Abstract
The purpose of this manuscript is three fold: 1) to describe an update to DockoMatic that allows the user to generate cyclic peptide analog structure files based on protein database (pdb) files, 2) to test the accuracy of the peptide analog structure generation utility, and 3) to evaluate the high throughput capacity of DockoMatic. The DockoMatic GUI interfaces with the software program Treepack to create user defined peptide analogs. To validate this approach, DockoMatic produced cyclic peptide analogs were tested for three dimensional structure consistency and binding affinity against four experimentally determined peptide structure files available in the Research Collaboratory for Structural Bioinformatics database. The peptides used to evaluate this new functionality were alpha-conotoxins ImI, PnIA, and their published analogs. Peptide analogs were generated by DockoMatic and tested for their ability to bind to X-ray crystal structure models of the acetylcholine binding protein originating from Aplysia californica. The results, consisting of over 300 simulations, demonstrate that DockoMatic predicts the binding energy of peptide structures to within 3.5 kcal*mol−1, and the orientation of bound ligand compares to within 1.8 Å root mean square deviation (rmsd) for ligand structures as compared to experimental data. Evaluation of high throughput virtual screening capacity demonstrated that Dockomatic can collect, evaluate and summarize the output of 10,000 AutoDock jobs in less than two hours of computational time, 100,000 jobs requires approximately 15 hours and 1,000,000 jobs is estimated to take up to a week.
Keywords: Peptide analog creation, high throughput screening, conotoxin, acetylcholine binding protein, DockoMatic, AutoDock
Introduction
The use of computational modeling strategies to predict desirable ligand binding interactions with biological receptors have become widespread due to the time and cost savings as compared to wet bench experimentation. DockoMatic is an intuitive graphical user interface (GUI) designed to facilitate job submission, and expand the capabilities of the widely used suite of automated docking tools collectively called, AutoDock.1–3 DockoMatic accepts protein database (pdb) files of ligands and receptors with corresponding grid coordinate files (gpf) that specify the experimentally determined or predicted ligand binding domain on the receptor. DockoMatic accepts text files containing lists of peptide ligands and receptors, and then automatically sets up and initiates processing of the associated set of AutoDock jobs. DockoMatic is able to run AutoDock jobs on a local machine, but the intended use for DockoMatic is high throughput screening on a computational cluster. DockoMatic can manage tens of thousands, and even hundreds of thousands, of simultaneous jobs running on a computational cluster.
DockoMatic allows the user to enter peptide pdb files as ligands, and it can also create linear peptide ligand structure pdb files from strings of single letter amino acid code as described previously.4 The peptide ligands that are entered into DockoMatic are prepared for submission to AutoDock. The number of amino acids in any given ligand is not restricted, but the torsional mobility of bonds within the ligand is a limitation of AutoDock. The maximum number of flexible bonds allowed by AutoDock, in both the ligand and the receptor is 32. This number can be increased at the expense of computational time, but is typically not required nor recommended.
The DockoMatic script was modified to allow the preparation of ligand pdb files for amino acid substitution by Treepack. This software combination provides an in silico method of site-directed mutagenesis for complex peptide and protein structures that incorporates experimentally determined tertiary structure. The user enters the desired peptide mutation into DockoMatic as a text file. DockoMatic performs an excision of amino acids surrounding the site of mutation from the pdb file of the peptide ligand. The excised segment consisting of a tripeptide pdb file is modified to remove amino acid side chain atoms consistent with the native peptide. The backbone designators are changed to model the desired mutation in a way that is in an acceptable format for Treepack. Treepack then adds new amino acid side chains based on the backbone identifiers. Adjustments are made for the Van der Waal radii of new atoms prior to the DockoMatic directed re-insertion of the excised peptide sequence back into the native peptide structure. The end result is the structure file representing the single point mutant of the original ligand. It deserves mention that this process also works well for double and triple mutants. The combination of Treepack and DockoMatic allows energetically optimized amino acid substitution to be performed on an experimentally determined peptide structure template in an automated fashion. Figure 1 shows the steps that are controlled by DockoMatic (4 A–C, E) and the critical mutation that is accomplished by TreePack (4 D).
Figure 1.
DockoMatic mediated Treepack ligand site directed amino acid substitution pdb file creation. From DockoMatic initiated command to produce ligand.pdb:K4W, the following five steps take place: A) the residue of interest (lys4) and the two surrounding amino acids (asp3 and cys5) are copied into a new pdb file; B) the side chain atoms of the excised tripeptide are stripped from the analog pdb file, the backbone atoms and the beta carbon atom are retained; C) the amino acid at the point of mutation is replaced to create the peptide analog (lys4→trp4); D) the analog tripeptide file is submitted to Treepack, which uses the backbone atoms in concert with beta carbon atoms to form point vectors for the new side chains; E) the desired side chains are then extracted from the Treepack modified analog pdb file to be grafted back into the original ligand file.
