Abstract
There is currently a renewed focus aimed at understanding allosteric mechanisms at atomic resolution. This current interest seeks to understand how both changes in protein conformations and changes in protein dynamics contribute to relaying an allosteric signal between two ligand binding sites on a protein (e.g. active site and allosteric site). Both NMR, by monitoring protein dynamics directly, and hydrogen/deuterium exchange, by monitoring solvent accessibility of backbone amides, offer insights into protein dynamics. Unfortunately, many allosteric proteins exceed the size limitations of standard NMR techniques. Although hydrogen/deuterium exchange as detected by mass spectrometry (H/DX-MS) offers an alternative evaluation method, any application of hydrogen/deuterium exchange requires that the property being measured functions in both H2O and D2O. Due to the promising future H/DX-MS has in the evaluation of allosteric mechanisms in large proteins, we demonstrate an evaluation of allosteric regulation in D2O. Exemplified using phenylalanine inhibition of rabbit muscle pyruvate kinase, we find that binding of the inhibitor is greatly reduced in D2O, but the effector continues to elicit an allosteric response.
Keywords: hydrogen/deuterium exchange, mass spectrometry, allosteric regulation, allostery, pyruvate kinase
The original definition of allosteric regulation focused on how a protein binds a ligand differently when a second ligand is or is not prebound [1, 2]. This definition neither limits the mechanism of allostery to a change in conformation, nor excludes a role for change in protein dynamics in this mechanism [3]. Nonetheless, several decades of research on allosteric proteins was dominated by fitting data to a “model,” the primary model being a two-state mechanism in which two interconverting conformations had different affinities for the respective ligands [4]. Recently developed techniques (primarily NMR) that can monitor protein dynamics have served as a spark to reinvigorate interest in how changes in protein dynamics contribute to the allosteric mechanism [5–8].
Despite selective labeling techniques, there continues to be an upper limitation to the size of protein that can be studied by NMR. In contrast, hydrogen/deuterium exchanges detected by mass spectrometry (H/DX-MS) does not have a restricted use due to the size of the protein of interest. Therefore, H/DX-MS holds a promising future in the study of allosteric proteins [9, 10], as well as the evaluation of whole protein contributions to other functions such as catalysis [11, 12].
Due to this potential, we have developed both hardware [13] and software advances to facilitate the use of H/DX-MS. One key hardware feature that had previous received little advancement was the temperature/timing control of the exchange reaction, the pepsin cleavage (used as initial fragmentation of the sample), and column separation of cleaved peptides. To address this deficiency, a temperature controlled box with automated valves was developed to house the immobilized pepsin column and the HPLC reverse phase column used in conjunction with a rapid elution for partial separation of cleaved peptides [13]. A primary deficiency in data analysis was an automated technique to extract ionic envelopes for peptides of interest. Therefore, we have now developed a software suite (available at http://www.kumc.edu/mspc/) that automatically completes this task. This software development greatly reduces the time necessary to evaluate H/DX-MS data outputs.
