Abstract
Rationale
Recent studies suggest an important role of autophagy in protection against αB-crystallin-based (CryABR120G) desmin-related cardiomyopathies (DRC) but this has not been demonstrated in a different model of cardiac proteinopathy. Mechanisms underlying the response of cardiomyocytes to proteotoxic stress remain incompletely understood.
Objective
First, to determine whether and how the autophagic activity is changed in a mouse model of desminopathy; second, to investigate the role of p62 in the protein quality control of cardiomyocytes.
Methods and Results
Using an autophagosome reporter and determining changes in LC3-II protein levels in response to lysosomal inhibition, we found significantly increased autophagic flux in mouse hearts with transgenic overexpression of a DRC-linked mutant desmin. Similarly, autophagic flux was increased in cultured neonatal rat ventricular myocytes (NRVMs) expressing a mutant desmin. Suppression of autophagy by 3-methyladenine increased, whereas enhancement of autophagy by rapamycin reduced, the ability of a comparable level of mutant desmin overexpression to accumulate ubiquitinated proteins in NRVMs. Furthermore, p62 mRNA and protein expression was significantly upregulated in cardiomyocytes by transgenic overexpression of the mutant desmin or CryABR120G both in intact mice and in vitro. p62 depletion impaired aggresome and autophagosome formation, exacerbated cell injury, and decreased cell viability in cultured NRVMs expressing the misfolded proteins.
Conclusions
Autophagic flux is increased in desminopathic hearts and, as previously suggested in CryABR120G-based DRC, this increased autophagic flux serves as an adaptive response to overexpression of misfolded proteins. p62 is upregulated in mouse proteinopathic hearts. p62 promotes aggresome formation and autophagy activation and protects cardiomyocytes against proteotoxic stress.
Keywords: p62/SQSTM1, autophagy, aggresome, ubiquitin, desmin related cardiomyopathy
INTRODUCTION
Protein quality control (PQC) functions to support protein folding, segregate and remove terminally misfolded proteins, thereby preventing abnormal proteins from damaging the cell.1 The removal of abnormal proteins is primarily done by the ubiquitin-proteasome system (UPS), the major protein degradation pathway in the cell.1 Macroautophagy (commonly referred to as autophagy) engulfs a portion of cytoplasm, often including organelles, in a membrane-bound compartment for degradation by lysosomes.2 Through self-digestion of portions of cytoplasm, non-selective autophagy provides fuel for energy supply during starvation whereas the selective autophagy disposes of damaged organelles and perhaps aberrant protein aggregates for intracellular quality control.3, 4 The two degradation systems appear to collaborate with each other in PQC.5 It has been shown that increased expression of misfolded proteins in cardiomyocytes causes proteasome functional insufficiency,6, 7 aggresome formation,8 and perhaps autophagic activation.9 However, very little is known about the molecular underpinnings of these responses in cardiomyocytes during proteotoxic stress, a condition that is often seen in common forms of heart disease.8, 10
Proteinopathies are caused by increased expression of misfolded proteins and featured by the presence of aberrant protein aggregates in the affected cells. Desmin-related cardiomyopathy (DRC) is a family of cardiomyopathy caused by genetic mutations of desmin, αB-crystallin (CryAB), and other desmin partner proteins.11 The most prominent pathological feature of DRC is the presence of aberrant desmin-positive aggregates in muscle cells, which qualifies DRC as a bona fide cardiac proteinopathy. Several transgenic (tg) mouse models of DRC were created by cardiomyocyte-restricted tg overexpression of human DRC-linked mutant genes, including a 7 amino acid (R172~E178) deletion mutation of the desmin gene (D7-des) and a missense mutation (R120G) of CryAB (CryABR120G).8, 12, 13 These transgenic mice provide excellent intact animal models for studying cardiomyocyte responses to proteotoxic stress.6–8 Most, if not all, of the neurodegenerative disease belong to proteinopathies. Autophagy is up-regulated and appears to facilitate the removal of aberrant aggregates in neurodegenerative diseases.14, 15 Studies on autophagy in cardiac proteinopathy have begun to emerge. Hill and colleagues reported a robust increase of autophagosomes in CryABR120G mouse hearts.9 However, an increase in autophagosomes does not always represent increased autophagic activity. Based on ultrastructural studies, Robbins’ laboratory suspected that the autophagy activity was decreased in the heart of a slightly different CryABR120G tg mouse model.16, 17 Hence, the change of autophagic activities in mouse hearts with CryABR120G-based DRC remains controversial. Nevertheless, blunting autophagy via beclin1 haploinsufficiency was shown to accelerate disease progression in the CryABR120G-based DRC mice.9 However, it remains unclear whether autophagy enhancement would attenuate cardiac proteinopathy although an increase in autophagy appeared to associate with the Bcl2 overexpression induced delay in the premature-death of CryABR120G tg mice.16 Moreover, although mutations in either CryAB or desmin genes can cause DRC, the pathogenic pathways are not necessarily identical. Despite of sharing DRC characteristics in terms of the presence of desmin aggregates in cardiomyocytes, D7-des-based and CryABR120G-based DRC mice do display diverse phenotypes.12, 13 Therefore, in the first part of this study, we sought to investigate changes in the autophagic activity in the heart of a mouse model of cardiac desminopathy and the impact of altered autophagic activities on the removal of misfolded proteins in cardiomyocytes.
Recently, it has been reported that protein aggregates can trigger cardiomyocyte autophagy;10 however, the molecular events that mediate the activation of autophagy by aberrant protein aggregates in cardiomyocytes remain undefined. The finding of the late-onset neurodegeneration in p62 null mice suggests the involvement of p62 in PQC.18 Moreover, several p62 knockdown and knockout experiments further reveal that p62 is required for aggresome formation in metabolically stressed conditions.19, 20 Aggresome formation is an important cytoprotective process in proteinopathies. Notably, several lines of evidence from non-cardiac cells suggest that p62 may mediate the activation of autophagy by protein aggregates. p62 has also been identified as a common component in protein aggregates in neurodegenerative diseases.21 Meanwhile, p62 directly interacts with LC3 (microtubule-associated protein 1 light chain 3) and the interaction is required for recruiting autophagosomes.19 Hence, p62 has been proposed to be a mediator between the aberrant aggregates and autophagosomes in brains and livers.4 However, the understanding on this function of p62 remains incomplete. A direct interaction between p62 and LC3 has been established but p62 is not required for autophagosome formation in HeLa cells under basal condition or upon starvation-induced autophagic activation.22 Moreover, p62 depletion increased the induction of cell death by poly-glutamine;23 however, it was also reported that liver dysfunction caused by Atg7 knockout is alleviated by p62 deficiency.20 Hence, the role of p62 in PQC appears to be cell type-dependent. The change and pathophysiological significance of p62 has not been explored in cardiomyocytes. Therefore, we have examined the role of p62 in cardiomyocytes in response to the overexpression of DRC-linked misfolded proteins. Our data show that autophagic activity is increased in desminopathy and this increase helps remove ubiquitinated proteins. We have also found that p62 is upregulated in DRC mouse hearts and serves as a crucial mediator aggresome formation and autophagy activation by misfolded proteins in cardiomyocytes.
METHODS
(A full description of Methods can be found in online supplement.)
Transgenic mice
The protocol for the care and use of animals in this study was approved by University of South Dakota Institutional Animal Care and Use Committee. The GFP-LC3 transgenic (tg) mouse model was generously donated by Dr. N. Mizushima.24 FVB/N tg mice with cardiac-specific overexpression of D7-des or CryABR120G have been described.12, 13
Neonatal rat ventricular myocytes (NRVMs) culture and adenoviral gene delivery
NRVMs culture was performed as previously described.25 The generation of recombinant adenoviruses encoding a 469 amino acid, 53kDa mutated form of the mouse desmin gene with human DRC-associated A360P/N393I compound missense mutations (Ad-MT-des),26 the mouse wild type desmin gene (Ad-WT-des), a missense (R120G) mutant form of the mouse CryAB gene (Ad-CryABR120G),7 or the LacZ gene (Ad-β-gal) were previously described.27 Adenoviral infection of cultured NRVMs for gene delivery was done as previously described.28
Transmission electron microscopy (TEM),12 western blot analyses,29 and semi-quantitative reverse transcription (RT-)-PCR were performed as previously described.30
siRNA transfection
To knock down the target gene expression, 2×106 NRVMs were plated in each 60-mm dish. The Lipofectamine™-2000 transfection reagent (Invitrogen, Carlsbad, CA) was used for siRNA transfection following the manufacturer’s protocol. siRNA transfection was started at 48–72 hours after the cells were plated. Six hours after the transfection, the siRNA-containing medium was replaced with the regular medium. To achieve sustained knock-down a second round of siRNA transfection was required and performed 3 days after the first transfection.
LDH assay
LDH activity in the collected medium was measured using a cytotoxicity detection kit (Roche, Indianapolis, IN) following the manufacturer′s protocols. LDH release from NRVMs to culture media of an experimental group was evaluated as a percentage of the mean LDH activity of the corresponding control group.
MTT assay
The MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide) assay, an index of cell viability and cell growth, is based on the ability of viable cells to reduce MTT from a yellow water soluble dye to a dark blue insoluble formazan product.31 Briefly, 1×106 NRVMs were plated in the 6-well plate. After the experimental treatments were performed, MTT dye (Sigma, 500µg/ml) was added to each well of the plate and the plates were incubated at 37°C for 2 hours. At the end of the incubation, the dye solution was completely removed, 400µl solvent solution (1 volume of 1N HCl in 9 volume of anhydrous isopropanol) was added and the absorbance was determined at 570nm in a Tecan plate reader. Cell viability of the experimental group was determined as a percentage of the reading of the control group.
