Summary
The RNA binding protein Sam68 is implicated in various cellular processes including RNA metabolism, apoptosis and signal transduction. Here we identify a role of Sam68 in TNF-induced NF-κB activation and apoptosis. We found that Sam68 is recruited to the TNF receptor and its deficiency dramatically reduces RIP recruitment and ubiquitylation. It also impairs cIAP-1 recruitment and maintenance of recruited TRAF2 at the TNF receptor. In its absence, activation of the TAK1-IKK kinase complex is defective, greatly reducing signal transduction. Sam68 is also found as a part of the TNF-induced cytoplasmic caspase-8-FADD complex. RIP is not recruited to this complex in Sam68 knockout cells and caspase activation is virtually absent. These findings delineate previously unknown functions for Sam68 in the TNF signaling pathway where it acts as a signaling adaptor both in the membrane-associated complex I and in the cytoplasmic complex II, regulating both NF-κB activation and apoptosis.
Introduction
TNFα is a potent cytokine that plays an important role in the inflammatory response, immunity, cell replication and differentiation as well as apoptosis. The majority of the known functions of TNF occur through TNF receptor1 (TNFR1) triggering, which initiates either a pro-survival pathway, mainly through the activation of the transcription factor NF-κB, or a pro-apoptotic pathway, through the activation of caspases. TNF signaling also triggers MAP kinases, which activate the JNK, p38 and ERK pathways and contribute to the biological effects TNF (Locksley et al., 2001) (Wajant et al., 2003).
The NF-κB family of transcription factors is composed of five members: p65, RelB, c-Rel, p105 (and its processed form, p50), and p100 (and its processed form, p52). In the resting state, NF-κB proteins exist as homo- or hetero-dimers in the cytoplasm bound to the inhibitory proteins of the IκB family. Activation occurs by signal-induced phosphorylation, ubiquitylation and proteasomal degradation of the IκB proteins, releasing the bound NF-κB proteins to translocate to the nucleus where they initiate transcription. IκB’s are phosphorylated by an upstream kinase complex mainly composed of IKK1, IKK2 and NEMO, which is activated following the recruitment of the complex to the TNFR (Vallabhapurapu and Karin, 2009).
TNF stimulation of cells leads to formation of two signaling complexes; an early complex in the membrane functioning in NF-κB activation and a later complex in the cytoplasm involved in apoptosis (Micheau and Tschopp, 2003). The membrane complex I is mainly composed of TNFR, TRADD, RIP, TRAF2, cIAPs and IKKs and is formed within minutes after TNF stimulation. Several studies suggest that K63-linked ubiquitylation of RIP by the E3 ligases TRAF2 and cIAPs in this complex is a crucial determinant of the activation of NF-κB by TNFR1 (Ea et al., 2006); (Alvarez et al., 2010) (Varfolomeev et al., 2008). A polyubiquitin chain on RIP can bind NEMO and TAB2 and may stabilize the binding of the recruited IKK complex and TAK1/TAB2/TAB3 complex to the TNFR (Ea et al., 2006). Subsequent phosphorylation of the kinases is thought to occur by activation induced by close proximity. In line with the delayed induction of apoptosis after TNF stimulation, the cytoplasmic complex II is formed hours later. It is devoid of TNFR, but contains several other components like TRADD, RIP and TRAF2 that are recruited to the TNFR as well as caspase8 and FADD. Caspase8 activation in the complex II instigates the apoptotic process (Micheau and Tschopp, 2003) (Wang et al., 2008).
Although the above model was thought to be complete, recent discoveries like the role of linear polyubiquitin chains that function as molecular patches stabilizing the protein complexes recruited to the TNFR (Haas et al., 2009) and the importance of the lipid Sphingosine-1-phosphate as a cofactor in TRAF2 and RIP ubiquitylation (Alvarez et al., 2010) shows that the nature of these signaling pathways is yet to be unraveled. While studying TNF signaling in our laboratory, by chance we came across the surprising finding that TNF stimulation of cells causes dramatic modulation of the cytoplasmic levels of a protein called Sam68. This led us to consider the novel possibility that Sam68 might be involved in TNF signaling.
Sam68 or KHDRBS1 (KH domain containing, RNA binding, signal-transduction associated 1), a specific target of the Src tyrosine kinase in mitosis also contains multiple proline rich regions that bind to SH3 and WW domain-containing proteins (Taylor and Shalloway, 1994) (Lukong and Richard, 2003). Its function has been implicated in T-cell receptor (Fusaki et al., 1997) and insulin receptor signaling where it acts as a docking molecule bridging several signaling molecules to the respective receptor’s proximity and it was suggested to act as an adaptor in signal transduction (Najib et al., 2005). However, most of its function has been attributed to its RNA binding property by which it regulates metabolism, nuclear export and the stability of RNA. In line with its functional diversity, Sam68 exists both in the cytoplasm and nucleus and undergoes various post-translational modifications, which modulate its function (Lukong and Richard, 2003). Until now, Sam68 has not been implicated in cytokine receptor signaling.
Here we explore the role of Sam68 in TNFR signaling and show that Sam68 is required for both NF-κB activation and cell death induction by TNFR. Sam68 binds to the TNFR upon TNF stimulation and is a necessary component of both the early complex I and the late complex II generated following TNFR stimulation.
Results
NF-κB activation induced by TNF but not IL-1 is greatly reduced in Sam68 knockout cells
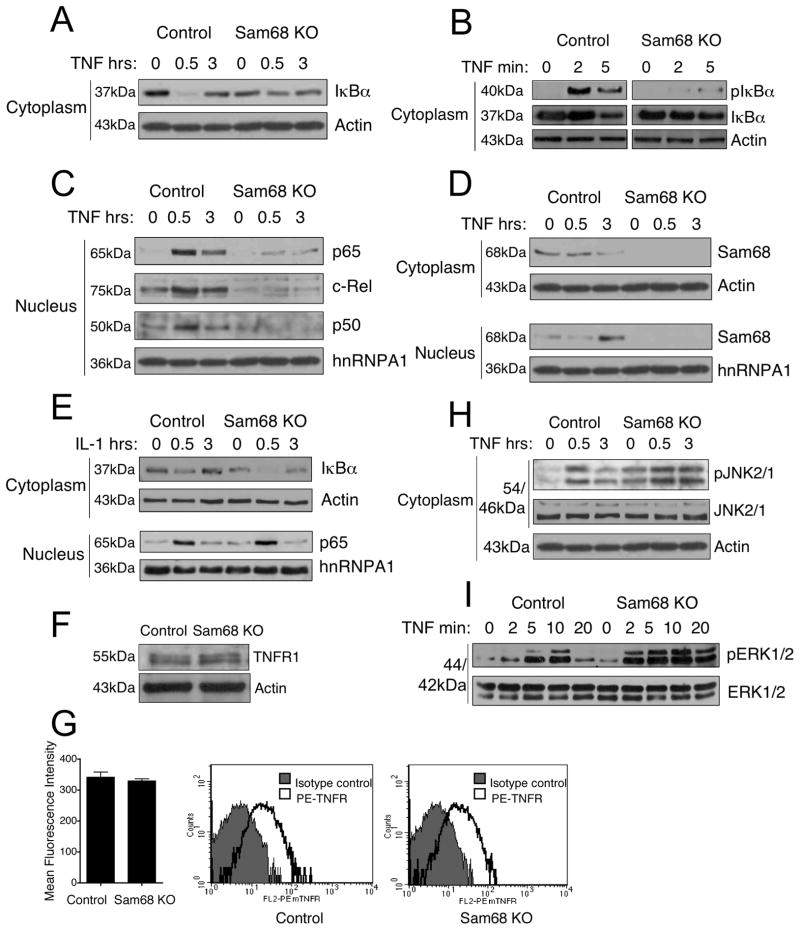
These studies of the role of Sam68 protein in TNF signaling were prompted by the chance observation that prolonged TNF stimulation of Jurkat cells caused a dramatic reduction in the cytoplasmic levels of Sam68 (Supplemental Figure S1). We therefore obtained wild type and Sam68 knockout mouse embryonic fibroblast (MEF) cells (Richard et al., 2005) and first assessed the activation of their NF-κB pathway by TNF as assayed by IκBα degradation and nuclear translocation of NF-κB proteins. We found that a deficiency of Sam68 resulted in greatly reduced TNF-induced IκBα degradation in the cytoplasm (Figure 1A). We also found an almost complete blockage of IκBα phosphorylation in Sam68 knockout cells (Figure 1B) and a dramatic reduction in nuclear translocation of the p65, c-Rel and p50 subunits of NF- κB (Figure 1C). In addition, consistent with the impaired NF-κB activation, Sam68 knockout cells also showed decreased levels of IκBα protein in the cytoplasm (compare Figure 1A lane 1 and lane 4). We examined Sam68 levels under these conditions and as we saw in Jurkat cells, MEF cells also showed decreased Sam68 levels in the cytoplasm following long term TNF stimulation. This was paralleled by a corresponding increase in its nuclear level showing that Sam68 is a TNF responsive protein (Figure 1D). To study whether there is any general defect in NF-κB activation in Sam68 knockout cells, we treated the cells with IL-1β, another proinflammatory cytokine. IL-1β-induced NF-κB activation was intact in these cells as measured by IκBα degradation and nuclear translocation of p65 (Figure 1E) suggesting that the NF-κB activation pathway is functional in these cells and Sam68 plays a role specifically in TNF signaling. We also confirmed that the observed defect in NF-κB signaling in Sam68 knockout cells is not due to an altered expression of TNFR (Figure 1F and 1G).