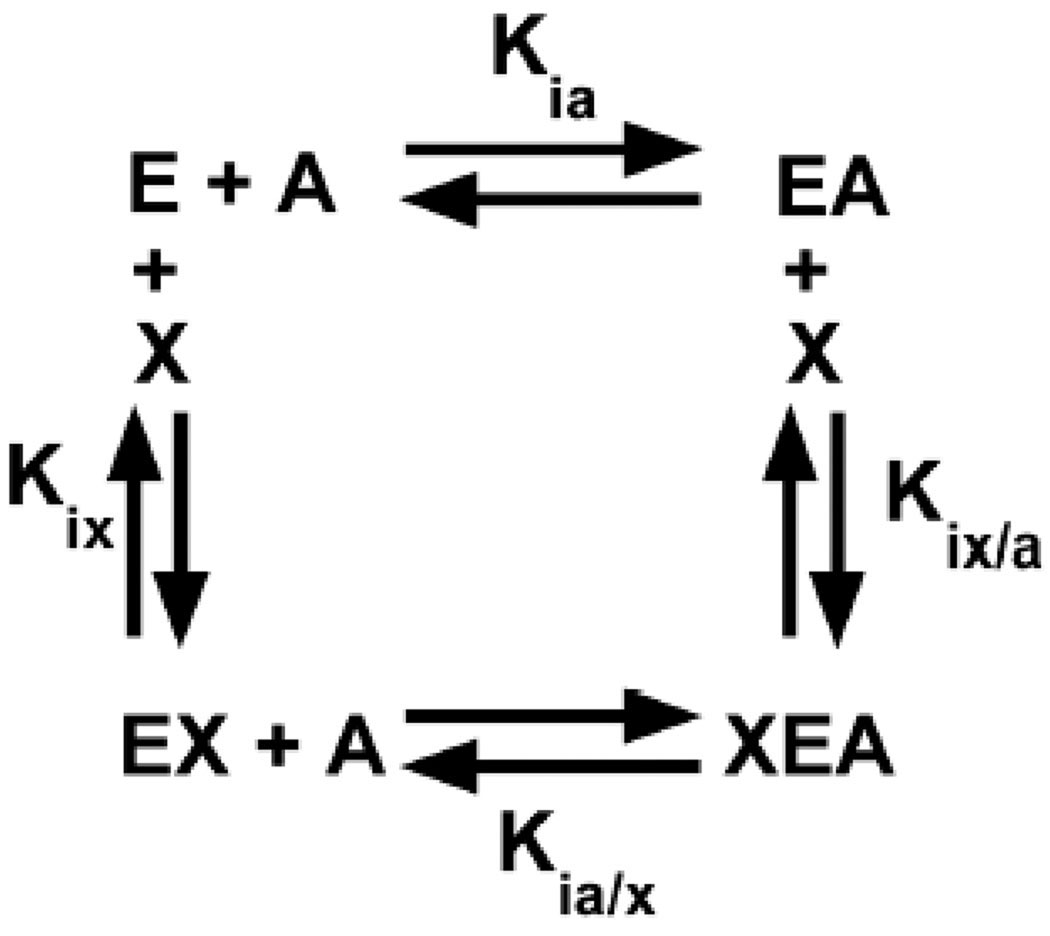
Even though we have made contributions to facilitate the use of H/DX-MS, the target of our interest in the application of H/DX-MS is the evaluation of allosteric regulation in proteins. This application is based on a linked-equilibrium view of allostery that considers the four enzyme complexes (i.e. 1: free enzyme, 2: the enzyme-substrate complex, 3: the enzyme-effector complex and 4: the ternary complex between enzyme, substrate and effector) in an energy cycle (Fig 1) [2, 14, 15]. H/DX-MS has the potential to identify regions of the protein that undergo changes in solvent accessibility when comparing the four enzyme complexes. A linked-equilibrium analysis is driven by the addition of ligands at concentrations that will saturate the respective binding site on the protein, both in the absence and in the saturating presence of the second ligand [2, 14–16]. Therefore, confidence in interpreting any structure/function study including H/DX-MS relies on the level of confidence that the ligand concentrations used are appropriate to obtain the enzyme complexes of interest.
Fig 1.
A thermodynamic energy cycle for an allosteric system representing the enzyme (E) binding substrate (A) and/or allosteric effector (X).
Functions of proteins can be modified in D2O as compared to the same function in H2O. These changes can be due to a number of reasons including altered protein hydration [17], altered solvent hydrophobicity [18], and altered protonation rates (i.e. solvent isotopic effect considered by enzymologists). Any one of these effects could modify how effector and/or substrate binds to the protein and/or the ability of the allosteric mechanism to function within the protein. It then becomes imperative to H/DX studies to confirm that allosteric regulation functions in D2O as well as in H2O and to demonstrate that the concentration of ligands used modulates the enzyme to the proper enzyme complex. Therefore, before H/DX-MS can be used, allostery must be monitored in D2O as well as H2O. Here we use the phenylalanine inhibition of the affinity of rabbit muscle pyruvate kinase (M1-PYK) for its substrate, phoshoenolpyruvate (PEP), to demonstrate a comparison of an evaluation of allostery in D2O vs. H2O.
Materials and Methods
Intrinsic protein fluorescence of rabbit M1-PYK (Roche) was titrated with phenylalanine or alanine over a concentration range of phoshoenolpyruvate (PEP) as previously described [16], only at 24°C. Buffer (50mM Tris, 10mM MgCl2, 0.1mM EDTA, and 500mM KCl) and ligand stocks were adjusted to pH 9.0 for H2O solutions and to a meter reading of 8.6 in D2O solutions to correct for effect of deuterium on glass electrodes [19].
Results and Discussion
Allostery is defined as how the substrate binds differently in the absence vs. presence of the effector [2]. This can be restated as how the effector binds differently in the absence vs. presence of the substrate. Therefore, the change in intrinsic protein fluorescence of M1-PYK upon binding of phenylalanine was used to monitor the affinity of the protein for this inhibitor. This affinity is denoted as Kapp-Phe, rather than Kd because changes in fluorescence intensity are not always directly proportional to ligand-protein complex formation [20]. This Kapp-Phe value changes upon the addition of PEP [16], consistent with the allosteric inhibition monitored by other techniques [21].