Statistical Analysis
All quantitative data were presented as mean ± S.D. Differences between groups were evaluated for significance using Student’s t-test for unpaired two group comparison or one-way or two-way ANOVA followed by the Scheffé's test when appropriate. The p-value <0.05 is considered statistically significant.
RESULTS
Autophagic flux is increased in desminopathic mouse hearts
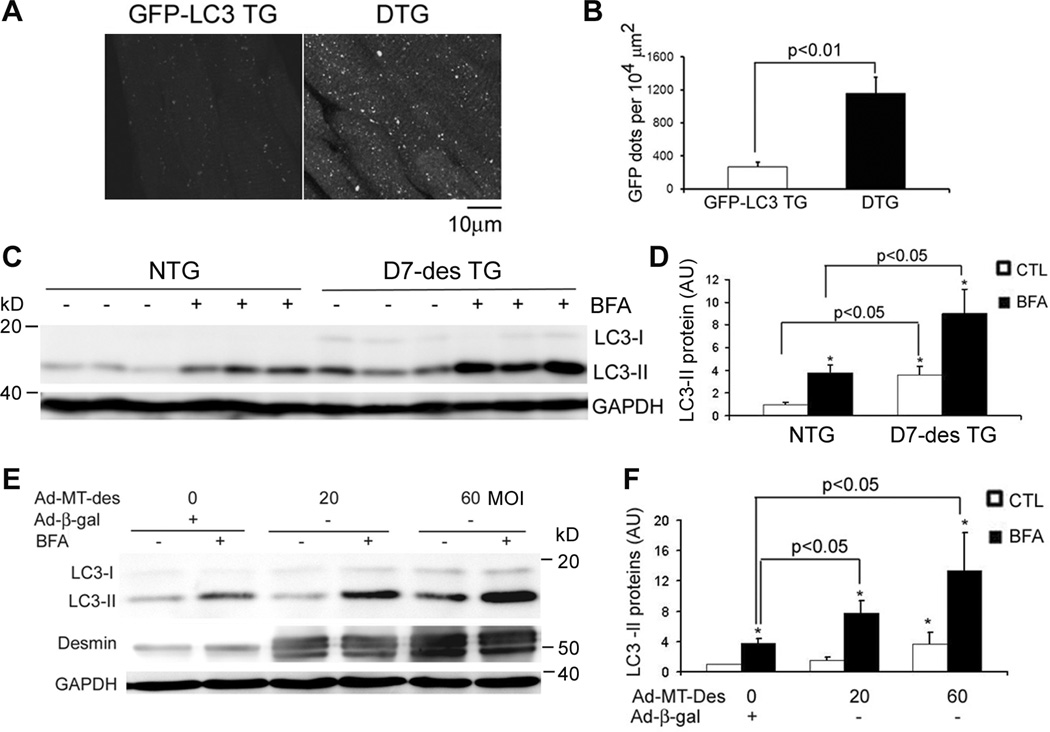
Alterations in the autophagic activity in desminopathic hearts have not been reported. Hence, we investigated autophagy activities in the heart of D7-des tg mice, a well characterized model of cardiac desminopathy. LC3 is a mammalian homologue of yeast autophagy-related gene (Atg) 8. Native LC3 (LC3-I) is diffusely distributed in the cytoplasm. Upon autophagic activation, LC3-I is lipidated to form LC3-II. LC3-II integrates to the autophagosomal membrane and thereby displaying a punctate distribution that marks the presence of autophagic vacuoles. Therefore, LC3-II is considered a marker of autophagosomes.32 Accordingly, GFP-LC3 tg mice have been successfully used in a number of studies to detect in vivo changes in autophagic vacuoles.24, 33, 34 We first monitored the abundance of autophagic vacuoles in D7-des hearts by cross-breeding heterozygous D7-des mice with GFP-LC3 mice. Fluorescence confocal microscopy of myocardial sections revealed that the number of GFP-LC3 puncta in cardiomyocytes was substantially greater in the hearts of GFP-LC3/D7-des double tg mice, compared with that of the littermate GFP-LC3 single tg mice (Figure 1A, B). Western blot analysis also showed that the endogenous LC3-II levels were markedly elevated in D7-des mouse hearts (Figure 1C, D). These findings indicate that autophagic vacuoles are increased in D7-des hearts.
Figure 1.
Expression of a DRC-linked mutant desmin increases autophagic flux in mouse hearts and NRVMs. A and B, Confocal microscopic analysis of GFP-LC3 distribution in ventricular myocardium. D7-des tg mice were cross-bred with GFP-LC3 mice. The resultant GFP-LC3::D7-des double tg (DTG) mice and their littermate GFP-LC3 single tg mice were analyzed at 2 months of age. The representative images (A) and the quantitative analysis of the number of GFP-LC3 puncta (B) are presented. The GFP dot data (B) were quantified from the 3 randomly selected fields per section, 3 representative sections per heart, and 3 hearts per group. C and D, Increases in autophagic flux in the D7-des tg mouse heart. D7-des tg and NTG littermate mice at 2 months were intravenously injected with one dose of bafilomycin A1 (BFA, 6µmol/kg) or vehicle control (CTL) at 3 hours before the hearts were harvested for western blot analyses of LC3. Representative images (C) and a summary of LC3-II densitometry data (D) are presented. *p<0.05 vs. CTL. E and F, Autophagic flux was increased in NRVMs expressing DRC-linked mutant desmin (MT-Des). Cultured NRVMs were infected with Ad-MT-Des or Ad-β-gal (as control). Six days after infection, the cells were treated with BFA (100nM) or DMSO for 3 hours before being harvested for western blot analysis for the indicated proteins. Representative images (E) and a summary of densitometry data (F) are presented. *p<0.05 vs. CTL; #p<0.05 vs. Ad-β-gal/CTL; N=3/group in all cases. AU = arbitrary units.
Increases in the prevalence of autophagic vacuoles can be caused by either an increase in autophagosome formation or a decrease in the lysosome-mediated removal. To identify the potential cause, the concept of autophagic flux, which is measured by comparing autophagosome prevalence in the presence and absence of lysosomal inhibition, has recently been introduced and considered a better indicator of autophagic activities.35, 36 We measured the autophagic flux in D7-des hearts. Further accumulation of LC3-II in D7-des mouse hearts upon lysosomal inhibition by BFA was observed (Figure 1C, D), indicating a significant increase in autophagic flux in D7-des mouse hearts. Consistent with an increased autophagic activity, the protein level and enzyme activity of cathepsin D, a bona fide lysosomal protease, were significantly increased in D7-des tg hearts (Supplementary Figure I). In the D7-des::GFP-LC3 double tg hearts, the majority of GFP-LC3 puncta stain positive for cathepsin D (Supplementary Figure II), identifying them as autolysosomes. The increase in autophagy activities in D7-des mice is restricted to the heart where D7-des was over-expressed as both LC3-II abundance and the autophagic flux in the liver were comparable between D7-des tg and Ntg littermate mice (Supplementary Figure III).
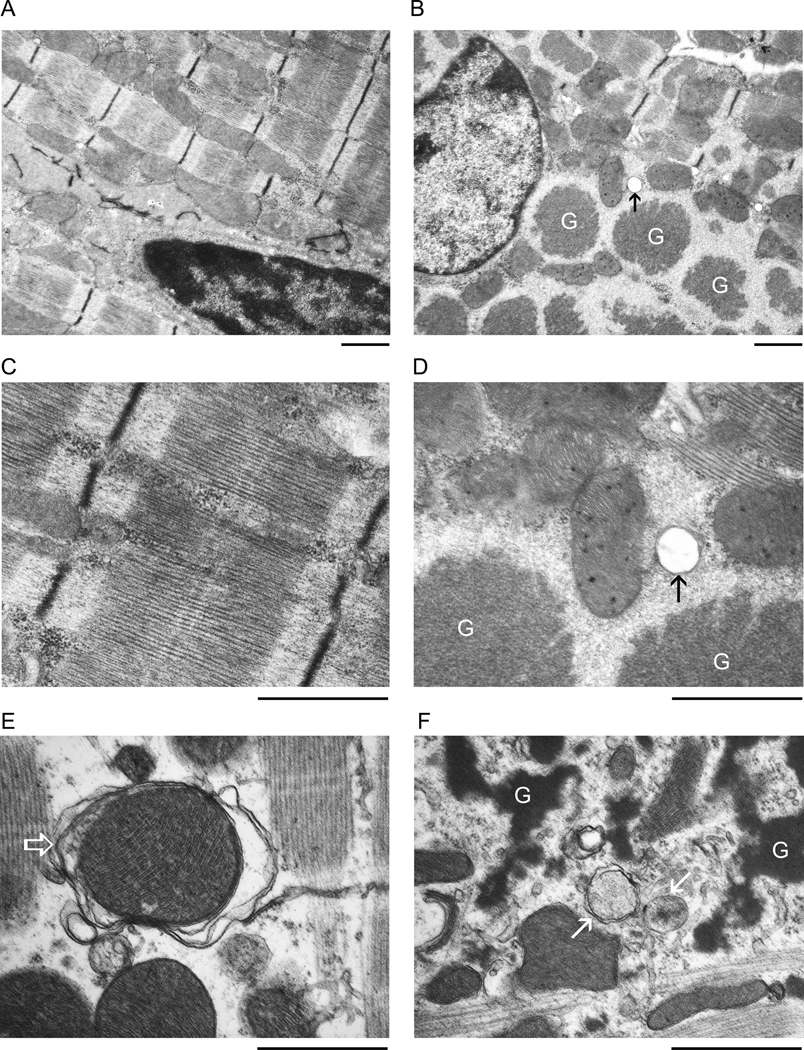
According to a recent report,37 LC3 may also be incorporated into protein aggregates independently of autophagy. To determine whether autophagic structures are increased in D7-des tg mouse hearts, we performed transmission electron microscopy (TEM). At the baseline, autophagic vacuoles were not observed in the cardiomyocytes of the Ntg hearts (Figure 2A, 2C) but were observed, albeit rare, in the cardiomyocytes of D7-des tg hearts (Figure 2B, 2D, arrow). After 2 hours of BFA treatment to inhibit the lysosome, the frequency with which autophagic structures were observed in the Ntg hearts was markedly increased (Figure 2E) but the frequency for autophagic structures of all stages (the early and the late stage) to be observed in the D7-des tg hearts was significantly greater (Figure 2F). The autophagosome observed in the Ntg heart often engulfs a mitochondrion (Figure 2E, open arrow) while the autophagic vacuoles (Figure 2F, arrows) in D7-des tg hearts are frequently found in the neighborhood of electron dense protein aggregates, are usually smaller, and usually do not contain mitochondria. Our TEM morphological data further confirm that autophagic flux is increased in D7-des tg mouse hearts.