Figure 1. TNF-induced NF-κB activation is attenuated in Sam68 knockout cells, while the IL-1 pathway is not affected.
(A) Control and Sam68 knockout (Sam68 KO) MEF cells were treated with 100ng/ml TNF for the indicated times and cytoplasmic extracts were immunoblotted with anti-IκBα antibody. (B) Total lysates from control and Sam68 KO cells were immunoblotted for phosphorylated IκBα (p IκBα) and IκBα. (C) Nuclear extracts from (A) were immunoblotted for p65, c-Rel, p50. (D) The membranes from (A) and (C) were reprobed for Sam68. (E) IL-1-induced NF-κB activation is normal in Sam68 KO cells. Cells were treated with 100ng/ml recombinant human IL-1 beta and analyzed as above. Actin and hnRNPA1 were used as cytoplasmic and nuclear loading controls respectively. (F) and (G) Sam68 KO cells express similar levels of TNFR as wild type cells. (F) Cells lysates were immunoblotted using anti-TNFR antibody. (G) Cells were analyzed by flow cytometry for surface expression of TNFR and Mean Fluorescence Intensity (MFI) represented as mean +/ SEM (left panel) and histogram plots (right panel) are shown. TNF-induced JNK and ERK activation does not require Sam68. (H) and (I) TNF-induced JNK and ERK activation is enhanced in Sam68 KO cells. (H) The membrane from (A) was reprobed using phosphorylated JNK (pJNK) and total JNK antibodies. (I) Cells were starved overnight in serum free medium to minimize basal ERK activation and then treated with TNF for indicated times. Cell lysates were immunoblotted using phosphorylated ERK (pERK) and total ERK antibodies. Actin was used as loading control. See also Figure S1.
In addition to NF-κB, TNF also activates MAP kinases, including the ERK and JNK pathways (Karin and Gallagher, 2009). We therefore examined whether Sam68 mediates TNF-induced JNK and ERK activation. JNK showed an enhanced basal level of phosphorylation but was induced over that level by TNF and did not fall back to the basal level as quickly (Figure 1H). Activation of ERK by TNF was intact but enhanced in Sam68 knockout cells (Figure 1I). These findings suggest that Sam68 deficiency has a much larger effect on TNF-induced NF-κB activation than on MAP kinase activation although the deficiency does somewhat alter control of MAP kinases.
Sam68 deficiency blocks TNF-induced NF-κB dependent gene expression
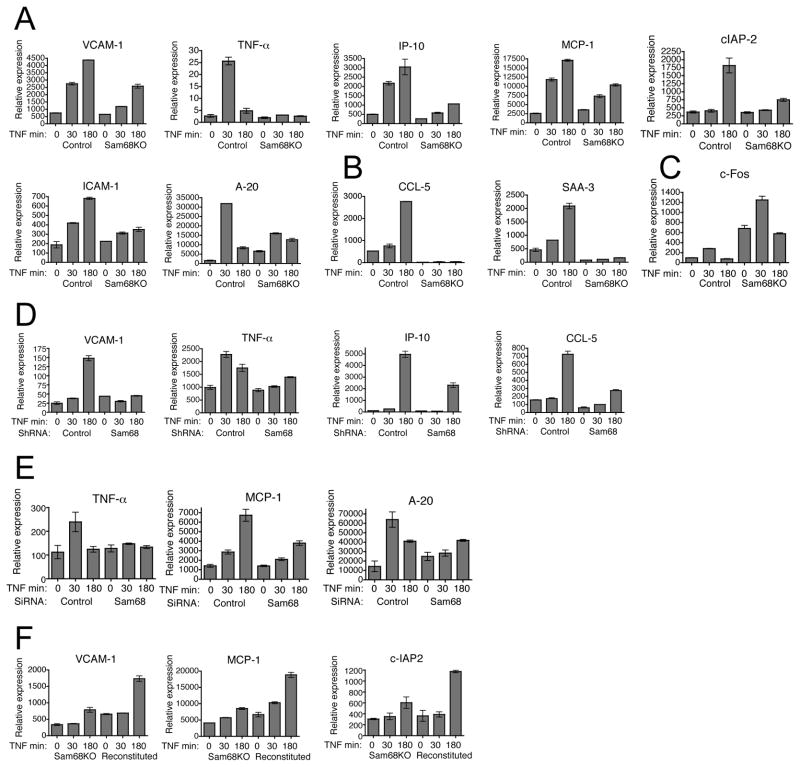
To gain further insight into the consequences of defective TNF-induced NF-κB signaling in Sam68 knockout cells, we assessed NF-κB dependent gene expression in these cells by quantitative RT-PCR. Cells were stimulated with TNF for 30 minutes to examine fast-responding genes and 3 hours for the slower ones. In line with the severely decreased nuclear translocation of NF-κB, overall expression of the NF-κB target genes was substantially reduced in Sam68 knockout cells as compared to the wild type cells. Interestingly, the NF-κB target genes studied showed different degrees of requirement of Sam68 for their expression (Figure 2A). Some genes like TNFα and IP-10 were reduced up to 5-fold and were largely dependent on Sam68 while several others like VCAM-1, c-IAP2, A20, ICAM-1 and MCP-1 showed about a 2-fold reduction in their expression level. These differences could reflect the quantitative requirement for NF-κB at different genes. Notably, Sam68 deficiency did not decrease the basal expression level of these genes and the observed reduction was entirely correlated to interference in TNF signaling (Figure 2A). Some NF-κB target genes like CCL5 and SAA3 showed a total dependence on Sam68 for both their basal and TNF-induced levels and their expression was completely blocked in Sam68 knockout cells (Figure 2B). We also checked the expression of a JNK dependent gene, c-Fos, in Sam68 knockout cells. In line with the enhanced JNK activation in these cells, basal c-Fos expression was found elevated and TNF stimulation resulted in a further increase in its expression (Figure 2C). These findings suggest that Sam68 is a critical component for effective and complete TNF signaling leading to NF-κB-induced gene expression.
Figure 2. TNF-induced NF-κB dependent gene expression in greatly reduced in Sam68 knockout cells.
(A) Control and Sam68 KO cells were treated with 10ng/ml of TNF for the indicated times. Quantitative real time PCR was performed in triplicate and the relative abundance of each transcript was calculated with respect to the expression of ribosomal protein L32. (B) Expression of CCL-5 and SAA-3 and (C) c-Fos was studied as above. (D) Control and Sam68 suppressed HeLa cells (E) MEF cells transfected with control or Sam68 siRNA and (F) Sam68 KO cells and Sam68 KO cells reconstituted with FLAG-Sam68 were treated and gene expression was studied as above. Statistical significance was determined by two-tailed unpaired student’s t test and data are represented as mean +/ SEM. See also Figure S2.