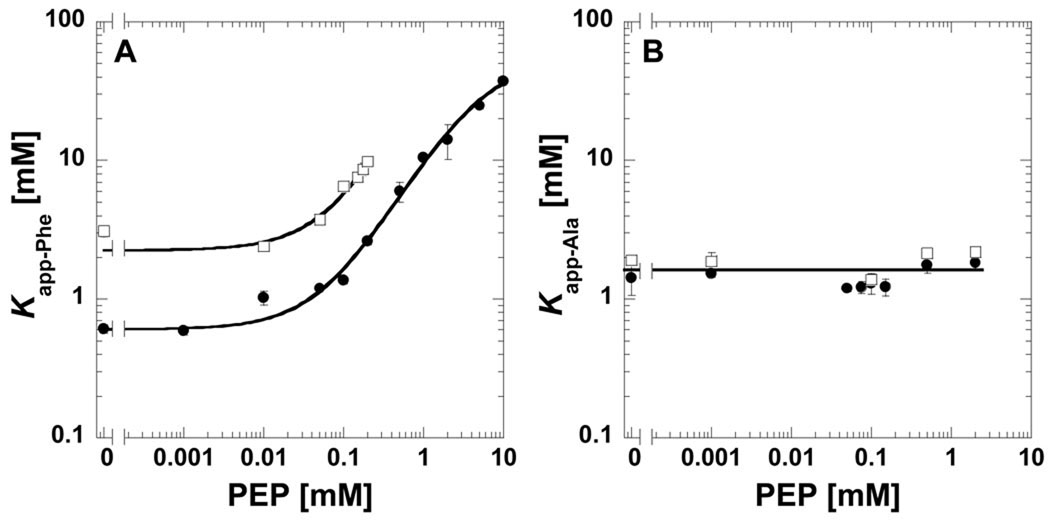
When D2O is used in place of H2O, the binding affinity for phenylalanine is greatly reduced (i.e. vertical displacement on the left hand y-axis in Fig 2A). We previously used a series of 40 amino acid analogues to map which chemical moieties in the effector are necessary for binding and which chemical moieties determine the magnitude of the allosteric response [21]. The L-2-aminopropanaldehyde substructure of phenylalanine is the minimal required for binding and the length of the hydrophobic side chain determines the magnitude of the allosteric response (In Fig 2A, the magnitude of the allosteric response is the difference between the upper and lower plateaus at low and high PEP, respectively). However, we also found that the length of the hydrophobic side chain influences effector affinity. Effector affinity is reduced at intermediate chain length (i.e. norvaline), compared to shorter chain lengths (e.g. alanine). As the length of the hydrophobic side chain is extended beyond norvaline (e.g. phenylalanine and norleucine), affinity returns to a level comparable to that of very short side chains (i.e. alanine). The data shown in Fig 2A are consistent with a hydrophobic contribution to phenylalanine binding to M1-PYK. Due to the increased hydrophobic nature of D2O as compared to H2O, there should be less of an energetic penalty for maintaining phenylalanine solvated in D2O, giving rise to the reduced affinity. In an effort to further verify this interpretation, we considered the impact of D2O when phenylalanine was replaced by alanine as the effector (Fig 2B). Alanine binds to the amino acid binding site of M1-PYK, but elicits only a minimal perturbation of PEP affinity [16, 21]. Consistent with an interpretation that assays in D2O reflect reduced phenylalanine affinity due to the removal of an energetic penalty for maintaining phenylalanine solvated by the more hydrophobic D2O, Kapp-Ala is the same in D2O and H2O (Fig 2B).
Fig 2.
Kapp-amino acid values for binding of A) phenylalanine or B) alanine determined by ligand induced changes in protein fluorescence plotted as a function of the concentration of PEP. Data collected in both H2O (●) and D2O (□). Lines represent trends in the data. When error bars are not apparent, they are smaller than the data point symbols.
Despite the reduced affinity of M1-PYK for phenylalanine, the allosteric mechanism continues to be functional, as is apparent from the upward curvature as PEP concentration is increased (Fig 2A). As presented as in Fig 2A, the indication for formation of the ternary (effector-enzyme-substrate) complex is the upper plateau at high PEP concentration [2, 14]. In Fig 2A, it is clear that where the upper plateau is starting to form at very high PEP concentrations in H2O, the reduced affinity of M1-PYK for phenylalanine in D2O prevents any formation of the upper plateau within the working concentration ranges of phenylalanine and PEP. In Fig 2A, lack of data at high PEP in D2O is in part due to the formation of a precipitation forms at high PEP/high Phe conditions. Reducing D2O from 100% may improve solubility limits, but would complicate data interpretation due to the mixed population of protonated/deuterated protein. Nonetheless, by monitoring allostery in 100% D2O, it is clear that an analysis of solvent accessibility of the ternary (substrate-enzyme-effector, when phenylalanine is the effector) using H/DX-MS will not be possible. However, the M1-PYK example included here highlights the need to monitor allosteric function in both H2O and D2O before initiating H/DX-MS studies.