Figure 2.
Representative electron micrographs. Perfusion-fixed ventricular tissues from 2-month-old D7-des Ntg (A, C, E) and tg (B, D, F) mice at the baseline (A~D) or 1 hour after the second BFA injection (E, F; 2 intraperitoneal injections with 1 hour in between, 3µmol/kg/injection) were used for electron microscopy. G: protein aggregates. Arrows or open arrow point to autophagosomes. Bar=1µm.
To further determine whether the autophagic activation is a direct consequence of expression of a misfolded desmin or a secondary response to protein overexpression, autophagic flux was also evaluated in cultured NRVMs expressing DRC-linked MT-des or WT-des. Cultured NRVMs were infected with Ad-MT-des, Ad-WT-des, or Ad-β-gal as a control. Six days after the infection, cells were treated with BFA or vehicle control for 3 hours. LC3-II abundance under basal condition as well as with lysosomal inhibition was increased in NRVMs expressing MT-des in a dose-dependent manner, compared to the controls (Figure 1E, F). However, at a level comparable to MT-des overexpression, WT-des overexpression did not increase autophagic flux in cultured NRVMs (Supplementary Figure IV), suggesting that the mutation/misfolding of the transgenic protein is responsible for the increased autophagic flux. Taken together, these data indicate that autophagy can be up-regulated in cardiomyocytes by the expression of mutant desmin in both intact mice and in vitro cultures.
Manipulating autophagic activities mitigates the accumulation of ubiquitinated proteins in MT-des expressing NRVMs
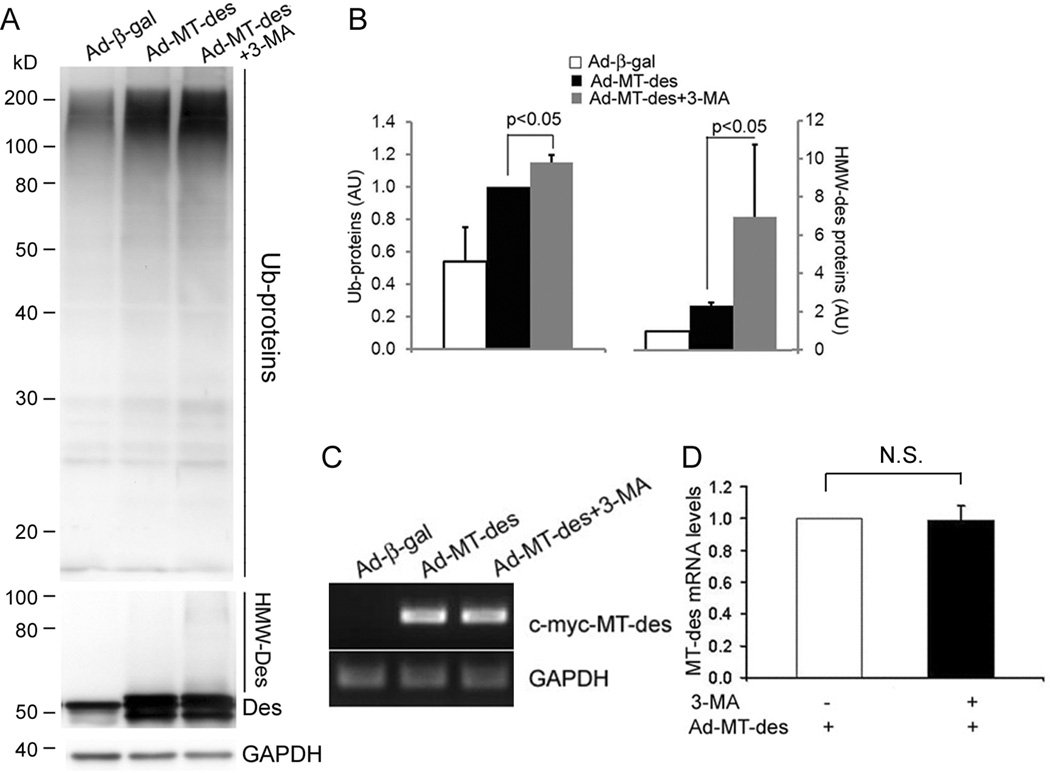
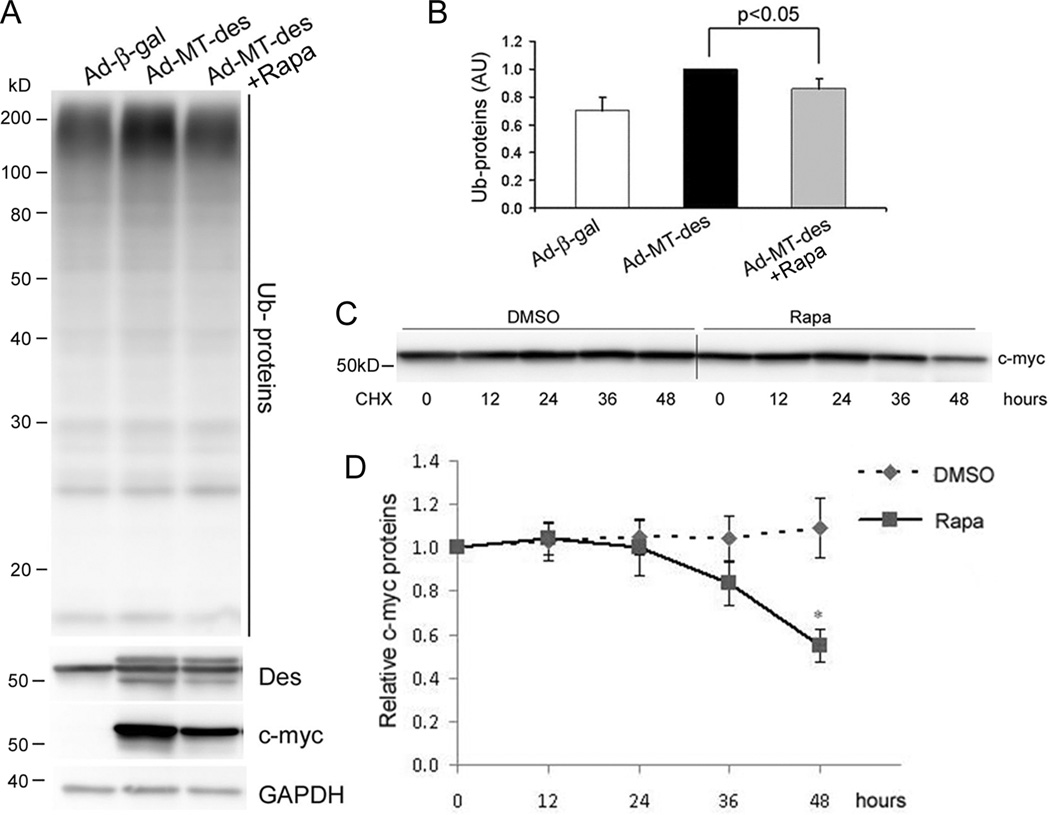
Accumulation of ubiquitinated proteins in the cell is a major feature of PQC inadequacy seen in proteinopathy.1 To determine the role of autophagy in the removal of misfolded proteins in cardiomyocytes, we tested the effects of reducing and enhancing autophagy on MT-des overexpression induced accumulation of ubiquitinated proteins in cultured NRVMs. Inhibition of autophagy was achieved by administration of 3-methyladenine (3-MA), a specific inhibitor of the class III phosphatidylinositol 3-kinase (PI3K) pathway, which is involved in the formation of autophagosomes and initiation of autophagy.35 Rapamycin, an mTOR inhibitor, was used to enhance autophagy. By monitoring changes in GFP-LC3 distribution and the abundance of endogenous LC3-II, the expected autophagy inhibition and enhancement effects respectively by 3-MA and rapamycin were confirmed in cultured NRVMs (Supplementary Figure V). Consistent with previous reports,7 the accumulation of the high molecular weight ubiquitinated proteins were observed in NRVMs expressing MT-des. Compared with vehicle control, the treatment of 3-MA significantly increased the accumulation of total ubiquitinated proteins (Figure 3A, B). The MT-des transcript levels were comparable between the 3-MA and the control treatment groups (Figure 3C, D) but the high molecular weight desmin species (HMW-Des) which are likely the ubiquitinated forms of desmin, were markedly increased by 3-MA (Figure 3A). By contrast, rapamycin treatment significantly decreased the accumulation of total ubiquitinated proteins (Figure 4A, B) caused by comparable MT-des expression at the mRNA level (Supplementary Figure VI). Moreover, cycloheximide chase experiments revealed that rapamycin treatment substantially shortened the half-life of the MT-des proteins expressed in NRVMs (Figure 4C, D).