To examine if the observed defect in NF-κB activation in Sam68 knockout cells might be a cell type-specific phenomenon, or caused by secondary genetic alterations in immortalized 3T3 fibroblastic cells, we generated HeLa cells where approximately 70% of Sam68 expression was suppressed using lentiviral-mediated shRNA expression (Supplemental Figure S2A). These cells showed a diminished expression of several TNF-induced NF-κB dependent genes, as found in the Sam68 knockout MEF cells (Figure 2D). We also tried transient siRNA mediated suppression of Sam68 in MEF cells. Even though Sam68 suppression was not efficient in MEF cells (Supplemental Figure S2B), a significant decrease in the TNF-induced expression of several genes was observed (Figure 2E) further demonstrating the requirement of Sam68 for TNF-induced NF-κB-dependent gene expression.
We attempted rescue of the defective TNF-induced NF-κB pathway activation by reconstituting the Sam68 knockout cells with a FLAG-tagged Sam68 cDNA. Since lentiviral transduction gave only weak expression, we transfected the cells by nucleofection and picked clones that expressed relatively high amounts of FLAG-Sam68 at about 50% of the quite high endogenous Sam68 protein level (Supplemental Figure S2C). Although a partial rescue, the ectopically expressed Sam68 could indeed increase the expression of NF-κB target genes (Figure 2F).
Sam68 deficiency blocks TNF-induced apoptosis
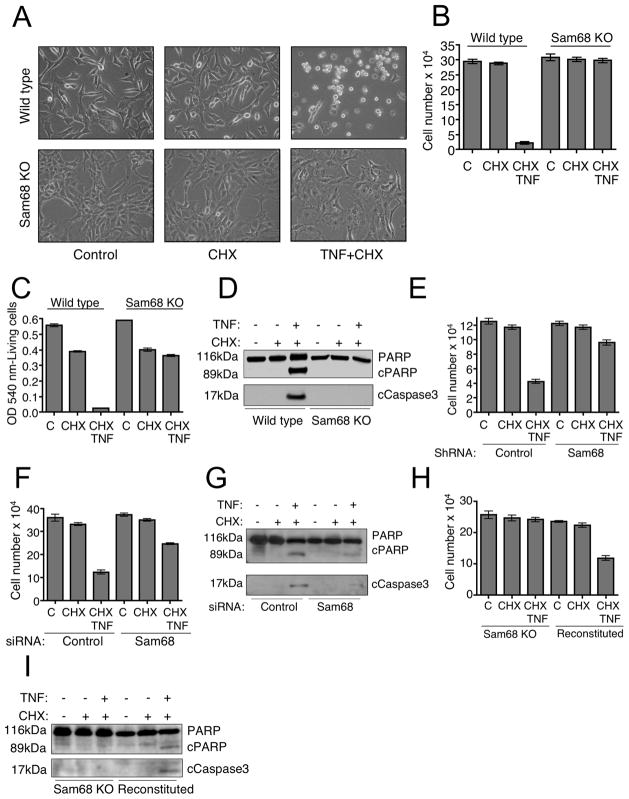
To study the role of Sam68 in TNF-induced apoptosis, we treated wild type and Sam68 knockout cells with TNF and cycloheximide. Apoptosis of wild type cells occurred by 3-4 hours after the treatment while no death of Sam68 knockout cells could be observed up to 12 hours. We analyzed cells by microscopy (Figure 3A), trypan blue exclusion staining for the absolute number of living cells (Figure 3B) and also quantified death by neutral red uptake assay (Figure 3C). To confirm that the observed death was indeed apoptosis, we analyzed the cell extracts for caspase3 activation and cleavage of the caspase substrate, PARP. Immunoblotting showed cleaved caspase3 and cleaved PARP in the wild type cells treated with TNF and cycloheximide, while no caspase3 activation or PARP cleavage were observed in Sam68 knockout cells (Figure 3D). To exclude the possibility that lack of TNF-induced apoptosis of Sam68 knockout MEF cells is not due to some cell-intrinsic defect, we tried to reproduce the phenomenon in shRNA Sam68 expressing HeLa cells. Similar to the knockout cells, Sam68-suppressed HeLa cells also showed resistance to apoptosis induced by TNF plus cycloheximide (Figure 3E). Excluding the possibility of cellular defects due to long term loss of Sam68, transient suppression of Sam68 by siRNA also reduced TNF induced apoptosis (Figure 3F) and cleavage of caspase3 and PARP (Figure 3G). Sam68 knockout cells reconstituted with Sam68 regained the ability to activate the apoptotic pathway following TNF plus cycloheximide treatment further implicating Sam68 in this process (Figure 3H and 3I).
Figure 3. Sam68 is required for cycloheximide-induced TNF-dependent apoptosis.
(A) Control and Sam68 KO cells were seeded in 6 well plates and treated in triplicates with either cycloheximide (CHX) (5μg/ml) alone or CHX and TNF (100ng/ml) for 6 hours. Cells were visually examined on a Nikon Diaphot 300 phase contrast microscope at 200X final magnification and photographed using a SPOT digital camera and software. Data represented is one out of five independent experiments performed. Cell death was quantified either by (B) counting absolute cell numbers following trypan blue exclusion of dead cells or by (C) neutral red assay. (D) Cells were treated as in (A) and harvested just before the appearance of apoptotic morphology by 2.5–3.5 hours. Cell lysates were immunoblotted using anti-PARP and anti-cleaved caspase3 antibodies. cPARP and cCaspase3 represents cleaved forms of these proteins (E) Control and Sam68 suppressed HeLa cells were treated as above for 8 hours and cell death was quantified by counting. One out of three independent experiments performed is shown. (F) Wild type MEF cells transfected with control or Sam68 siRNA were treated as above and cell death was quantified by counting and (G) lysates were immunoblotted as above. (H) Sam68 KO cells and Sam68 KO cells reconstituted with FLAG-Sam68 were treated as above and cell death was quantified by counting and (I) lysates were immunoblotted as above. Data showing cell death in (B) (C) (E) (F) and (H) are represented as mean +/ SEM.
Sam68 is not required for the Fas-induced extrinsic or stress-induced intrinsic pathway of apoptosis
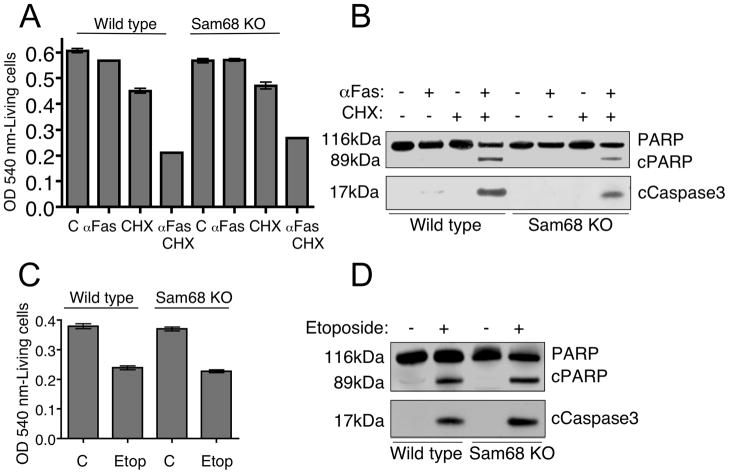
To investigate the role of Sam68 in apoptosis induced by factors other than TNF, we examined apoptosis induced by Fas, DNA damage and oxidative stress. Fas is a prototypic death receptor that induces apoptosis by the extrinsic pathway (Strasser et al., 2009). To study Fas-induced death, we treated the wild type and Sam68 knockout cells with agonistic anti-Fas antibody and cycloheximide. We found that unlike TNF, Fas stimulation induced apoptosis of both wild type and Sam68 knockout cells (Figure 4A) and induced cleavage of caspase3 and PARP (Figure 4B). Thus, the extrinsic apoptotic pathway as well as caspase activation is intact in Sam68 knockout cells and Sam68 function is, again, shown to be specific for TNF signaling.
Figure 4. Sam68 is not required for Fas-induced or genotoxic stress- induced apoptosis.