Footnotes
Acknowledgments
This work was supported by NIH grant DK78076 (to A.W.F.).
References
- 1.Monod J, Changeux JP, Jacob F. Allosteric proteins and cellular control systems. J Mol Biol. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- 2.Fenton AW. Allostery: an illustrated definition for the 'second secret of life'. Trends Biochem Sci. 2008;33:420–425. doi: 10.1016/j.tibs.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui Q, Karplus M. Allostery and cooperativity revisited. Protein Sci. 2008;17:1295–1307. doi: 10.1110/ps.03259908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monod J, Wyman J, Changeux JP. On the Nature of Allosteric Transitions: a Plausible Model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 5.Lipchock JM, Loria JP. Nanometer propagation of millisecond motions in V-type allostery. Structure. 2010;18:1596–1607. doi: 10.1016/j.str.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 7.Swain JF, Gierasch LM. The changing landscape of protein allostery. Curr Opin Struct Biol. 2006;16:102–108. doi: 10.1016/j.sbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Kern D, Zuiderweg ER. The role of dynamics in allosteric regulation. Curr Opin Struct Biol. 2003;13:748–757. doi: 10.1016/j.sbi.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Frantom PA, Zhang HM, Emmett MR, Marshall AG, Blanchard JS. Mapping of the allosteric network in the regulation of alpha-isopropylmalate synthase from Mycobacterium tuberculosis by the feedback inhibitor L-leucine: solution-phase H/D exchange monitored by FT-ICR mass spectrometry. Biochemistry. 2009;48:7457–7464. doi: 10.1021/bi900851q. [DOI] [PubMed] [Google Scholar]
- 10.Laine O, Streaker ED, Nabavi M, Fenselau CC, Beckett D. Allosteric signaling in the biotin repressor occurs via local folding coupled to global dampening of protein dynamics. J Mol Biol. 2008;381:89–101. doi: 10.1016/j.jmb.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Dangott LJ, Fitzpatrick PF. Regulation of phenylalanine hydroxylase: conformational changes upon phenylalanine binding detected by hydrogen/deuterium exchange and mass spectrometry. Biochemistry. 49:3327–3335. doi: 10.1021/bi1001294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S, Sura GR, Dangott LJ, Fitzpatrick PF. Identification by hydrogen/deuterium exchange of structural changes in tyrosine hydroxylase associated with regulation. Biochemistry. 2009;48:4972–4979. doi: 10.1021/bi9004254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villar MT, Miller DE, Fenton AW, Artigues A. SAIDE: A Semi-Automated Interface for Hydrogen/Deuterium Exchange Mass Spectrometry. Proteomica. 2010;6:63–69. [PMC free article] [PubMed] [Google Scholar]
- 14.Reinhart GD. Quantitative analysis and interpretation of allosteric behavior. Methods Enzymol. 2004;380:187–203. doi: 10.1016/S0076-6879(04)80009-0. [DOI] [PubMed] [Google Scholar]
- 15.Weber G. Ligand binding and internal equilibria in proteins. Biochemistry. 1972;11:864–878. doi: 10.1021/bi00755a028. [DOI] [PubMed] [Google Scholar]
- 16.Fenton AW, Williams R, Trewhella J. Changes in small-angle X-ray scattering parameters observed upon binding of ligand to rabbit muscle pyruvate kinase are not correlated with allosteric transitions. Biochemistry. 2010;49:7202–7209. doi: 10.1021/bi100147w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makhatadze GI, Clore GM, Gronenborn AM. Solvent isotope effect and protein stability. Nat Struct Biol. 1995;2:852–855. doi: 10.1038/nsb1095-852. [DOI] [PubMed] [Google Scholar]
- 18.Jelinska-Kazimierczuk M, Szydlowski J. Isotope Effect on the Solubility of Amino Acids in Water. Journal of Solution Chemistry. 1996;25:1175–1184. [Google Scholar]
- 19.Glasoe PK, Long FA. Use of glass electrodes to measure acidities in deuterium oxide. Journal of Physical Chemistry. 1960;64:188–189. [Google Scholar]
- 20.Oberfelder RW, Lee JC. Measurement of ligand-protein interaction by electrophoretic and spectroscopic techniques. Methods Enzymol. 1985;117:381–399. doi: 10.1016/s0076-6879(85)17023-0. [DOI] [PubMed] [Google Scholar]
- 21.Williams R, Holyoak T, McDonald G, Gui C, Fenton AW. Differentiating a Ligand's Chemical Requirements for Allosteric Interactions from Those for Protein Binding. Phenylalanine Inhibition of Pyruvate Kinase(,) Biochemistry. 2006;45:5421–5429. doi: 10.1021/bi0524262. [DOI] [PubMed] [Google Scholar]