Figure 3.
Inhibition of autophagy by 3-methyladenine (3-MA) promotes the ability of MT-des expression to accumulate ubiquitinated proteins (Ub-proteins). A and B, NRVMs were infected with Ad-MT-des. Starting one day after infection, cells were treated daily with 3-MA (1.5mM) or DMSO for 4 days. The level of Ub-proteins, desmin (Des), and GAPDH were measured using western blot analyses. Ub-proteins and the high molecular weight Des (HMW-Des) in MT-des expressing cells were markedly increased by 3-MA. All Ub-proteins in the lane as illustrated in the representative image (A) are included in the densitometry. In each cohort, the densitometric reading of the Ad-MT-des lane was defined as 1AU and used to normalize other lanes on the same image. A summary of quantitative data from 3 repeats is presented (B). C and D, Representative images (C) and a summary of densitometry data (D) of semi-quantitative RT-PCR analyses for the mRNA levels of transgenic MT-des. GAPDH was analyzed for loading control. N.S., not significant; t-test, n = 3.
Figure 5.
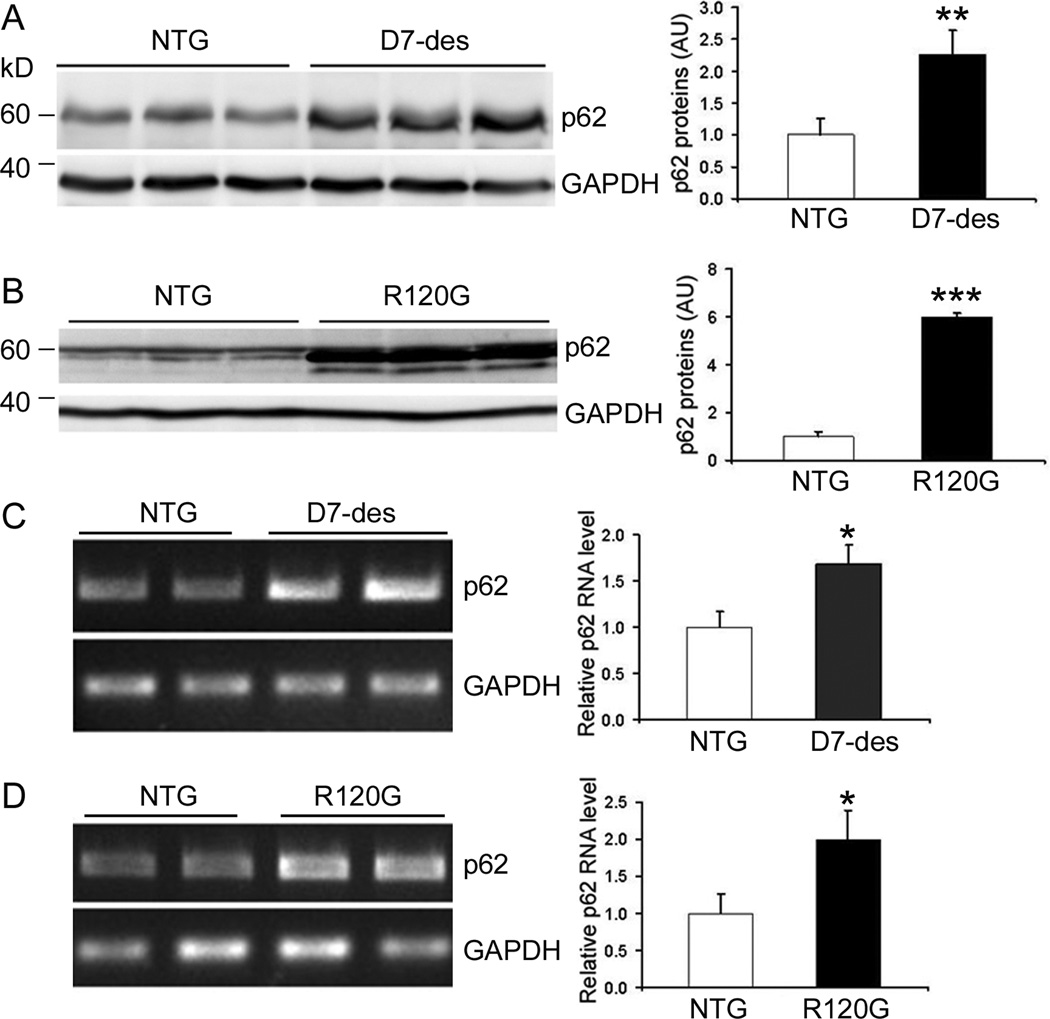
The levels of the steady state p62 protein and mRNA are increased in the hearts of mouse models of cardiac proteinopathy. Ventricular myocardium was collected from 2-month-old D7-des, 3-month-old CryABR120G (R120G), and their NTG littermates for extraction of total proteins (A and B) or total RNA (C and D). A and B, Western blot analysis for p62 protein. C and D, Semi-quantitative RT-PCR analyses of p62 mRNA levels in D7-des (C) and R120G (D) mouse hearts. The representative images are presented on the left and a summary of densitometric data are shown on the right of each panel. GAPDH was included for loading controls. * p<0.05, **p<0.01, ***p<0.001 vs. NTG, t-test, n=3~4 mice/group.
Figure 4.
Enhancement of autophagy by rapamycin decreases the ability of MT-des expression to accumulate ubiquitinated proteins (Ub-proteins). A and B, NRVMs were infected with Ad-MT-des one day before the initiation of the daily rapamycin (Rapa, 200nM) or DMSO treatment. After 4 daily treatments, the cells were harvested for western blot analyses of the indicated proteins. The representative image (A) and a summary of quantitative data of 3 repeats (B) are presented. The quantification of Ub-proteins was performed as in Figure 3B. C and D, Cycloheximide (CHX) chase assay for c-myc-tagged MT-des. CHX (10µM) was used to block protein synthesis. MT-des protein levels at the indicated time points after CHX treatment were measured using western blot analyses for the c-myc tag. A representative image is shown in C. The relative levels of myc-MT-des at post-CHX time points to that of the 0 hour time point were summarized in D. *p < 0.05 vs. the DMSO group, t-test, n = 4.
These data are consistent with the hypothesis that the activation of autophagy facilitates the removal of MT-des in cardiomyocytes and enhancing autophagy improves PQC in cardiomyocytes.
p62 is upregulated in proteinopathic mouse hearts
Based on the findings from studying non-cardiac cells,38 p62 was purported to play a critical role in mediating the activation of autophagy by ubiquitinated proteins but this has not been tested in cardiac proteinopathy. Therefore, we first examined p62 transcript and protein expression and protein distribution in the hearts of D7-des and CryABR120G tg mice, two well established mouse models of cardiac proteinopathy. Compared with their respective littermate Ntg controls, both CryABR120G and D7-des tg mouse hearts showed marked increases in both the transcript and the protein levels of p62 (Figure 5). Immunofluorescence staining revealed that the upregulation of p62 was limited in cardiomyocytes (Supplementary Figure VII). Furthermore, western blot analyses showed that p62 protein levels were also markedly increased in cultured NRVMs expressing CryABR120G or MT-des (Supplementary Figure VIII), suggesting that upregulation of p62 in proteinopathic hearts is specifically induced by expression of misfolded proteins rather than a secondary response to cardiac dysfunction.
We next examined the distribution of p62 in relation to autophagic vacuoles and ubiquitinated protein aggregates in D7-des tg hearts using immunofluorescence confocal microscopy. p62 formed small speckles in NTG mouse hearts; whereas in D7 mouse hearts, p62 formed large punctate structures with dimensions consistent with autophagosomes. To monitor autophagosomes in D7-des tg hearts, we cross-bred D7-des mice with GFP-LC3 mice. The co-localization among p62, ubiquitin, and GFP-LC3 was observed in D7-des mouse hearts (Supplementary Figure VII). These findings suggest a potential functional relationship between p62 and ubiquitin-positive aggregates as well as between p62 and autophagosomes in cardiomyocytes.
p62 depletion decreases LC3-II in cardiomyocytes
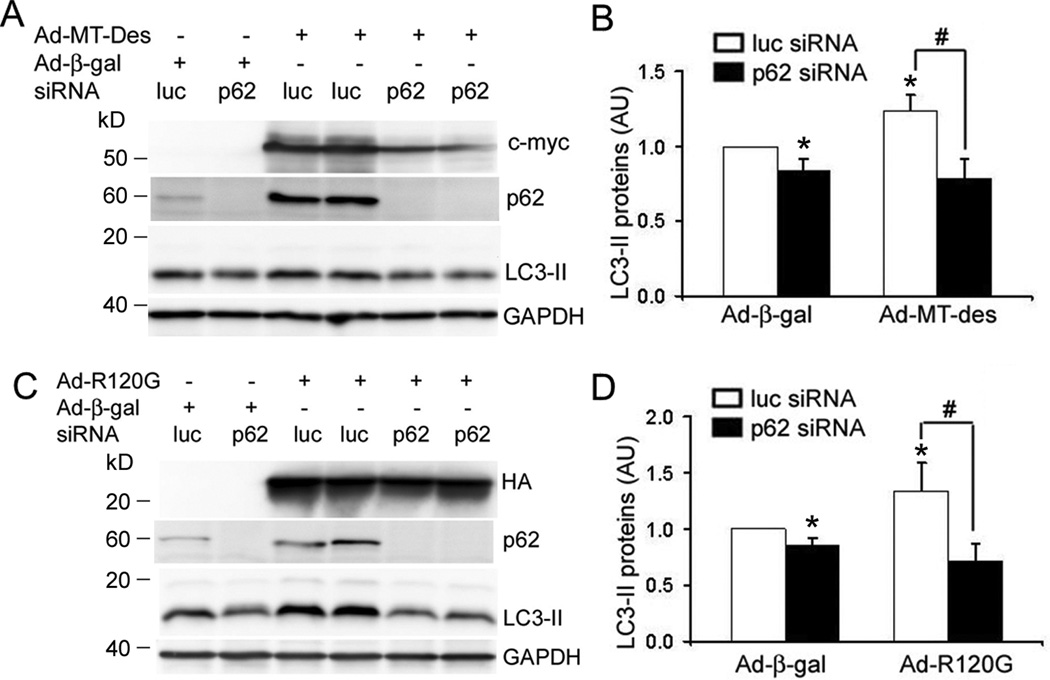
To test the functional role of p62 in the cell models of cardiac proteinopathy, we performed p62 knock down experiments in NRVMs to test the impact of p62 depletion on LC3 lipidation which renders LC3-I to LC3-II and represents a critical event in autophagosome formation.32 We achieved a very efficient p62 knockdown in cultured NRVMs via two consecutive transfections of p62 siRNA at a dose of 160pmol for 2×106 cells with an interval of 72 hours. Three days after the second p62 siRNA transfection, p62 proteins in the cells were reduced to a level undetectable by western blot analysis (Figure 6). The protein abundance of LC3 was assessed in NRVMs with p62 depletion. We found that p62 knockdown reduced LC3-II levels under basal condition. Expression of MT-des or CryABR120G significantly increased LC3-II abundance in NRVMs. This increase was blunted by p62 knockdown (Figure 6). These results suggest that p62 is essential to the formation of at least a significant portion of autophagosomes in cardiomyocytes both under basal condition and during overexpression of misfolded proteins.