(A) Cells were seeded as in Figure 3A, and treated with either cycloheximide (5μg/ml) alone or cycloheximide and anti-Fas antibody (2μg/ml) for 12 hours. Cell death was quantified by Neutral red assay. Data represented are one out of three independent experiments performed. (B) Cells were treated as in (A) and harvested just before the appearance of apoptotic morphology by 6–7 hours. Cell lysates were immunoblotted using anti-PARP and anti-cleaved caspase3 antibodies. (C) Cells were treated with etoposide (Etop) (200μM) for 24 hours and cell death was quantified by Neutral red assay. (A) and (C) Data are represented as mean +/ SEM. (D) Cells were treated as in (C) and harvested by 12 hours and immunoblotted as above. Data showing cell death in (A) and (C) are represented as mean +/ SEM. See also Figure S3.
To examine whether Sam68 plays a role in the intrinsic pathway of apoptosis, we studied death induction by genotoxic and oxidative stress (Roos and Kaina, 2006) (Dumont et al., 1999). To induce DNA damage we used etoposide, an anti-neoplastic chemical that inhibits topoisomerase I. Etoposide treatment induced apoptosis of both wild type and Sam68 knockout cells (Figure 4C) accompanied by activation of caspase3 and PARP cleavage (Figure 4D). Similarly, oxidative stress induced by hydrogen peroxide treatment also caused death induction in both wild type and Sam68 knockout cells (Supplemental Figure S3). These results demonstrate that DNA damage-induced and oxidative stress-induced apoptosis does not require Sam68 and emphasize the unique dependence of TNF signaling on Sam68 for inducing apoptosis.
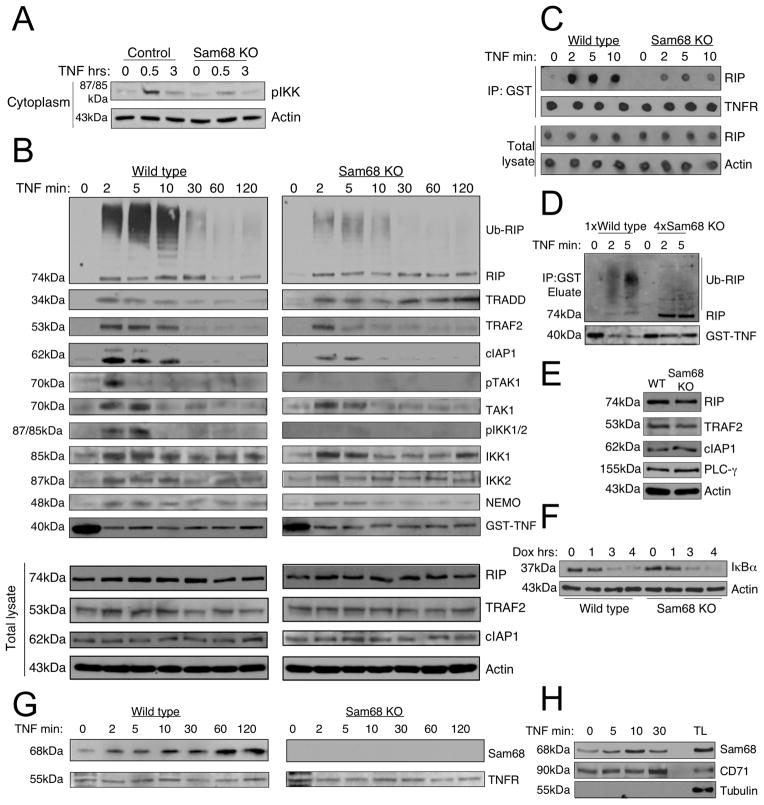
Sam68 is part of complex I of TNFR signaling and it is required for RIP recruitment, its ubiquitylation and kinase activation
TNF-induced signal transduction is initiated by the recruitment of several cytoplasmic molecules to the TNFR leading to their activation (Zhang et al., 2000) (Ramakrishnan et al., 2004). TNF stimulation generates two distinct signaling complexes, one dedicated to NF-κB activation (complex I) and the other for apoptosis (complex II) (Micheau and Tschopp, 2003). The defects in both IκBα phosphorylation and caspase activation in Sam68 knockout cells suggest that Sam68 might play roles in both complexes.
First we examined the activation status of IKK as a possible cause for defective TNF-induced IκBα phosphorylation in Sam68 knockout cells and found that IKK phosphorylation was greatly decreased in these cells (Figure 5A). To understand how Sam68 functions in TNF-induced IKK activation, we performed a detailed comparative study of the recruitment of signaling proteins to the membrane-associated TNFR complex I in wild type and Sam68 knockout cells following TNF stimulation for various times. TRADD recruitment to the TNFR was independent of Sam68 (Figure 5B). Interestingly, in Sam68 knockout cells, much higher TRADD levels were seen at later time points suggesting that it is stabilized in the complex by the lack of Sam68. Previous studies have shown that TNF stimulation rapidly induces recruitment and ubiquitylation of RIP in the receptor complex and this process is crucial for TNF-induced NF-κB activation (Zhang et al., 2000) (Ea et al., 2006). We examined RIP in the TNFR complex and found greatly decreased RIP recruitment and its ubiquitylation in Sam68 knockout cells (Figure 5B). However, the amounts of unmodified RIP recruited seemed comparable in wild type and Sam68 knockout cells indicating that Sam68 deficiency specifically affects the modified RIP fraction.
Figure 5. Sam68 is required for signaling from complex I of TNFR.
(A) Cells were treated as in Figure 1A and cytoplasmic extracts were immunoblotted with anti-phospho IKK (pIKK) antibody. (B) Kinetic analysis of recruitment of signaling molecules to TNFR. Cells were treated with GST-TNF for the indicated times and the receptor-associated complex immunoprecipitated through the GST-tag using glutathione sepharose beads (top 11 panels) and total lysates (bottom 4 panels) were immunoblotted using the indicated antibodies. The data presented is representative of seven independent experiments. (C) Eluate from Glutathione beads (top panels) and total lysates (bottom panels) were analyzed for total RIP levels by dot blotting. TNFR and Actin were used as loading controls. (D) Eluate from glutathione beads approximately normalized for total RIP levels in wild type and Sam68 knockout cells by loading 4 times the sample from the latter was analyzed by western blotting for RIP and GST-TNF. (E). Total cell lysates from wild type and Sam68 knockout cells were analyzed for indicated proteins. (F) Cells were treated with 10μg/ml doxorubicin and IκBα degradation was studied at indicated times. (G) Cells were treated and immunoprecipitated as above and Sam68 recruitment was assessed by immunoblotting. TNFR shows the uniformity of immunoprecipitation. See also Figure S4. (H) Sam68 is present in association with cell membrane. MEF cells were treated with TNF as above and membrane fractions were immunoblotted with anti-Sam68, anti-CD71 and tubulin antibodies. Total cell lysate was loaded as a control for protein expression. See also Figure S4.
We also compared the total recruited RIP levels in the TNFR in wild type and Sam68 knockout cells by dot blotting and observed an approximately 4-5-fold reduction in its level (Figure 5C). To examine whether Sam68 deficiency also affects RIP ubiquitylation at TNFR, we normalized the recruited RIP level in wild type and Sam68 knockout cells by dot blotting (Supplemental Figure S4A) and analyzed it by western blotting. Interestingly, we observed an increase in unmodified RIP in Sam68 knockout cells suggesting that the recruited RIP in these cells is poorly ubiquitylated (Figure 5D).
We showed that the defect in recruitment of RIP is not a result of a lack of its expression by showing that the level of mRNA (Supplemental Figure S4B) and protein (Figure 5E) was only slightly reduced in the Sam68 knockout cells. We also examined the possibility of RIP degradation in knockout cells as a cause of its poor recruitment and could not observe any TNF induced RIP degradation by western blotting or pulse-chase assay or any elevation in its level following proteasomal inhibition (Supplemental Figure S4C and S4D). Although the level of RIP in Sam68 knockout cells was slightly lower than that in wild type cells, it was fully functional in causing IκB degradation induced by doxorubicin (Figure 5F), which depends on RIP (Hur et al., 2003).
