCGAATTCCTGA GACTGAAGATGTT), corresponding to amino acid residues 661–666 of SUR2A, and S2LΔ3 (ACAGAATTCAGCCTGGAAGGTACCAC), corresponding to amino acid residues 643–648. The antisense primer was ALΔ1 (GTTGGAATTCGAATGTCAATATTGG), corresponding to amino acid residues 685–693. The plasmid pRSUR2A was used as a template. The 110 and 160 bp products amplified by the PCR using two sets of primers of S2LΔ2/ALΔ1 and S2LΔ3/ALΔ1 were purified, digested with EcoRI, and cloned into the EcoRI site of pSLΔ1 to yield pSLΔ2 and pSLΔ3, respectively. In the plasmid pSLΔ2, the segment encoding amino acid residues 616–658 of the SUR2A subunit was deleted, and the codons CCT and GCT for Pro660 and Ala661 were replaced with the codons ATT (Ile) and CCT (Pro), respectively. In the plasmid pSLΔ3, the segment encoding amino acid residues 616–640 of the SUR2A subunit was deleted, and the codon AAG for Lys642 was replaced with the codon ATT (Ile).
SLM1, SLM2 and SLM3. Genetic mutations of the loop-NBD of SUR2A were further performed with PCR using a sense primer and three kinds of antisense primers. The sense primer was S2LM1 (CTCCTTAAGCTGTATGCCTGGG), corresponding to amino acid residues 501–508 of SUR2A. The antisense primers, corresponding to amino acid residues 652–663, were ALM1 (AGTCTCAGCAGGCCGA AGAGCCGCCGCCTGCTCG), ALM2 (AGTCTCAGCAGGCCGAAG AGCCCGCGCCTGCTCG) and ALM3 (AGTCTCAGCAGGCCGAAG ACGCGCCGCCTGCTCG). ALM1 contained two point mutations (GC) replacing the first and second codons CGG and CGT coding for Arg656 and Arg657 of SUR2A by GCG and GCT coding for Ala; ALM2 contained a point mutation (GC) replacing the first and second codons CGT coding for Arg657 by GCT coding for Ala; and ALM3 contained a point mutation (GC) replacing the first and second codons CGG coding for Arg656 by GCG coding for Ala. The plasmid pRSUR2A was used as a template. In PCR, three sets of primers, namely S2LM1/ALM1, S2LM1/ALM2 and S2LM1/ALM3, were utilized to produce 490 bp products, which were then purified, digested with AflII and DdeI, and cloned into the AflII–EcoRI site of pRSUR2A together with the 90 bp DdeI–EcoRI fragment from pRSUR2A, in order to yield pSLM1, pSLM2 and pSLM3, respectively.
STM1. A genetic mutation of the C-terminus containing NBD-2 of SUR2A was also carried out with PCR using a set of sense (S2TM1, ATAGCTGCCGCTGTCTCCTCTATTATG) and antisense (M13 primer M4, GTTTTCCCAGTCACGAC; Takara, Japan) primers. The primer S2TM1, corresponding to amino acid residues 1499–1507, contained two point mutations (GC) replacing the first and second codons of CAC and CGT coding for His1501 and Arg1502 by GCC and GCT coding for Ala. The 250 bp products amplified by the PCR were purified, digested with BseRI and XhoI, and cloned into the SalI–XhoI site of pRSUR2A together with the 430 bp SalI–BseRI fragment excised from pRSUR2A, in order to yield pSTM1.
S1LM1. A genetic mutation of the intracellular loop containing NBD-1 of SUR1 was performed with PCR using the sense primer S1M1 (CTGGGCCCGCTGCAGGCACTGACTCCC) and the antisense primer A1M1 (CCCCACGATCATGGTCAGCTG), corresponding to amino acid residues 660–668 and 707–713 of SUR1, respectively. The primer S1M1 contained two point mutations (GC) replacing the first and second codons of AGA coding for Arg665 by GCA coding for Ala. The 50 bp KpnI–EcoRI fragment form pBluescript SK(–) (Stratagene) was cloned into the HindIII site of pSPA2 (Nakamura et al., 1994) to produce pSPA3. The 4.8 kb SalI–SpeI fragment containing the entire coding sequence of SUR1 was excised from pRSUR1 and recombined into the EcoRV site of pSPA3 to produce pSPSUR1. To delete internal ApaI and XhoI sites, the plasmid pSPSUR1 was cleaved by AatII, partially digested with XhoI, blunted, and circularized to produce pSUR1AX. The 160 bp products amplified by the PCR were purified, digested with ApaI and BstXI, and cloned into the ApaI–XhoI site of pSUR1AX together with the 490 bp BstXI–XhoI fragment excised from pRSUR1, in order to yield pS1LM1.