Figure 6.
p62 Knockdown decreases LC3-II in cardiomyocytes. NRVMs were infected with Ad-HA-CryABR120G (Ad-R120G), Ad-Myc-MT-des, or Ad-β-gal as indicated on day 0. The cells were also transfected with either luciferase siRNA (Luc-siRNA, as control) or p62 siRNA using the Lipofectamine reagent on day1 and day 4. On day 7, the cells were harvested for western blot analyses of the indicated proteins. p62 Knockdown significantly reduced LC3-II levels in both the control and the MT-des or CryABR120G (R120G) expressing cells. The representative image (A and C) and a summary of quantitative data from 4 repeats (B and D) are presented. * p<0.05 vs. β-gal/Luc-siRNA; # p<0.05; two-way ANOVA followed by the Scheffé's test.
p62 is required for efficient formation of aggresomes in NRVMs expressing MT-des or CryABR120G
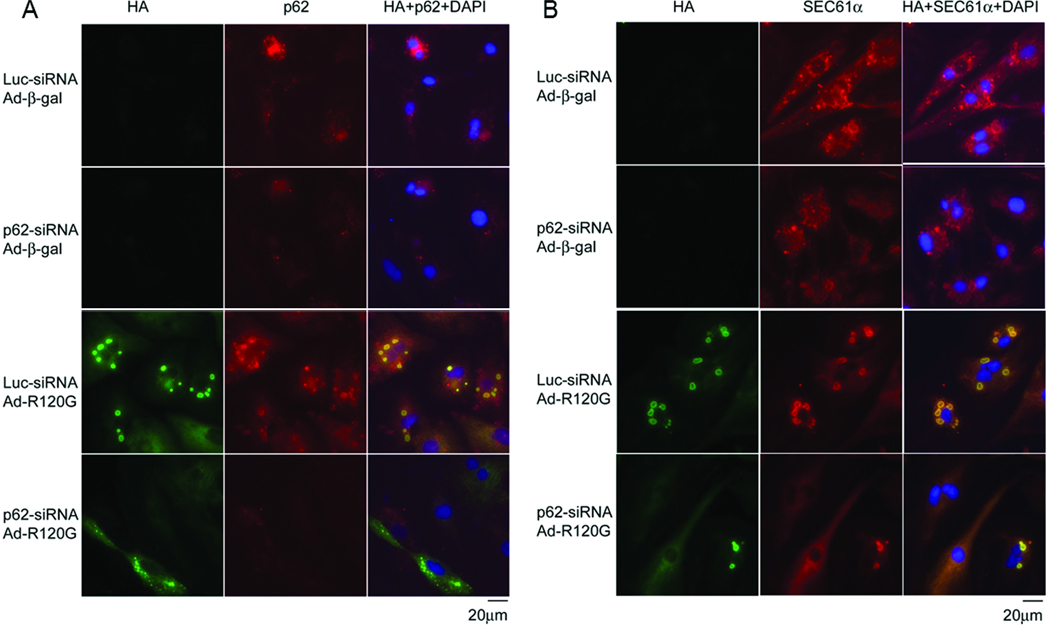
Aggresome formation is considered a protective response to increased expression of misfolded proteins in the cell.39 Transgenic expression of CryABR120G has been shown to cause aggresome formation in cardiomyocytes in vivo and in vitro.8, 40 Aggresomes always contain elevated levels of ubiquitin and SEC61α.39, 41 SEC61α is a normal constituent of endoplasmic reticulum (ER).41 To determine a potential obligatory role of p62 in the formation of aggresomes in cardiomyocytes, we assessed the prevalence of aggresomes resulting from overexpression of misfolded proteins in cultured NRVMs. Immunofluorescence images showed that p62 formed small speckles in the perinuclear region in the control cardiomyocytes. Consistent with previous reports,8 NRVMs expressing HA-CryABR120G and treated with control siRNA showed numerous large perinuclear HA-positive aggregates (Figure 7A). These aggregates showed elevated levels of ubiquitin (Supplementary Figure IX) and SEC61α (Figure 7B), indicative of aggresomes. Interestingly, both p62 and SEC61α were concentrated in these aggresomes. However, after p62 knockdown, Ad-CryABR120G infection at the same multiplicity of infection (MOI) induced substantially fewer and/or smaller aggresomes in NRVMs and aggresomes were formed in fewer CryABR120G cells (Figure 7 and Supplementary Figure IX). Similar results were also obtained from NRVMs expressing Myc-MT-des (Supplementary Figure X–XII). These data reveal the necessity of p62 in the formation of aggresomes in cardiomyocytes expressing misfolded proteins.
Figure 7.
p62 is required for efficient formation of aggresomes in NRVMs expressing CryABR120G. NRVMs were treated as described in Figure 6. On day 7 after Ad-HA-CryABR120G infection, cultured NRVMs were fixed in 2% paraformaldehyde and used for indirect double immunofluorescence labeling for the HA-tag of HA-CryABR120G (green) and for p62 (red, A) or SEC61α (red, B). Nuclei were stained blue with DAPI. HA-labeled aggregates, induced by HA-CryABR120G expression, co-localize with p62 (A) and SEC61α (B). To show the normal distribution of SEC61α, the images of the groups infected with Ad-β-gal were obtained by a longer exposure (1.5s) compared to the Ad-CryABR120G groups (400ms).
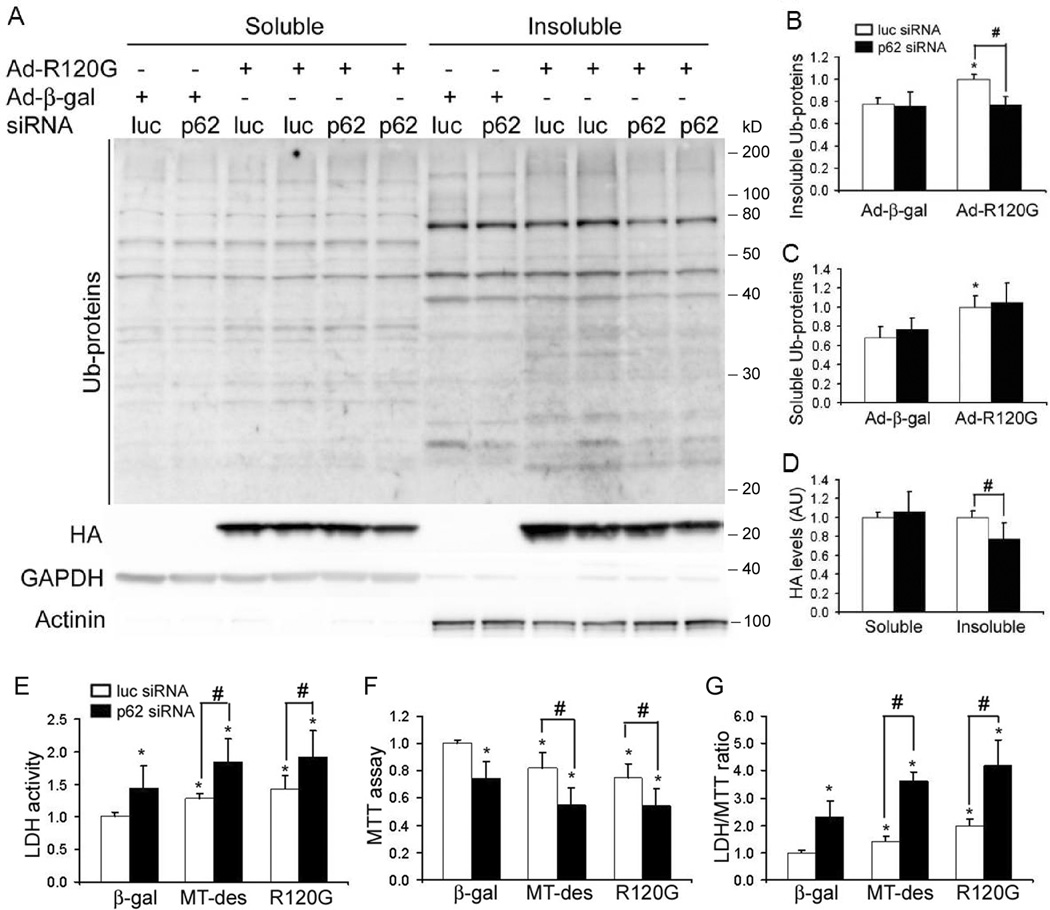
Given that aggresomes are insoluble, its chief components, including misfolded proteins and ubiquitin conjugates, are present in the insoluble fraction of the cell lysates. To further test the necessity of p62 in aggresome formation in a more quantitative manner, we assessed the effects of p62 depletion on the abundance of the overexpressed mutant proteins and the total ubiquitin conjugates in the Triton-X100 soluble and insoluble fractions of cardiomyocytes. Western blot analyses revealed that expression of CryABR120G (Figure 8) or MT-desmin (Supplementary Figure XIII) in NRVMs both accumulated ubiquitinated proteins in the soluble and insoluble fractions. Overexpression of CryABR120G (Figure 8A and 8B) or MT-des (Supplementary Figure XIIIA and XIIIB) induced accumulation of ubiquitinated proteins in the insoluble fraction was significantly attenuated by p62 knockdown. Moreover, HA- CryABR120G and Myc-Mt-des protein levels in the insoluble fraction were both reduced with p62 depletion (Figure 8A and 8D, Supplementary Figure XIIIA and XIIID). However, the mutant protein levels in the Triton-X100 soluble fraction responded differently with p62 depletion. With p62 depletion, soluble HA-CryABR120G proteins were not altered, whereas Myc-MT-des protein levels in Triton-X100 soluble fractions were decreased (Figure 8A and 8C, Supplementary Figure XIIIA and XIIIC). The difference might be due to the different functions and distributions of these two proteins in the cell.42 Taken together, these data show that p62 plays an important role in the recruitment of soluble species of misfolded proteins into the insoluble aggresomes and, therefore, was critical in aggresome formation.