Seeing the dramatic change in ubiquitylated RIP levels, we looked for TRAF2 and cIAP1, which are thought to be the E3 ligases for RIP (Alvarez et al., 2010) (Varfolomeev et al., 2008). At 2 minutes post TNF stimulation, similar amounts of TRAF2 recruitment were seen in wild type and Sam68 knockout cells. Although this initial TRAF2 recruitment occurred independently of Sam68, TRAF2 rapidly dissociated from the receptor complex in Sam68 knockout cells suggesting a role for Sam68 in maintaining the recruited TRAF2 in the TNFR complex (Figure 5B). Consistent with a role for TRAF2 in recruiting cIAP1 to the TNF receptor (Shu et al., 1996), cIAP1 recruitment was also greatly decreased in Sam68 knockout cells (Figure 5B). We also examined the total protein levels of TRAF2 and cIAP1 in wild type and Sam68 knockout cells to demonstrate that their decreased recruitment was not due to lack of their expression (Figure 5B bottom panels and 5E). Phosphorylation of TAK1 and IKK1/IKK2 was not detectable in Sam68 knockout cells suggesting that the kinase activation depends on RIP recruitment and ubiquitylation. However, in contrast to the previous report (Ea et al., 2006), the reduced ubiquitylated RIP did not affect recruitment of TAK1 or the IKK complex to the TNFR (Figure 5B). These results suggest that Sam68 is required for RIP recruitment and ubiquitylation, maintaining proper TRAF2 and cIAP1 levels at the activated TNFR and TAK1-IKK kinase activation following TNF stimulation but it is not actually required for the recruitment of these kinases.
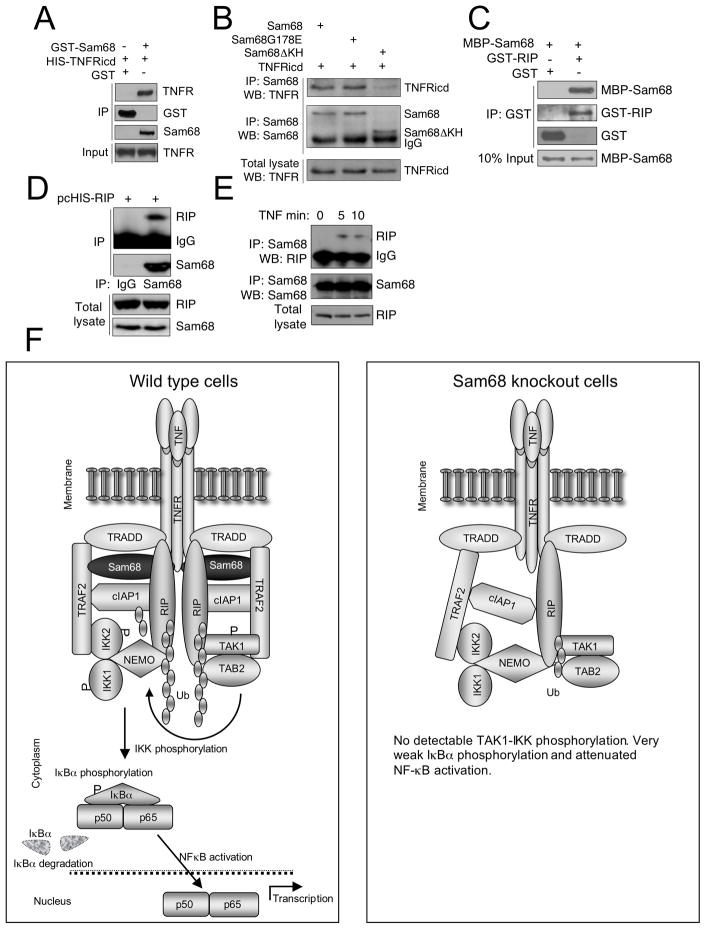
Because Sam68 deficiency affected multiple aspects in the recruitment and activation of signaling proteins linked to TNFR, we explored the possibility that Sam68 itself might be a physical component of this complex. We looked for Sam68 in the TNFR complex and found that it was bound to the TNFR at a low level in unstimulated cells and its recruitment steadily increased with time of TNF stimulation (Figure 5G). The uniformity of the immunoprecipitation was confirmed by immunoblotting for TNFR (Figure 5G bottom panel). We reconfirmed Sam68 recruitment to the TNFR also in HeLa cells along with ubiquitylated RIP and TRAF2 (Supplemental Figure S4E). We confirmed that Sam68 indeed binds to the TNFR by reverse immunoprecipitation of TNFR through Sam68 and observed a ligand-dependent increase in their binding (Supplemental Figure S4F). As additional evidence for the presence of Sam68 in association with the cell membrane, we did membrane fractionation of cells treated with TNF. As previously shown (Huot et al., 2009), we also saw Sam68 in the membrane fraction where its level increased during early TNF signaling. The fractions were free of cytoplasmic contamination as shown by the lack of tubulin and they contained the membrane marker transferrin receptor (CD71) (Figure 5H). The presence of Sam68 in the membrane fraction and its weak binding to TNFR in unstimulated cells suggested that Sam68 might bind directly to the TNFR. To test this possibility, we performed an in vitro binding assay and found direct binding of recombinant GST-tagged Sam68 and HIS-tagged TNFR-intracellular domain (TNFRicd) (Figure 6A). We also confirmed the binding of Sam68 to the TNFR by transient expression in HeLa cells. In an attempt to define the TNFR binding region in Sam68, we tested the binding of a deletion mutant of Sam68 lacking the KH domain (amino acids 157-256) or a point mutant (G178E) and found that the KH domain was required for its binding to the TNFR (Figure 6B). However, the KH domain, when expressed alone was unable to bind TNFR suggesting that it is a structural component required for binding and not the binding domain itself (Supplemental Figure S5A). We also examined the binding of Sam68 to RIP and observed that they bind directly in an in vitro binding assay (Figure 6C) and overexpressed RIP was specifically immunoprecipitated through endogenous Sam68 in 293T cells (Figure 6D). More importantly, we could immunoprecipitate RIP through Sam68 from MEF cells following TNF stimulation indicating that these proteins associate in a TNF-induced signaling complex (Figure 6E).
Figure 6. Sam68 binds to TNFR and RIP.
(A, B) KH domain of Sam68 is required for its binding to TNFR. (A) In vitro binding assay of recombinant GST- Sam68 and HIS-TNFRicd. (B) HeLa-shRNA-Sam68 cells were transfected with the indicated plasmids. Sam68 immunoprecipitate was probed for TNFRicd (top panel) and Sam68 (middle panel). The bottom panel shows the expression level of TNFRicd in the cell lysates. See also Figure S5. (C, D, E) Sam68 binds to RIP. (C) In vitro binding assay of recombinant MBP-Sam68 to GST-RIP. (D) 293T cells were transfected with pcHIS-RIP and immunoprecipitation was done with anti-Sam68 or rabbit IgG as control. (E) MEF cells were treated with TNF for the indicated time points, Sam68 was immunoprecipitated and probed for RIP binding. (F) Model of Sam68 role in membrane-associated TNFR complex. TNF triggering results in the recruitment of signaling proteins to the TNFR. Connection between the proteins shows their ability to bind directly. Sam68 and ubiquitin chains on RIP assist their proper orientation. TAK1 is activated by induced proximity. It phosphorylates the IKK complex, which then phosphorylates IκB leading to its ubiquitylation and degradation, releasing NF-κB to move to the nucleus. Phosphorylation is represented by ‘P’. Ub indicates Ubiquitin chains. In Sam68 knockout cells, the recruitment, orientation and activation of recruited molecules are defective.
Since Sam68 is a protein known to be phosphorylated (Taylor and Shalloway, 1994), we tested whether it is phosphorylated by TNF during the signaling process. While treatment of starved cells with 20% serum enhanced tyrosine phosphorylation of Sam68 (Supplemental Figure S5B), TNF stimulation failed to do so (data not shown). To detect whether TNF induces serine, threonine or tyrosine phosphorylation of Sam68, we performed in vivo orthophosphate-labeling. Although degradation of IκBα was seen, no radio-phosphate incorporation in Sam68 could be detected following TNF stimulation (Supplemental Figure S5C).
Figure 6F summarizes our understanding of the role of Sam68 in TNF-induced signal transduction. We see no alteration in Sam68 and it has no obvious catalytic function so we model it as a pure adaptor, holding bound proteins in the complex in a configuration that leads to ubiquitylation and phosphorylation of downstream elements and ultimately NF-κB activation.