Subcloning and mutagenesis procedures were verified by restriction enzyme analysis and DNA sequencing. DNA fragments containing either the entire coding sequence of mutant SUR or that of Kir6.2 were recombined into the dual expression vectors derived from pTracer-CMV as described above.
Electrophysiology
COS-7 cells were transfected with the above plasmids by using a LipofectAMINE PLUS reagent (Life Technologies) to allow visual identification of the transfected cells with GFP fluorescence. Unitary K+ currents were recorded by the inside-out configuration of the patch–clamp technique, using a voltage–clamp amplifier (Axopatch 1D, Axon Instruments) (Furukawa et al., 1998). The pipette solution contained 140 mM KCl, 3 mM MgCl2, 2 mM CaCl2 and 5 mM HEPES–KOH pH 7.2. The bath (intracellular) solution contained 140 mM KCl, 3 mM MgCl2, 1 mM EGTA and 5 mM HEPES–KOH pH 7.2, and the nominal concentration of free Mg2+ in this solution was kept constant at 2.8 mM. Currents were low-pass filtered at 10 kHz, digitized at 2 kHz using a Digidata 1200 Interface (Axon Instruments), and analyzed on a computer using pCLAMP software (Axon Instruments). Electrophysiological measurements were performed at 20–24°C, and the holding potential was –50 mV. Gβγ2 complex (Asano et al., 1993) and Goα (Asano et al., 1988) purified from bovine brain were added into bath solution at the final concentration indicated in the text. The vehicle solutions were composed of 20 mM Tris–HCl pH 8, 0.5 mM EDTA, 0.5 mM dithiothreitol, 0.6% sodium cholate, 50 mM NaCl and 50 mM NaPO4, and diluted by >30 000 times in the bath solution. They showed no effect on the channel activities (n = 10). For the pre-treatment with neomycin, COS-7 cells were incubated with 2 mM neomycin sulfate for >4 h prior to the measurement. H7 (100 µM) or wortmannin (50 µM) was added into the bath solution.
Data analysis
The threshold for judging the open state was set at half amplitude of the single channel currents. The open probability (Po) was calculated based on Sánchez et al. (1998), and data were expressed as NPo (N is the number of channels in the patch), since the number of channels in each individual patch was unknown. We assumed that N remained constant throughout the experiment. To lessen individual variations, the channel activities under the influence of ATP were represented, as a first approximation, by an NPo ratio (relative NPo, NPoex/NPoi), where NPoex is NPo obtained from the experiment with ATP to study effects of Gβγ2, and NPoi is an initial value of NPo without ATP. To characterize the single channel activity, NPo was calculated in 5 s intervals during 30 s recording and then averaged. Concentration–response curves were fitted to the Hill equation: relative NPo = NPoex/NPoi = 1/{1 + ([ATP]/Ki)nH}, where Ki is the inhibition constant, and nH is the Hill coefficient. Statistical data were represented by the mean ± SEM.
Gβγ2 binding
The 1.0 kb PvuII–HincII fragment (encoding amino acid residues 616–963 of SUR2A), the 710 bp PvuII–BamHI fragment (encoding amino acid residues 616–851), the 230 bp PvuII–EcoRI fragment (encoding amino acid residues 616–691) and the 820 bp EcoRI–HincII fragment (encoding amino acid residues 691–963) were excised from the plasmid pRSUR2A (see above), and fused in-frame to the GST coding sequence in pGEX-2T (Amersham Pharmacia Biotech) to produce GST fusion proteins of the intracellular loop containing NBD-1 of SUR (SL1 to SL4), respectively (see Figure 7). Similarly, to produce GST fusion proteins of the C-terminus containing NBD-2 of SUR (ST1 to ST5), the 820 bp XmnI–XhoI fragment (encoding amino acid residues 1281–1545 of SUR2A), the 580 bp SalI–XhoI fragment (encoding amino acid residues 1360–1545), the 280 bp Csp45I–XhoI fragment (encoding amino acid residues 1461–1545), the 240 bp XmnI–SalI fragment (encoding amino acid residues 1281–1360) and the 310 bp SalI–Csp45I fragment (encoding amino acid residues 1360–1461) were excised from pRSUR2A and fused in-frame.