Figure 8.
p62 depletion decreases the insoluble ubiquitinated (Ub−) proteins and the insoluble CryABR120G protein in NRVMs overexpressing CryABR120G (R120G) and exacerbates cell injury, and reduces cell survival in cultured NRVMs overexpressing CryABR120G or MT-des. NRVMs were treated as described in Figure 6. A ~ D, Cells were lysed with a lysis buffer containing 1% Triton X-100. Triton X-100 soluble and insoluble fractions of the cell lysates were used for western blot analysis for the abundance of ubiquitinated proteins or HA-tagged CryABR120G. GAPDH and α-actinin were probed as loading controls for proteins in the soluble and the insoluble fractions, respectively. Representative images (A) and a summary of quantitative data (B, C and D) are presented. E ~ G, The following assays were performed on day 7. E, LDH activities in the cultured medium were measured to assess cell injury. F, Cell viability was assessed by the MTT assay. Cell viability was deceased by expressing CryABR120G or MT-des, and was further reduced by p62 knockdown. G, Changes in the LDH/MTT ratio. * p<0.05, vs. β-gal/luc-siRNA; # p<0.05; n=4.
p62 deficiency sensitizes cell injury induced by expressing MT-des and CryABR120G
Our findings demonstrate that p62 regulates both aggresome formation and autophagy activation in cardiomyocytes. Given that both aggresome formation and autophagy activation may protect the cell from toxic proteins in cardiomyocytes,8, 9 we sought to determine whether p62 plays a protective role in cells expressing mutant proteins. Cell injury and cell viability were assessed respectively by LDH leakage and the MTT assay. Knocking down p62 significantly increased cytotoxicity under basal conditions as evidenced by a significant increase of LDH activities in the culture medium and decreased cell viability in the NRVM cultures transfected by p62-siRNA, compared with those transfected by the control siRNA (Luc-siRNA). LDH activities in cultured media were also elevated by expression of CryABR120G in NRVMs. Strikingly, after p62 depletion, the elevation of LDH activity induced by CryABR120G expression was more than doubled (from a 43% increase to a 92% increase; Figure 8E). In agreement with the LDH data, the MTT assays revealed that p62 knockdown reduced cell viability under basal conditions. CryABR120G expression caused a significant loss of cell viability. Notably, after p62 knockdown, the same extent of CryABR120G overexpression caused a twice greater loss of cell viability (from a 25% loss to a 46% loss; Figure 8F). Moreover, LDH activities in the medium were normalized by the number of viable cells of the same dish as measured by the MTT assay. The normalized LDH activities (i.e., LDH/MTT ratio) further confirmed that p62 knockdown enhanced cell injury and decreased cell viability in NRVMs (Figure 8G). Similar results were also obtained from in NRVMs expressing MT-des (Figure 8E~ 8G). The significantly decreased cell viability and increased cell injury caused by p62 deficiency indicate a protective role of p62 in the PQC of cardiomyocytes.
DISCUSSION
Two reported studies have investigated autophagy in cardiac proteinopathy and both used mouse models of CryABR120G–based DRC.9, 16 However, the two reports do not appear to agree with each other regarding whether autophagic activity is increased or not in the DRC mouse hearts. Moreover, DRC can be caused by expression of other misfolded proteins such as mutations in the desmin gene;13 but no study has been reported on autophagy in the heart with DRC caused by a mutant gene other than CryABR120G. More importantly, the mechanism underlying autophagic activation in cardiac proteinopathy is unknown. In attempt to address some of these unanswered questions, we have found in the present study that (1) autophagic flux is increased in the heart of a mouse model of desminopathy, (2) lysosomal function is also up-regulated in desminopathic mouse hearts, (3) autophagy helps to clear ubiquitinated proteins in cells expressing human DRC-linked misfolded proteins, and (4) p62 plays a crucial role in the formation of aggresomes as well as the activation of autophagy in cardiomyocytes expressing misfolded proteins. These new findings provide strong evidence that autophagic flux is increased in desminopathic hearts and establish an important role of p62 in the PQC in cardiomyocytes. Our study suggests that p62 can potentially be a therapeutic target for improving PQC in the heart.
1. Autophagy is upregulated in cardiac proteinopathies
A recent study on cultured SH-SY5Y cells, a cell line derived from human neuroblastoma, suggests that activation of autophagy is not a universal response to all misfolded proteins. It shows that aggresomes formed by ectopic expression of a DRC-linked mutant desmin do not trigger autophagy and are not cleared by autophagy.43 In the present study, however, we have collected multiple lines of compelling evidence supporting the notion that autophagy is upregulated in mutant desmin-based proteinopathy, both in cultured cardiomyocytes and in the heart of intact mice. Both endogenous LC3 and overexpressed GFP-LC3, commonly used autophagosome markers, are markedly increased in D7-des mouse hearts. Cathepsin D expression levels and enzyme activities are upregulated in D7-des mouse hearts (Supplementary Figure S1). The superimposing immunolabeling of cathepsin D and GFP-LC3 puncta indicates the formation of autolysosome (Supplementary Figure II). Furthermore, the additional increases in both LC3-II protein levels and TEM detected autophagic vacuoles in the heart after lysosomal inhibition demonstrate that autophagic flux is significantly increased in D7-des mouse hearts. These data indicate that the increase in autophagosomes in D7-des heart is not due to diminished lysosomal function but is rather caused by increased autophagosome formation. The increased autophagic flux is unlikely a secondary response to heart malfunction because our results also show an increased autophagic flux in cultured cardiomyocytes expressing MT-des.
Using TEM, Maloyan et al failed to detect any increase in autophagic vacuoles in the heart of CryABR120G tg mice under basal condition.16 Indeed, we also found in our TEM study that autophagic vacuoles can be found but are very rare in the D7-des mouse hearts at baseline. However, the situation is dramatically changed by a short-term lysosomal inhibition. At 2 hours after intraperitoneal injection of BFA, autophagic vacuoles become detectable in the Ntg control mouse hearts and are markedly more prevalent in D7-des tg mouse hearts (Figure 2). These data suggest that the entire process from the formation of an autophagosome to the complete removal of the autophagosome by lysosomes is extremely fast in cardiomyocytes both under the normal condition and during desminopathy. Notably, there is an apparent discrepancy between the 4-fold increase of LC3-II proteins and the very modest increase in autophagic vacuoles detected in the hearts of D7-des mice under basal condition. This discrepancy suggests that a portion of the upregulated LC3-II proteins in D7-des cardiomyocytes is not associated with typical autophagosomes, consistent with a recent report.37 Nevertheless, paralleling increases in both LC3-II protein levels and autophagic vacuoles were observed both in D7-des tg and Ntg hearts after lysosomal inhibition. Our TEM study observed all stages of autophagic vacuoles in the D7-des hearts. These vacuoles tend to be smaller than those observed in the control hearts. As previously reported,13 the large and electron-dense protein aggregates in D7-des tg cardiomyocytes are always surrounded by and continued with amorphous less-dense materials. The latter appear to be an intermediate species of small protein aggregates that are at a stage either before merging with the aggresomes or immediately after breaking off the dense aggresomes. Another interesting and perhaps important TEM feature of the autophagic vacuoles detected in the cardiomyocytes of D7-des tg mice is that they often appear in the vicinity of the electron-dense aggresomes but show no intent to engulf the large and electron-dense aggregates (or aggresomes). Rather, the content engulfed in the early stage autophagosome appears to be those intermediate species of amorphous aggregates (Figure 2F). This observation suggests that autophagy removes the misfolded proteins in the vicinity of aggresomes rather than directly remove the aggresome per se. The aggresome constituent may need to be broken down to smaller and less-dense materials before being engulfed and removed by autophagosomes. It is also possible that the loose and small protein aggregates have the opportunity to be removed by autophagy before merging with the aggresome.