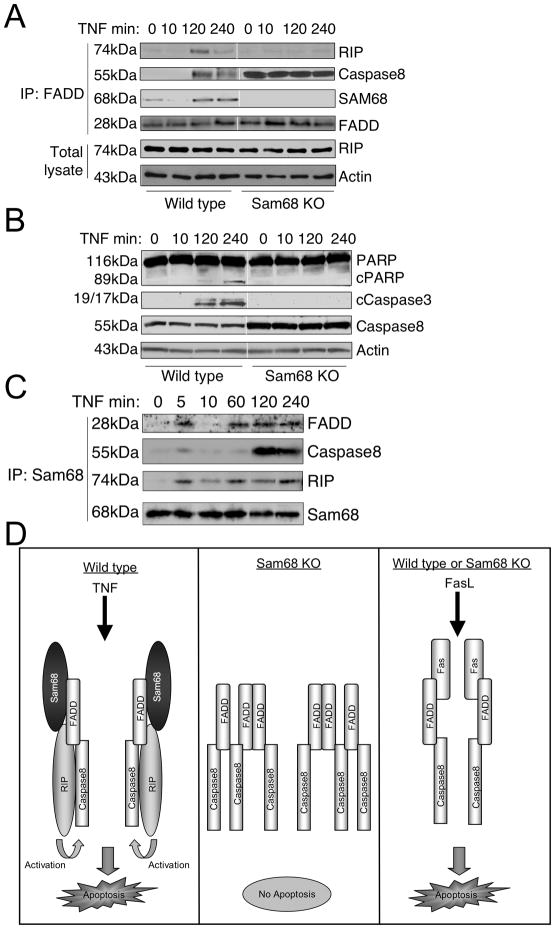
Sam68 is required for RIP recruitment to complex II and induction of apoptosis
To examine the mechanism of Sam68 action in TNF-induced apoptosis, we analyzed the FADD/caspase8-containing cytoplasmic complex II in wild type and Sam68 knockout cells. As previously reported (Micheau and Tschopp, 2003), long time TNF treatment resulted in a FADD/caspase8 complex, which, in wild type cells, also contained RIP (Figure 7A left panel). Though Sam68 knockout cells had sufficient expression of RIP (Figure 7A bottom panels), its binding to complex II was not detected in the absence of Sam68. Moreover, Sam68 knockout cells contained elevated levels of caspase8 (Figure 7B) and showed constitutive binding of caspase8 to FADD (Figure 7A right panel). However, this FADD/caspase8 complex was inactive, as we could not observe any caspase3 activation or PARP cleavage in Sam68 knockout cells when treated with TNF and cycloheximide (Figure 3D) or even with high concentrations of TNF (Figure 7B).
Figure 7. Sam68 is required for signaling from complex II of TNFR.
(A-C) Sam68 is associated with the late complex II formed after TNFR stimulation. (A) Cells were treated with TNF (1μg/ml) for the indicated times. Complex II was immunoprecipitated through FADD and analyzed for the presence of RIP, caspase8 and Sam68. Bottom panels show RIP levels in the lysate and actin as control (B) Total lysates from (A) were immunoblotted using anti-PARP, anti-cleaved caspase3, caspase8 and actin antibodies. (C) Cells were treated as in (A) and Sam68 immunoprecipitates were analyzed for the presence of FADD, caspase8 and RIP. (D) Model of Sam68’s role in the cytoplasmic FADD-caspase8 complex. Sam68 links RIP to the FADD-caspase8 complex. RIP causes caspase activation. In the absence of Sam68, FADD and caspase8 forms inactive aggregates. Fas-induced cell death does not require Sam68 and occurs as apoptosis caused by direct binding of the FADD-caspase8 complex to the Fas receptor and its activation.
To examine in another way whether Sam68 is a part of complex II, we tried to immunoprecipitate this complex from wild type cells through Sam68. As shown in Figure 7C, following prolonged TNF stimulation, we could precipitate the complex containing caspase8, FADD and RIP through Sam68 indicating that this complex is associated with Sam68. We also observed an early relatively weak association of FADD and caspase8 with Sam68, which fell by 10 minutes and later appeared as part of complex II. Caspase8 appeared in the complex after FADD recruitment suggesting that Sam68 binds to FADD before the complete complex II is formed (Figure 7C). In agreement with the role of RIP in both NF-κB activation and apoptosis, it was found associated with Sam68 throughout the TNF stimulation period. Figure 7D summarizes our understanding of the complexes formed with and without Sam68 that lead to the Sam68 dependence for TNFR-induced apoptosis and contrasts this with Fas-induced apoptosis.
Discussion
We report here our observations that Sam68 is a previously unappreciated TNFR binding protein that is required quite specifically for proper NF-κB activation and apoptosis induction, but not for JNK or ERK activation by the TNFR. Sam68 appears uninvolved in the closely related IL-1 or Fas signaling complexes that also lead to NF-κB activation and apoptosis, respectively. Sam68 is key to the appropriate organization at the cell membrane of the complex that forms around TNFR following TNF stimulation.
Sam68 facilitates the recruitment of RIP and maintenance of recruited TRAF2 and cIAP1 in the TNFR and allows them to catalyze the RIP ubiquitylation that precedes TAK1-IKK-IκB phosphorylation and NF-κB activation. Sam68 is also required for RIP recruitment to the cytoplasmic FADD-caspase8 complex that is required for caspase activation. Our results uncover the previously unrecognized role of Sam68 as a signaling adaptor both in the membrane-associated complex I (see model in Figure 6F) and in the cytoplasmic complex II (see model in Figure 7D) generated following TNF stimulation.
Role of Sam68 in TNF-induced membrane proximal events and NF-κB activation
Two major effects on TNFR signaling in Sam68 knockout cells were associated with TRAF2 and RIP. Consistent with this observation, although Sam68 was essential for TNF-induced NF-κB activation, its deficiency did not affect IL-1 signaling, which acts independently of TRAF2 and RIP (Verstrepen et al., 2008). We also examined RIP-dependent NF-κB pathway to verify that the defects in Sam68 knockout cells were not due to defective RIP function. NF-κB activation as a consequence of Doxorubicin-mediated DNA damage was normal in these cells further proving the specific involvement of Sam68 in TNF signaling (Figure 5F). TNF-induced activation of MAP kinase pathways, JNK and ERK, requires TRAF2 (Devin et al., 2003) but the role of RIP in these pathways is debated (Devin et al., 2003) (Kelliher et al., 1998). Our results indicate that Sam68 is not required for MAP kinase activation by TNF. It could be that the initial recruitment of TRAF2 to the TNFR, which is not affected by Sam68 deficiency, is sufficient for the initiation of MAP kinase activation. However, Sam68 seems to have a regulatory role in keeping MAP kinase activation under control because in its absence basal JNK activation is significantly increased and TNF-induced JNK activation is sustained to even 3 hours post-stimulation. Also, TNF-induced ERK activation is much stronger in Sam68 knockout cells than the wild type cells. These results also suggest that TRAF2 is functional in these cells.
As previously observed (Ea et al., 2006), our results also suggest that RIP recruitment and ubiquitylation is required for TNF-induced NF-κB activation. The ubiquitin chains on RIP have been suggested to be a tether, linking TAK1 and the IKK complex to the TNFR because expression of the non-ubiquitylatable mutant RIP K377A in cells compromised the recruitment of this kinase complex to the TNFR as well as NF-κB activation. But this mutation also affected the recruitment of unmodified RIP to the TNFR and there is the possibility of a structural defect in RIP caused by this mutation. In contrast, we noticed a specific reduction only in the ubiquitylated fraction of RIP and the recruitment of unmodified RIP was not affected in Sam68 knockout cells. Similarly to our results, a deficiency of TRADD also decreases ubiquitylated RIP in the TNFR complex without affecting the recruitment of unmodified RIP. In this case, TRAF2 recruitment is completely absent which might account for the lack of RIP ubiquitylation and activation of the NF-κB and MAP kinase pathways (Chen et al., 2008). Although cIAPs were shown to induce direct ubiquitylation of RIP in vitro and were suggested to be the E3 ligase for RIP (Varfolomeev et al., 2008), a role for TRAF2 still remains crucial because cIAP recruitment to the TNFR is mediated by TRAF2 (Shu et al., 1996). Consistent with this interpretation, we saw a reduced recruitment of cIAP1 in Sam68 knockout cells (Figure 5B). Because Sam68 knockout cells showed an increase in the mRNA level of A20, which has been suggested to modulate RIP ubiquitylation and levels (Vallabhapurapu and Karin, 2009), we also examined the possibility that alteration in the RIP level is due to elevated A20 levels. Though A20 protein level was also increased in Sam68 knockout cells (Supplemental Figure S6A), this did not increase the amount of A20 recruited to the TNFR (Supplemental Figure S6B). Moreover, partial suppression of A20 by siRNA did not enhance TNF induced RIP ubiquitylation in Sam68 knockout cells (Supplemental Figure S6C and S6D).