The 740 bp BspHI–EcoRI fragment (encoding amino acid residues 169–390 of Kir6.2) from pVGK1 (see above) was fused in-frame in pGEX-2T to yield GST fusion proteins of the C-terminus of Kir6.2 (KT1). To produce GST fusion proteins of the N-terminus of Kir6.2 (KN1), the sense (SKN1, GGAATTCTGTCCCGAAAGGGCATTAT) and antisense (AKN1, GGTGAATTCGAGCAGCGTGTGGGG) primers corresponding to amino acid residues 2–8 and 69–73 of Kir6.2 were synthesized, and PCR was carried out using pVGK1 as a template (see above). The 220 bp EcoRI fragment (encoding amino acid residues 2–73) from the 230 bp product amplified by PCR was cloned into the EcoRI site of pGEX-2T.
Eleven kinds of fusion protein were expressed in E.coli DH5α, solubilized, and immobilized on glutathione–Sepharose 4B beads (Amersham Pharmacia Biotech), according to the procedure described previously (Furukawa et al., 1998). Approximately 3 µg of GST fusion proteins bound to beads were used in each binding assay. Association of Gβγ2 complex (3 µg) purified from bovine brain (Asano et al., 1993) with these GST fusion proteins bound to beads was determined by immunoblot analysis using the antibody M-14 (Santa Cruz; diluted at 1:8000) against Gβ1 and the peptide antigen sc-261P for M-14 (Santa Cruz), according to the procedure of Furukawa et al. (1998). The GST fusion proteins were tested more than three times for their ability to bind Gβγ2.
Acknowledgments
Acknowledgements
We are grateful to Dr Susumu Seino for providing us with rat SUR2A cDNA. We also thank Dr Mitsunobu Yoshii for critical reading of the manuscript. This investigation was supported in part by research grants from the Ministry of Education, Science and Culture of Japan to T.N. (#02557013, #08680855 and #12680766) and the foundation from Advanced Research Laboratory, Hitachi, Ltd to T.N.
References
- Aguilar-Bryan L. et al. (1995) Cloning of the β cell high-affinity sulfonylurea receptor: a regulator of insulin secretion. Science, 268, 423–426. [DOI] [PubMed] [Google Scholar]
- Asano T., Kamiya,N., Morishita,R. and Kato,K. (1988) Immunoassay for the βγ subunits of GTP-binding proteins and their regional distribution in bovine brain. J. Biochem., 103, 950–953. [DOI] [PubMed] [Google Scholar]
- Asano T., Morishita,R., Matsuda,T., Fukada,Y., Yoshizawa,T. and Kato,K. (1993) Purification of four forms of the βγ subunit complex of G proteins containing different γ subunits. J. Biol. Chem., 268, 20512–20519. [PubMed] [Google Scholar]
- Asano T., Shinohara,H., Morishita,R., Norota,I., Kato,K. and Endoh,M. (1995) The G-protein Go in mammalian cardiac muscle: localization and coupling to A1 adenosine receptors. J. Biochem., 117, 183–189. [DOI] [PubMed] [Google Scholar]
- Ashcroft F.M. (1988) Adenosine 5′-triphosphate-sensitive potassium channels. Annu. Rev. Neurosci., 11, 97–118. [DOI] [PubMed] [Google Scholar]
- Babenko A.P., Gonzalez,G. and Bryan,J. (1999) Two regions of sulfonylurea receptor specify the spontaneous bursting and ATP inhibition of KATP channel isoforms. J. Biol. Chem., 274, 11587–11592. [DOI] [PubMed] [Google Scholar]
- Baukrowitz T., Schulte,U., Oliver,D., Herlitze,S., Krauter,T., Tucker,S.J., Ruppersberg,J.P. and Fakler,B. (1998) PIP2 and PIP as determinants for ATP inhibition of KATP channels. Science, 282, 1141–1144. [DOI] [PubMed] [Google Scholar]
- Cantí C., Page,K.M., Stephens,G.J. and Dolphin,A.C. (1999) Identification of residues in the N terminus of α1B critical for inhibition of the voltage-dependent calcium channel by Gβγ. J. Neurosci., 19, 6855–6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham D.E. and Neer,E.J. (1993) New roles for G-protein βγ-dimers in transmembrane signalling. Nature, 365, 403–406. [DOI] [PubMed] [Google Scholar]
- De Waard M., Liu,H., Walker,D., Scott,V.E.S., Gurnett,C.A. and Campbell,K.P. (1997) Direct binding of G-protein βγ complex to voltage-dependent calcium channels. Nature, 385, 446–450. [DOI] [PubMed] [Google Scholar]