The soluble misfolded proteins are mainly degraded by the proteasome, whereas the aggregated forms resist proteasomal degradation.44 This resistance to UPS degradation is probably caused by the difficulty of aggregated proteins to enter the narrow opening of the 20S proteasome barrel.7, 44 Moreover, aberrant protein aggregation, in turn, overwhelms and impairs proteasome function,7, 26 potentially disrupting other important cellular processes.45 In contrast, autophagy has a more remarkable capability of degrading aggregated proteins.46 We have previously reported that cardiac proteasome function is insufficient in both D7-des-based and CryABR120G-based DRC mice.6, 7 Therefore, the upregulated autophagic flux observed in cardiac proteinopathy suggests a likely collaborative relationship between the UPS and autophagy in handling misfolded proteins in the heart.
2. Autophagy removes ubiquitinated proteins in cardiomyocytes expressing MT-des
Detrimental and protective roles of an increased autophagic activity in cardiac dysfunction have both been reported.33, 34 The role of autophagy in a disease may depend on disease stages and the nature of the stress that causes the disease.
Here we have tested, and obtained results consistent with, the hypothesis that autophagy helps remove the DRC-linked MT-des proteins in cardiomyocytes, thereby protects against desminopathy. Terminally misfolded proteins in the cell, when escaped from the surveillance from chaperones and the UPS, trigger aberrant protein aggregation which can damage the cell by a number of mechanisms including impairing the proteasome.1, 26 Accumulation of ubiquitinated proteins in the form of protein aggregates is a common feature of cells under proteotoxic stress. Under comparable levels of MT-des overexpression, suppression of autophagy by administration of 3-MA significantly reduces, whereas enhancement of autophagy by rapamycin accelerates, the ability of the cultured cardiomyocytes to eliminate ubiquitinated proteins and MT-des proteins. Activation of autophagy by rapamycin significantly decreased the half-life of MT-des whereas suppression of autophagy increased the HMW species of MT-des expressed in cultured cardiomyocytes (Figures 3, 4). These data suggest that autophagy functions to remove ubiquitinated proteins and the MT-des proteins in cardiomyocytes overexpressing MT-des. Consistent with our findings, Tannous et al demonstrated a protective effect of autophagy on the pathogenesis of CryABR120G in mouse hearts by a loss-of-function approach.9 Hence, in DRC caused by either a mutant CryAB or a mutant desmin, it appears that autophagy is activated and plays an essential role in maintaining protein homeostasis in the cardiomyocytes.
3. p62 is upregulated in cardiac proteinopathy and facilitates aggresome formation and autophagy activation
Recently, p62 is purported to act as an adaptor molecule linking ubiquitinated proteins to the autophagic machinery.23 It was shown in Hela cells that p62 directly interacts with LC3 and is degraded by autophagy.19 Here we report that both the transcript and protein levels of p62 are significantly upregulated in the heart of two mouse models of cardiac proteinopathy. The upregulation was recapitulated in cultured NRVMs. p62 has also been identified as a common component in the inclusion bodies seen in a number of neurodegenerative diseases.21 Our study has extended this prospect to cardiac proteinopathy. We found that p62 is enriched in the ubiquitin-positive aggregates in D7-des mouse hearts and co-localizes with autophagosome marker GFP-LC3, prompting us to hypothesize that p62 provides a key link between protein aggregation and selective autophagy in cardiomyocytes. To the best of our knowledge, this is the first study on the functional role of p62 in cardiac disease.
Under normal conditions, p62 distributes as cytosolic speckles but under conditions of metabolic stress, p62 redistributes into aggregates.19, 20, 47 Here we found that p62 co-localizes with the mutant proteins in the large aggregates in cultured cardiomyocytes expressing CryABR120G or MT-des. The aggregates also contain ubiquitin and SEC61α and most of them are perinuclear, identifying them as aggresomes.8 Notably, consistent with what we showed previously in mouse hearts,6, 12, 13 here we observed that overexpression of CryABR20G or MT-desmin in cultured NRVMs also led to formation of large protein aggregates that are not perinuclear. By definition, these non-perinuclear aggregates may not be considered as aggresomes. Importantly, our study shows that p62 depletion impairs the formation of both aggresomal and non-aggresomal large aggregates from the overexpressed misfolded proteins in cardiomyocytes. Accordingly, the insoluble fractions of ubiquitin conjugates are reduced by p62 depletion. This is in agreement with the previous observations in non-cardiac cells and tissues. Depletion of p62 diminishes the formation of ubiquitin-positive inclusions following puromycin treatment in Hela cells or in Atg7-deficient mouse livers and brains.19, 20 These lines of evidence suggest an important role of p62 in the cells under proteotoxic stress through promoting the formation of aggresomes. It is generally believed that aggresomes restrict the intracellular distribution of misfolded proteins. Aggresome formation sequesters toxic soluble oligomers into the less harmful insoluble aggregates; thereby protecting the cell.46 Given the similarity of their composition to aggresomes, non-perinuclear large aggregates, which also help sequester the misfolded proteins, may conceivably not only make misfolded proteins less toxic but also more accessible to selective autophagy. This is because autophagy machinery is not limited to the perinuclear location.
An important way by which aggresome formation protects the cell is to facilitate the delivery of dispersed small protein aggregates to the autophagic machinery for bulk degradation.41 p62 is a proven substrate of, and constitutively degraded by, selective autophagy.19, 23 Therefore, it is likely that by interacting with LC3-II, p62 incorporation into aggresomes recruits the phagophore and thereby triggers autophagic degradation of the aggresome. Supporting this hypothesis, our study shows that p62 knockdown significantly decreased LC3-II formation in cardiomyocytes both under basal condition and during overexpression of misfolded proteins. p62 is not required for the non-selective autophagy because starvation induced autophagy is not affected in p62 deficient mice.20 Hence, our study shows an important functional role of p62 in the selective autophagy in cardiomyocytes.
As a further demonstration of a protective role for p62-mediated aggresome formation and autophagic activation in cardiomyocytes in response to increased proteotoxic stress, we have observed that p62 depletion increases cell injury and decreases cell viability in NRVMs under basal condition and during overexpression of misfolded proteins, as evidenced by the increased leak of LDH to the culture media and the decreased number of viable cells (Figure 8).
Collectively, our study shows that p62 is important for aggresome formation as well as the activation of selective autophagy in cardiomyocytes in response to proteotoxic stress. Based on our data and previous reports, the following model is proposed for the role of p62 in the response of cardiomyocytes to the increase in misfolded proteins in the cytosol: (1) ubiquitinated misfolded proteins escape from proteasome-mediated degradation and form the relative small and soluble oligomeric aggregates which are active and toxic to the cell or alternatively, the misfolded proteins escaped from the UPS surveillance forms the oligomers first and are then ubiquitinated; (2) the toxic oligomers are packed into the less harmful aggresomes in a p62-dependent manner; and (3) aggresome-associated p62 recruits autophagic machinery to the aggresome and triggers the selective autophagy to remove the aggregated misfolded proteins.
Novelty and Significance.
What Is Known?
Increased autophagosomes were observed in CryABR120G-based desmin-related cardiomyopathy (DRC) mouse hearts but autophagic flux has not been determined in a model of cardiac proteinopathy.
p62 is purported to mediate the formation of inclusion bodies and activate selective autophagy but this remains to be demonstrated in cardiomyocytes.
The role of p62 in protein quality control in the cell appears to be cell type dependent.
What New Information Does This Article Contribute?
The first report of an increased autophagic flux in the heart of a mouse model of proteinopathy and the first evidence that enhancing autophagy facilitates the removal of misfolded proteins in cardiomyocytes.
p62 expression is upregulated in the heart of DRC mouse models, representing the first study of p62 in the heart.
The first demonstration that p62 is required for aggresome formation and activation of selective autophagy in cardiomyocytes under the proteotoxic stress and protects cardiomyocytes from proteotoxic stress.
Understanding the mechanism by which cardiomyocytes handle increased expression of misfolded proteins, an inevitable consequence and cause of increased cardiac stress, is essential to delineating the pathogenesis of a large subset of heart disease. This is because aberrant aggregation and accumulation of misfolded proteins in cardiomyocytes have been observed in failing human hearts resulting from common forms of heart disease. However, the molecular mechanisms of protein quality control, in cardiomyocytes in particular, remain incompletely delineated. Using intact mice and/or cultured cardiomyocytes, this study reveals for the first time that (1) autophagic flux is adaptively increased in a mouse model of cardiac proteinopathy; (2) p62 is transcriptionally upregulated in cardiomyocytes and the heart by overexpression of human DRC-linked misfolded proteins; and (3) p62 appears to play a pivotal and protective role in cardiomyocytes in mediating two major defending mechanisms against increased misfolded proteins: aggresome formation and the activation of selective autophagy. These mechanistic findings suggest that autophagy and p62 can potentially be key targets for developing therapeutic strategies to improve cardiac protein quality control.
Supplementary Material
Acknowledgements
We thank the Imaging Core of the Division of Basic Biomedical Sciences for assistance in confocal microscopy analyses, Ms. Andrea Jahn for outstanding technical assistance in maintaining mouse colonies and genotype determination, and Mr. Suleman Said for technical support on TEM sample preparation.
Sources of Founding
This work was in part supported by NIH grants R01HL085629 and R01HL072166 and American Heart Association grants 0740025N (to X. W.), 0625738Z (to H.S.), 0815571G (to Q. Z.), and 11PRE5730009 (to M.J.R.). The Imaging Core is supported by a NIH grant (5P20RR015567).