Our results suggest that ubiquitylated RIP is not essential for the recruitment or maintenance of the TAK1/IKK kinase complex in the TNFR but it is required for kinase activation. If indeed the ubiquitylation of RIP was crucial to recruiting the TAK1 and IKKs to the TNFR as proposed (Ea et al., 2006), we should have seen a corresponding decrease in the level of recruited kinases in parallel with the decreased ubiquitylated RIP levels. In fact, in a study using RIP and TRAF2 knockout cells, it has been shown that TRAF2 mediates recruitment of the IKK complex to the TNFR and RIP is not essential for this process (Devin et al., 2000). Other studies have shown that unmodified RIP has the potential to bind NEMO directly and this interaction could stabilize the recruited IKK complex (Zhang et al., 2000). Considering these results, it is conceivable that the recruitment of the IKK complex is not affected in Sam68 knockout cells because once the complex is initially recruited through TRAF2, it can continue in the receptor-bound complex through its interaction with unmodified RIP even though the bound TRAF2 level goes down. Interestingly, by sequence analysis, we found multiple TRAF2 binding sites in Sam68 and it was found to bind TRAF2 in transient transfection experiments (Supplemental Figure S6E). It will be of future interest to study the importance of the TRAF2 binding sites in Sam68 in the context of TNF signaling.
Unlike TCR stimulation, which induces tyrosine phosphorylation of Sam68 (Fusaki et al., 1997), we could not observe any phosphorylation of Sam68 after TNF stimulation (Supplemental Figure S5B and S5C). However, phosphorylation is not a prerequisite to involvement in signaling as it has been shown previously that the adaptor protein TRADD is also not phosphorylated by TNF during initial signaling events (Jiang et al., 1999). Our data suggests that like TRADD, Sam68 also binds to the TNF receptor directly. Interestingly, as in the RIP knockout cells (Devin et al., 2000), Sam68 knockout cells also show significantly enhanced binding of TRADD to the TNFR at later time points of TNF stimulation (Figure 5B), which suggests their competitive occupancy of the TNFR.
Sam68 lacks any functional domain like an E3 ubiquitin ligase or a kinase (Lukong and Richard, 2003). But, in support of an adaptor role, it contains several proline rich domains and a tyrosine rich region that is phosphorylated, which may facilitate protein-protein interactions with SH3- and SH2-domain containing proteins. Sam68 also binds non-SH3-, SH2-domain proteins like Cbl and JAK3 by unknown mechanisms (Fusaki et al., 1997). As an adaptor, Sam68 may facilitate the recruitment of signaling molecules and seems likely to control the signaling events sterically. Its absence may cause defective recruitment of the proteins that bind to it and may result in rearrangement of signaling complex hindering proper activation of its components. The TNF inducible binding of Sam68 to TNFR and RIP and its dynamic dissociation from and association with the FADD complex clearly implies a direct signaling role for Sam68. However, because we saw that TNF induces late nuclear translocation of Sam68, currently we are studying the nuclear role of Sam68 in relation to late gene induction by NF-κB.
Role of Sam68 in the TNF-induced cytoplasmic complex mediating apoptosis
The results reported here also show that Sam68 is an integral part of the TNF-induced death-inducing complex II. Although the cells have sufficient caspase8 and FADD in the cytoplasm these proteins do not interact to initiate the apoptotic process (Wang et al., 2008). We found that Sam68 binds to FADD in the cytoplasm in unstimulated cells, and this Sam68-FADD binding might prevent unproductive FADD-caspase8 binding in basal state. Interestingly, TNF stimulation dissociates the basal cytoplasmic FADD-Sam68 complex with a corresponding increase of Sam68 in the membrane-associated complex during early signaling. FADD-Sam68 binding reappears again at the time of complex II formation indicating the possibility of a TNF-induced translocation of Sam68 between different complexes (Figure 7A, 5C). Consistent with the idea of Sam68 playing a safeguarding role in blocking basal FADD-caspase8 binding, we found that caspase8 was constitutively bound to FADD in Sam68 knockout cells and formed an inactive complex likely due to lack of RIP recruitment (Figure 7A, 7B). Thus, Sam68 seems to play a structural role in regulating the proper assembly of FADD-caspase8 complex for its activation (Figure 7D, model).
Both of the complexes generated following TNF stimulation contain RIP (Micheau and Tschopp, 2003), which seems a likely candidate for carrying over the death message from the receptor to complex II. Consistent with this idea, RIP has been shown to bind caspase8 directly (Shikama et al., 2003). In addition, RIP detached from the TNFR is suggested to have a role in activating the caspase8-FADD complex, likely by phosphorylation, because its function depends on intact kinase domain (Wang et al., 2008). In agreement with this proposed function of RIP, our observation that RIP was absent from the FADD-associated complex in the absence of Sam68 provides an explanation for the inactivity of this complex. Consistent with the previously proposed models (Micheau and Tschopp, 2003) (Strasser et al., 2009) as well as our results presented here, we did not see any significant defect in Fas-induced apoptosis where the caspase activation occurs by direct binding of FADD-caspase8 to the Fas receptor. However, the apoptosis induction by Fas was slightly lower in Sam68 knockout cells, probably due to the presence of inactive FADD-caspase8 complex in these cells and the extra signaling intensity that this complex may demand for its activation. Similar to the Fas pathway, the stress-induced intrinsic pathway of apoptosis also apparently functioned normally in the absence of Sam68.
Our findings on Sam68 as a required factor for TNF-induced NF-κB activation and apoptosis reveals yet another complexity involved in this extensively studied signaling pathway. As shown here, apart from the enzymatic components, structural components like Sam68 are also crucial in TNF signaling complex assembly and activation. TNFα is a key effector of innate immune responses and deregulation of TNFα-NF-κB pathway is implicated in several immune and inflammatory disorders including cancer. Understanding the function of new proteins that are involved in this pathway expands our knowledge of its molecular regulation and provides potential targets for therapy.
Experimental Procedures
Please also see supplemental experimental procedures.
Lentiviral shRNA transduction, siRNA and plasmid Transfections
HeLa cells were transduced with lentivirus expressing shRNA against human Sam68 (sc-29476-V, Santacruz) and selected in 2μg/ml Puromycin. siRNAs were transiently transfected into MEF cells by nucleofection as described above. Sam68 knockout cells stably expressing FLAG tagged mouse Sam68 was generated by transfection using nucleofector device in MEF solution 1 and program A023 following manufacturer’s instruction (Lonza). Transient transfection of HeLa cells was performed using Lipofectamine 2000 reagent according to manufacturer’s instructions (Invitrogen). HEK-293T cells were transfected by calcium phosphate precipitation method.
Quantitative real time PCR
Quantitative real time PCR using cDNA corresponding to 10-20ng of total RNA was performed using SYBR GREEN PCR Master Mix (Kapa Biosystems) in a Real-Time PCR machine (Realplex, Eppendorf). The results obtained for individual genes were normalized to the ribosomal protein Rpl32. The primers used for PCR were designed from Primer Bank (http://pga.mgh.harvard.edu/primerbank/index.html) or Oligoperfect (Invitrogen) and their sequences are given in supplemental table. Difference in gene expression was statistically analyzed using two-tailed unpaired Student’s t test with PRISM. All the represented genes showed a P-value of <0.05. (Graphpad, La Jolla, CA).
In-vitro protein-binding assays
The purified proteins were incubated at 25°C for 10 min and then at 4°C for 1 hours in 100 μl of buffer containing 20mM HEPES pH 7.6, 5mM MgCl2, 150mM NaCl, and 0.5mM dithiothreitol (DTT), 0.1%Triton X-100, 10% Glycerol and complete protease inhibitor cocktail (Santacruz). The binding mixture was later diluted to 1 ml in the same buffer plus 1% Triton X-100 and 1mM EDTA, and was then subjected to immunoprecipitation at 4°C for 2 hours.