- Drain P., Li,L. and Wang,J. (1998) KATP channel inhibition by ATP requires distinct functional domains of the cytoplasmic C terminus of the pore-forming subunit. Proc. Natl Acad. Sci. USA, 95, 13953–13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne M.J. and Petersen,O.H. (1986) Intracellular ADP activates K+ channels that are inhibited by ATP in an insulin-secreting cell line. FEBS Lett., 208, 59–62. [DOI] [PubMed] [Google Scholar]
- Dunne M.J., Bullett,M.J., Li,G., Wollheim,C.B. and Petersen,O.H. (1989) Galanin activates nucleotide-dependent K+ channels in insulin-secreting cells via a pertussis toxin-sensitive G-protein. EMBO J., 8, 413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T. et al. (1998) Differential interactions of the C terminus and the cytoplasmic I-II loop of neuronal Ca2+ channels with G-protein α and βγ subunits: II. evidence for direct binding. J. Biol. Chem., 273, 17595–17603. [DOI] [PubMed] [Google Scholar]
- Gribble F.M., Tucker,S.J. and Ashcroft,F.M. (1997) The essential role of the Walker A motifs of SUR1 in K-ATP channel activation by Mg-ADP and diazoxide. EMBO J., 16, 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B. (1994) Modulation of ion-channel function by G-protein-coupled receptors. Trends Neurosci., 17, 531–536. [DOI] [PubMed] [Google Scholar]
- Inagaki N., Gonoi,T., Clement,J.P.,IV, Namba,N., Inazawa,J., Gonzalez,G., Aguilar-Bryan,L., Seino,S. and Bryan,J. (1995) Reconstitution of IKATP: an inward rectifier subunit plus the sulfonylurea receptor. Science, 270, 1166–1170. [DOI] [PubMed] [Google Scholar]
- Inagaki N., Gonoi,T., Clement,J.P.,IV, Wang,C.-Z., Aguilar-Bryan,L., Bryan,J. and Seino,S. (1996) A family of sulfonylurea receptors determines the pharmacological properties of ATP-sensitive K+ channels. Neuron, 16, 1011–1017. [DOI] [PubMed] [Google Scholar]
- Isomoto S., Kondo,C., Yamada,M., Matsumoto,S., Higashiguchi,O., Horio,Y., Matsuzawa,Y. and Kurachi,Y. (1996) A novel sulfonylurea receptor forms with BIR (Kir6.2) a smooth muscle type ATP-sensitive K+ channel. J. Biol. Chem., 271, 24321–24324. [DOI] [PubMed] [Google Scholar]
- Jois S.D.S., Hughes,R. and Siahaan,T.J. (1999) Comparison of the solution conformations of a cell-adhesive peptide LBE and its reverse sequence EBL. J. Biomol. Struct. Dyn., 17, 429–444. [DOI] [PubMed] [Google Scholar]
- Kakei M., Kelly,R.P., Ashcroft,S.J.H. and Ashcroft,F.M. (1986) The ATP-sensitivity of K+ channels in rat pancreatic B-cells is modulated by ADP. FEBS Lett., 208, 63–66. [DOI] [PubMed] [Google Scholar]
- Kirsch G.E., Codina,J., Birnbaumer,L. and Brown,A.M. (1990) Coupling of ATP-sensitive K+ channels to A1 receptors by G proteins in rat ventricular myocytes. Am. J. Physiol., 259, H820–H826. [DOI] [PubMed] [Google Scholar]
- Koster J.C., Sha,Q., Shyng,S.-L. and Nichols,C.G. (1999) ATP inhibition of KATP channels: control of nucleotide sensitivity by the N-terminal domain of the Kir6.2 subunit. J. Physiol., 515, 19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailov M.V. and Ashcroft,S.J.H. (2000) Interactions of the sulfonylurea receptor 1 subunit in the molecular assembly of β-cell KATP channels. J. Biol. Chem., 275, 3360–3364. [DOI] [PubMed] [Google Scholar]
- Mittl P.R.E., Deillon,C., Sargent,D., Liu,N., Klauser,S., Thomas,R.M., Gutte,B. and Grütter,M.G. (2000) The retro-GCN4 leucine zipper sequence forms a stable three-dimensional structure. Proc. Natl Acad. Sci. USA, 97, 2562–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F., Ogata,K., Shiozaki,K., Kameyama,K., Ohara,K., Haga,T. and Nukada,T. (1991) Identification of two novel GTP-binding protein α-subunits that lack apparent ADP-ribosylation sites for pertussis toxin. J. Biol. Chem., 266, 12676–12681. [PubMed] [Google Scholar]