Non-standard Abbreviations and Acronyms
- 3-MA
3-methyladenine
- BFA
bafilomycin A1
- β-gal
β-galactosidase
- CHX
cycloheximide
- CryAB
αB-crystallin
- des
desmin
- DRC
desmin-related cardiomyopathy
- GFP
green fluorescent protein
- LC3
microtubule-associated protein 1 light chain 3
- LDH
lactate dehydrogenase
- MT
mutant
- NRVMs
neonatal rat ventricular myocytes
- Ntg
non-transgenic
- PI3K
phosphatidylinositol 3-kinase
- PQC
protein quality control
- RT-PCR
reverse transcription (RT)-PCR
- tg
transgenic
- UPS
Ubiquitin-proteasome system
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Subject codes:
[145] Genetically altered mice
[137] Cell biology/structural biology
[107] Biochemistry and metabolism
Disclosure: None.
References
- 1.Wang X, Robbins J. Heart failure and protein quality control. Circ Res. 2006;99:1315–1328. doi: 10.1161/01.RES.0000252342.61447.a2. [DOI] [PubMed] [Google Scholar]
- 2.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, Pandey UB, Kaushik S, Tresse E, Lu J, Taylor JP, Cuervo AM, Yao TP. Hdac6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 5.Ding WX, Yin XM. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4:141–150. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Chen Q, Huang W, Horak KM, Zheng H, Mestril R, Wang X. Impairment of the ubiquitin-proteasome system in desminopathy mouse hearts. Faseb J. 2006;20:362–364. doi: 10.1096/fj.05-4869fje. [DOI] [PubMed] [Google Scholar]
- 7.Chen Q, Liu JB, Horak KM, Zheng H, Kumarapeli AR, Li J, Li F, Gerdes AM, Wawrousek EF, Wang X. Intrasarcoplasmic amyloidosis impairs proteolytic function of proteasomes in cardiomyocytes by compromising substrate uptake. Circ Res. 2005;97:1018–1026. doi: 10.1161/01.RES.0000189262.92896.0b. [DOI] [PubMed] [Google Scholar]
- 8.Sanbe A, Osinska H, Saffitz JE, Glabe CG, Kayed R, Maloyan A, Robbins J. Desmin-related cardiomyopathy in transgenic mice: A cardiac amyloidosis. Proc Natl Acad Sci U S A. 2004;101:10132–10136. doi: 10.1073/pnas.0401900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tannous P, Zhu H, Johnstone JL, Shelton JM, Rajasekaran NS, Benjamin IJ, Nguyen L, Gerard RD, Levine B, Rothermel BA, Hill JA. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2008;105:9745–9750. doi: 10.1073/pnas.0706802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tannous P, Zhu H, Nemchenko A, Berry JM, Johnstone JL, Shelton JM, Miller FJ, Jr, Rothermel BA, Hill JA. Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy. Circulation. 2008;117:3070–3078. doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldfarb LG, Dalakas MC. Tragedy in a heartbeat: Malfunctioning desmin causes skeletal and cardiac muscle disease. J Clin Invest. 2009;119:1806–1813. doi: 10.1172/JCI38027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Osinska H, Klevitsky R, Gerdes AM, Nieman M, Lorenz J, Hewett T, Robbins J. Expression of r120g-alphab-crystallin causes aberrant desmin and alphab-crystallin aggregation and cardiomyopathy in mice. Circ Res. 2001;89:84–91. doi: 10.1161/hh1301.092688. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Osinska H, Dorn GW, 2nd, Nieman M, Lorenz JN, Gerdes AM, Witt S, Kimball T, Gulick J, Robbins J. Mouse model of desmin-related cardiomyopathy. Circulation. 2001;103:2402–2407. doi: 10.1161/01.cir.103.19.2402. [DOI] [PubMed] [Google Scholar]
- 14.Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM. Extensive involvement of autophagy in alzheimer disease: An immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 15.Anglade P, Vyas S, Javoy-Agid F, Herrero MT, Michel PP, Marquez J, Mouatt-Prigent A, Ruberg M, Hirsch EC, Agid Y. Apoptosis and autophagy in nigral neurons of patients with parkinson's disease. Histol Histopathol. 1997;12:25–31. [PubMed] [Google Scholar]
- 16.Maloyan A, Sayegh J, Osinska H, Chua BH, Robbins J. Manipulation of death pathways in desmin-related cardiomyopathy. Circ Res. 2010;106:1524–1532. doi: 10.1161/CIRCRESAHA.109.212639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maloyan A, Robbins J. Autophagy in desmin-related cardiomyopathy: Thoughts at the halfway point. Autophagy. 2010;6 doi: 10.1161/CIRCRESAHA.109.212639. [DOI] [PubMed] [Google Scholar]
- 18.Ramesh Babu J, Lamar Seibenhener M, Peng J, Strom AL, Kemppainen R, Cox N, Zhu H, Wooten MC, Diaz-Meco MT, Moscat J, Wooten MW. Genetic inactivation of p62 leads to accumulation of hyperphosphorylated tau and neurodegeneration. J Neurochem. 2008;106:107–120. doi: 10.1111/j.1471-4159.2008.05340.x. [DOI] [PubMed] [Google Scholar]
- 19.Pankiv S, Hoyvarde Clausen T, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. P62/sqstm1 binds directly to atg8/lc3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;19:19. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 20.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata JI, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura SI, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 21.Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, Kleinert R, Prinz M, Aguzzi A, Denk H. P62 is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol. 2002;160:255–263. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. P62/sqstm1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang M, Li J, Huang W, Su H, Liang Q, Tian Z, Horak KM, Molkentin JD, Wang X. Proteasome functional insufficiency activates the calcineurin-nfat pathway in cardiomyocytes and promotes maladaptive remodelling of stressed mouse hearts. Cardiovasc Res. 2010;88:424–433. doi: 10.1093/cvr/cvq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Tang M, Mestril R, Wang X. Aberrant protein aggregation is essential for a mutant desmin to impair the proteolytic function of the ubiquitin-proteasome system in cardiomyocytes. J Mol Cell Cardiol. 2006;40:451–454. doi: 10.1016/j.yjmcc.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 27.Kumarapeli AR, Su H, Huang W, Tang M, Zheng H, Horak KM, Li M, Wang X. Alpha b-crystallin suppresses pressure overload cardiac hypertrophy. Circ Res. 2008;103:1473–1482. doi: 10.1161/CIRCRESAHA.108.180117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong X, Liu J, Zheng H, Glasford JW, Huang W, Chen QH, Harden NR, Li F, Gerdes AM, Wang X. In situ dynamically monitoring the proteolytic function of the ubiquitin-proteasome system in cultured cardiac myocytes. Am J Physiol Heart Circ Physiol. 2004;287:H1417–H1425. doi: 10.1152/ajpheart.01233.2003. [DOI] [PubMed] [Google Scholar]
- 29.Su H, Li J, Menon S, Liu J, Kumarapeli AR, Wei N, Wang X. Perturbation of cullin deneddylation via conditional csn8 ablation impairs the ubiquitin-proteasome system and causes cardiomyocyte necrosis and dilated cardiomyopathy in mice. Circ Res. 2011;108:40–50. doi: 10.1161/CIRCRESAHA.110.230607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei D, Li F, Su H, Tian Z, Ye B, Wei N, Wang X. Cop9 signalosome subunit 8 is required for postnatal hepatocyte survival and effective proliferation. Cell Death Differ. 2011;18:259–270. doi: 10.1038/cdd.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong JY, Rabkin SW. Palmitate-induced apoptosis in cardiomyocytes is mediated through alterations in mitochondria: Prevention by cyclosporin a. Biochim Biophys Acta. 2000;1485:45–55. doi: 10.1016/s1388-1981(00)00028-7. [DOI] [PubMed] [Google Scholar]
- 32.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. Lc3, a mammalian homologue of yeast apg8p, is localized in autophagosome membranes after processing. Embo J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: Roles of amp-activated protein kinase and beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 34.Zhu H, Tannous P, Johnstone JL, Kong Y, Shelton JM, Richardson JA, Le V, Levine B, Rothermel BA, Hill JA. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perry CN, Kyoi S, Hariharan N, Takagi H, Sadoshima J, Gottlieb RA. Novel methods for measuring cardiac autophagy in vivo. Methods Enzymol. 2009;453:325–342. doi: 10.1016/S0076-6879(08)04016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuma A, Matsui M, Mizushima N. Lc3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: Caution in the interpretation of lc3 localization. Autophagy. 2007;3:323–328. doi: 10.4161/auto.4012. [DOI] [PubMed] [Google Scholar]
- 38.Komatsu M, Ichimura Y. Physiological significance of selective degradation of p62 by autophagy. FEBS Lett. 2010;584:1374–1378. doi: 10.1016/j.febslet.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 39.Johnston JA, Ward CL, Kopito RR. Aggresomes: A cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanbe A, Osinska H, Villa C, Gulick J, Klevitsky R, Glabe CG, Kayed R, Robbins J. Reversal of amyloid-induced heart disease in desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2005;102:13592–13597. doi: 10.1073/pnas.0503324102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 42.Kumarapeli AR, Wang X. Genetic modification of the heart: Chaperones and the cytoskeleton. J Mol Cell Cardiol. 2004;37:1097–1109. doi: 10.1016/j.yjmcc.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 43.Wong ES, Tan JM, Soong WE, Hussein K, Nukina N, Dawson VL, Dawson TM, Cuervo AM, Lim KL. Autophagy-mediated clearance of aggresomes is not a universal phenomenon. Hum Mol Genet. 2008;17:2570–2582. doi: 10.1093/hmg/ddn157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verhoef LG, Lindsten K, Masucci MG, Dantuma NP. Aggregate formation inhibits proteasomal degradation of polyglutamine proteins. Hum Mol Genet. 2002;11:2689–2700. doi: 10.1093/hmg/11.22.2689. [DOI] [PubMed] [Google Scholar]
- 45.Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144:67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- 46.Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010;11:777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- 47.Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, Ashkenazi A. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.