Immunoprecipitations
For immunoprecipitation of the endogenous protein complexes, 20-25×106 control and treated cells were lysed in a rotator at 4°C for 30 minutes in the lysis buffer (1.0% Triton X-100, 20 mM HEPES (pH 7.6), 150 mM NaCl, 1 mM EDTA and complete protease inhibitor cocktail). To precipitate the TNFR complex, cells were treated with GST-TNF (1μg/ml) for various time points. For time ‘0’, 100ng/ml GST-TNF was added to the untreated cell lysate. TNFR complex was precipitated through glutathione sepharose beads (GE healthcare) for 3-4 hours at 4°C. For the immunoprecipitation of transiently expressed proteins, 2–3×106 cells were lysed as above and precipitated using relevant antibodies. All cell lysates were precleared with protein A/G sepharose beads and preimmune IgG to reduce nonspecific binding. The immunoprecipitates were washed thrice with the lysis buffer and boiled with LDS sample buffer (Invitrogen).
Supplementary Material
Acknowledgments
We thank Dr. Stephane Richard, McGill University, Canada, for kindly providing the Sam68 knock out and wild type MEF cells. We thank Drs. David Shalloway for Sam68 plasmids, Chee-Kwee Ea for expression vectors for TNF and TNFR as well as advice, Shengli Hao for several qPCR primers and members of Baltimore laboratory and Dr. Reshmi Parameswaran for helpful discussions. P.R. is supported by the National Institutes of Health grant 2R01GM039458 to D.B.
Footnotes
Supplemental data can be found online. It includes six figures, supplemental experimental procedures, supplemental references and supplemental table containing oligonucleotide sequences.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez SE, Harikumar KB, Hait NC, Allegood J, Strub GM, Kim EY, Maceyka M, Jiang H, Luo C, Kordula T, et al. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 2010;465:1084–1088. doi: 10.1038/nature09128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen NJ, Chio II, Lin WJ, Duncan G, Chau H, Katz D, Huang HL, Pike KA, Hao Z, Su YW, et al. Beyond tumor necrosis factor receptor: TRADD signaling in toll-like receptors. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12429–12434. doi: 10.1073/pnas.0806585105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devin A, Cook A, Lin Y, Rodriguez Y, Kelliher M, Liu Z. The distinct roles of TRAF2 and RIP in IKK activation by TNF-R1: TRAF2 recruits IKK to TNF-R1 while RIP mediates IKK activation. Immunity. 2000;12:419–429. doi: 10.1016/s1074-7613(00)80194-6. [DOI] [PubMed] [Google Scholar]
- Devin A, Lin Y, Liu ZG. The role of the death-domain kinase RIP in tumour-necrosis-factor-induced activation of mitogen-activated protein kinases. EMBO reports. 2003;4:623–627. doi: 10.1038/sj.embor.embor854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont A, Hehner SP, Hofmann TG, Ueffing M, Droge W, Schmitz ML. Hydrogen peroxide-induced apoptosis is CD95-independent, requires the release of mitochondria-derived reactive oxygen species and the activation of NF-kappaB. Oncogene. 1999;18:747–757. doi: 10.1038/sj.onc.1202325. [DOI] [PubMed] [Google Scholar]
- Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Fusaki N, Iwamatsu A, Iwashima M, Fujisawa J. Interaction between Sam68 and Src family tyrosine kinases, Fyn and Lck, in T cell receptor signaling. The Journal of biological chemistry. 1997;272:6214–6219. doi: 10.1074/jbc.272.10.6214. [DOI] [PubMed] [Google Scholar]
- Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, Rieser E, Feltham R, Vince J, Warnken U, Wenger T, et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell. 2009;36:831–844. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- Huot ME, Brown CM, Lamarche-Vane N, Richard S. An adaptor role for cytoplasmic Sam68 in modulating Src activity during cell polarization. Mol Cell Biol. 2009;29:1933–1943. doi: 10.1128/MCB.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur GM, Lewis J, Yang Q, Lin Y, Nakano H, Nedospasov S, Liu ZG. The death domain kinase RIP has an essential role in DNA damage-induced NF-kappa B activation. Genes & development. 2003;17:873–882. doi: 10.1101/gad.1062403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Woronicz JD, Liu W, Goeddel DV. Prevention of constitutive TNF receptor 1 signaling by silencer of death domains. Science. 1999;283:543–546. doi: 10.1126/science.283.5401.543. [DOI] [PubMed] [Google Scholar]
- Karin M, Gallagher E. TNFR signaling: ubiquitin-conjugated TRAFfic signals control stop-and-go for MAPK signaling complexes. Immunological reviews. 2009;228:225–240. doi: 10.1111/j.1600-065X.2008.00755.x. [DOI] [PubMed] [Google Scholar]
- Kelliher MA, Grimm S, Ishida Y, Kuo F, Stanger BZ, Leder P. The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity. 1998;8:297–303. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- Lukong KE, Richard S. Sam68, the KH domain-containing superSTAR. Biochimica et biophysica acta. 2003;1653:73–86. doi: 10.1016/j.bbcan.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- Najib S, Martin-Romero C, Gonzalez-Yanes C, Sanchez-Margalet V. Role of Sam68 as an adaptor protein in signal transduction. Cell Mol Life Sci. 2005;62:36–43. doi: 10.1007/s00018-004-4309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan P, Wang W, Wallach D. Receptor-specific signaling for both the alternative and the canonical NF-kappaB activation pathways by NF-kappaB-inducing kinase. Immunity. 2004;21:477–489. doi: 10.1016/j.immuni.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Richard S, Torabi N, Franco GV, Tremblay GA, Chen T, Vogel G, Morel M, Cleroux P, Forget-Richard A, Komarova S, et al. Ablation of the Sam68 RNA binding protein protects mice from age-related bone loss. PLoS genetics. 2005;1:e74. doi: 10.1371/journal.pgen.0010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends in molecular medicine. 2006;12:440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Shikama Y, Yamada M, Miyashita T. Caspase-8 and caspase-10 activate NF-kappaB through RIP, NIK and IKKalpha kinases. European journal of immunology. 2003;33:1998–2006. doi: 10.1002/eji.200324013. [DOI] [PubMed] [Google Scholar]
- Shu HB, Takeuchi M, Goeddel DV. The tumor necrosis factor receptor 2 signal transducers TRAF2 and c-IAP1 are components of the tumor necrosis factor receptor 1 signaling complex. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13973–13978. doi: 10.1073/pnas.93.24.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser A, Jost PJ, Nagata S. The many roles of FAS receptor signaling in the immune system. Immunity. 2009;30:180–192. doi: 10.1016/j.immuni.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SJ, Shalloway D. An RNA-binding protein associated with Src through its SH2 and SH3 domains in mitosis. Nature. 1994;368:867–871. doi: 10.1038/368867a0. [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annual review of immunology. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- Varfolomeev E, Goncharov T, Fedorova AV, Dynek JN, Zobel K, Deshayes K, Fairbrother WJ, Vucic D. c-IAP1 and c-IAP2 are critical mediators of tumor necrosis factor alpha (TNFalpha)-induced NF-kappaB activation. The Journal of biological chemistry. 2008;283:24295–24299. doi: 10.1074/jbc.C800128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen L, Bekaert T, Chau TL, Tavernier J, Chariot A, Beyaert R. TLR-4, IL-1R and TNF-R signaling to NF-kappaB: variations on a common theme. Cell Mol Life Sci. 2008;65:2964–2978. doi: 10.1007/s00018-008-8064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajant H, Pfizenmaier K, Scheurich P. Tumor necrosis factor signaling. Cell death and differentiation. 2003;10:45–65. doi: 10.1038/sj.cdd.4401189. [DOI] [PubMed] [Google Scholar]
- Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- Zhang SQ, Kovalenko A, Cantarella G, Wallach D. Recruitment of the IKK signalosome to the p55 TNF receptor: RIP and A20 bind to NEMO (IKKgamma) upon receptor stimulation. Immunity. 2000;12:301–311. doi: 10.1016/s1074-7613(00)80183-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.