- Nakamura K., Nukada,T., Haga,T. and Sugiyama,H. (1994) G protein-mediated inhibition of phosphoinositide metabolism evoked by metabotropic glutamate receptors in frog oocytes. J. Physiol., 474, 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols C.G., Shyng,S.-L., Nestorowicz,A., Glaser,B., Clement,J.P.,IV, Gonzalez,G., Aguilar-Bryan,L., Permutt,M.A. and Bryan,J. (1996) Adenosine diphosphate as an intracellular regulator of insulin secretion. Science, 272, 1785–1787. [DOI] [PubMed] [Google Scholar]
- Noma A. (1983) ATP-regulated K+ channels in cardiac muscle. Nature, 305, 147–148. [DOI] [PubMed] [Google Scholar]
- Olszewski K.A., Kolinski,A. and Skolnick,J. (1996) Does a backwardly read protein sequence have a unique native state? Protein Eng., 9, 5–14. [DOI] [PubMed] [Google Scholar]
- Pan P.K., Zheng,Z.F., Lyu,P.C. and Huang,P.C. (1999) Why reversing the sequence of the α domain of human metallothionein-2 does not change its metal-binding and folding characteristics. Eur. J. Biochem., 266, 33–39. [DOI] [PubMed] [Google Scholar]
- Reimann F., Tucker,S.J., Proks,P. and Ashcroft,F.M. (1999) Involvement of the N-terminus of Kir6.2 in coupling to the sulphonylurea receptor. J. Physiol., 518, 325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribalet B. and Eddlestone,G.T. (1995) Characterization of the G protein coupling of a somatostatin receptor to the K+ATP channel in insulin-secreting mammalian HIT and RIN cell lines. J. Physiol., 485, 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez J.A., Gonoi,T., Inagaki,N., Katada,T. and Seino,S. (1998) Modulation of reconstituted ATP-sensitive K+ channels by GTP-binding proteins in a mammalian cell line. J. Physiol., 507, 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyng S.-L. and Nichols,C.G. (1998) Membrane phospholipid control of nucleotide sensitivity of KATP channels. Science, 282, 1138–1141. [DOI] [PubMed] [Google Scholar]
- Taggart M.J. and Wray,S. (1998) Hypoxia and smooth muscle function: key regulatory events during metabolic stress. J. Physiol., 509, 315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao K., Yoshii,M., Kanda,A., Kokubun,S. and Nukada,T. (1994) A region of the muscarinic-gated atrial K+ channel critical for activation by G protein βγ subunits. Neuron, 13, 747–755. [DOI] [PubMed] [Google Scholar]
- Terzic A., Tung,R.T., Inanobe,A., Katada,T. and Kurachi,Y. (1994) G proteins activate ATP-sensitive K+ channels by antagonizing ATP-dependent gating. Neuron, 12, 885–893. [DOI] [PubMed] [Google Scholar]
- Tucker S.J., Gribble,F.M., Zhao,C., Trapp,S. and Ashcroft,F.M. (1997) Truncation of Kir6.2 produces ATP-sensitive K+ channels in the absence of the sulphonylurea receptor. Nature, 387, 179–183. [DOI] [PubMed] [Google Scholar]
- Tucker S.J., Gribble,F.M., Proks,P., Trapp,S., Ryder,T.J., Haug,T., Reimann,F. and Ashcroft,F.M. (1998) Molecular determinants of KATP channel inhibition by ATP. EMBO J., 17, 3290–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K., Inagaki,N. and Seino,S. (1997) MgADP antagonism to Mg2+-independent ATP binding of the sulfonylurea receptor SUR1. J. Biol. Chem., 272, 22983–22986. [DOI] [PubMed] [Google Scholar]
- Walker J.E., Saraste,M., Runswick,M.J. and Gay,N.J. (1982) Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J., 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Bogatkevich,G.S., Mukhin,Y.V., Benovic,J.L., Hildebrandt,J.D. and Lanier,S.M. (2000) Identification of Gβγ binding sites in the third intracellular loop of the M3-muscarinic receptor and their role in receptor regulation. J. Biol. Chem., 275, 9026–9034. [DOI] [PubMed] [Google Scholar]
- Zamponi G.W., Bourinet,E., Nelson,D., Nargeot,J. and Snutch,T.P. (1997) Crosstalk between G proteins and protein kinase C mediated by the calcium channel α1 subunit. Nature, 385, 442–446. [DOI] [PubMed] [Google Scholar]