The analog creation feature was tested on peptides isolated from the venom of snails of the genus Conus, referred to as conotoxins. A comparative study was performed between nuclear magnetic resonance (NMR) spectroscopy solution or X-ray crystal structures versus the DockoMatic created peptide analog structures and their binding to three different Aplysia californica acetylcholine binding protein (Ac-AChBP) structures, specifically alpha-conotoxins PnIA, ImI and analogs PnIA[A10L:D14K], ImI[R11E], ImI[R7L], and ImI[D5N]. Conotoxins are small, 10–30 amino acid peptides that are cystine rich (i.e. contain multiple cysteine residues joined by disulfide bonds), and tend to be highly constrained structurally. Conotoxins are among the most potent and selective ligands in their binding to myriad biological receptors, offering promise in the development of therapies for diseases including epilepsy, Parkinson's, Alzheimer's, schizophrenia, and many others.5,6 The α-conotoxins tested in this study are significant because of their selective binding affinity to ligand-gated ion channel receptors, specifically nicotinic acetylcholine receptors.7–9 The cysteine-rich sequence and cystine composition of conotoxins results in structure rigidity; a quality that is conducive to NMR structure elucidation. The structures of many conotoxins and their analogs are available in the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank.
Methods
File Preparation
As stated previously, the integration of DockoMatic and Treepack was validated by binding alpha-conotoxins of known structure (i.e. ImI and PnIA) to Ac-AChBP receptors. The specific ligands used for this study were NMR solution structures: 1) ImI[R11E], PDB code 1E74; 2) ImI[R7L], PDB code 1E75; 3) ImI[D5N], PDB code 1E76; and 4) PnIA, PDB code 1PEN. The receptor models used for this study consisted of X-ray crystal structures: 1) Ac-AChBP with ImI bound, PDB code 2BYP; 2) Ac-AChBP with ImI bound, PDB code 2C9T; and 3) Ac-AChBP with PnIA[A10L:D14K], PDB code 2BR8. The Ac-AChBP pdb files were manually cleaned to remove bound ligand and water molecules. The result of this cleaning procedure was a structure consisting of alpha and beta subunits containing the ligand binding domain where conotoxins are found to be present in crystal structures of ligand/receptor complexes. This structure was then used as the receptor for ligand binding studies.
File Submission
To access the analog utility in DockoMatic, the user enters the name of the native RCSB retrieved peptide pdb file, followed by a colon, and then the desired substitution. To generate the alpha-conotoxin ImI analog that substitutes an arginine amino acid in position eleven of the peptide with a glutamate, the user would enter: ImI.pdb:R11E. Each additional replacement is added and separated by colons (i.e. PnIA.pdb:A10L:D14K). The user may enter this type of analog specification by hand directly into DockoMatic, but DockoMatic also allows a user to load a file containing a list of peptide analogs specified in this format (the file format requires that each analog specification be separated by a carriage return). For example, a user might load a file containing the following list of analogs using PnIA as the structural template:
/home/username/conotoxins/PnIA.pdb
/home/username/conotoxins/PnIA.pdb:A10L
/home/username/conotoxins/PnIA.pdb:A10L:D14K
When the above text file is loaded, DockoMatic prepares three different ligands; one the original pdb file for alpha-conotoxin PnIA, and the other two are analogs using the original structure as a template. This approach allows high throughput virtual screening of ligand analogs.
Integration with Treepack
The new analog utility of DockoMatic was designed to enable combinatorial computational high throughput screening of peptide ligands against biological receptors. In the field of conotoxin research, this process offers great potential to facilitate drug discovery. DockoMatic and TreePack work together to perform amino acid side chain replacement as directed by user input to create the desired analog structure.11,12 Treepack is a software tool created to assist in homology modeling of receptors. Its performance is comparable to the widely used program Modeller; both programs take direction vectors from peptide backbone atoms and attach newly calculated amino acid side chains to the established template.13 The analog pdb file is read by DockoMatic. DockoMatic prepares a segment of the peptide structure surrounding the site of mutation for submission to the Treepack script. Treepack performs the amino acid side chain replacement and subsequent energy minimization of three dimensional orientations for the new atoms. Analog creation between DockoMatic and Treepack follows a five step process: A) the residue of interest and the two surrounding amino acids are copied into a new pdb file; B) the side chain atoms of the excised tripeptide are stripped from the analog pdb file, the backbone atoms and the beta carbon atom are retained; C) the amino acid at the point of mutation is replaced to create the peptide analog; D) the analog tripeptide file is submitted to Treepack, which uses the backbone atoms in concert with beta carbon atoms to form point vectors for the new side chains (except in the case of glycine which does not have a beta carbon atom); E) the desired side chains are then extracted from the Treepack modified analog pdb file to be grafted back into the original ligand file, adjusting the remaining atoms to account for atom numbering differences (see Figure 1). This process may be repeated as many times as is necessary depending on whether a single or multiple amino acid substitution is defined by the user. Treepack operates by first defining a bubble around the intended side chains to be modified. These bubbles are the parameters for the space available for the new side chain atoms. Treepack then minimizes the energy of the structure of the residue that is packed in the bubble.11,12 Threeside chains are submitted to Treepack from the original ligand pdb file to minimize the potential for atomic overlap when the new residue is grafted back into the original ligand file.
Result Analysis
The binding of conotoxin structures determined by NMR spectroscopy or X-ray crystallography to the Ac-AChBP receptor model in Autodock was compared to the same conotoxins with protein database analog files created by Treepack through the DockoMatic GUI. Application of the “results check” utility of DockoMatic automatically parses the most favorable results from the pdb reference files collected from all AutoDock jobs.4 By employing the result check feature, the most energetically favorable bound ligand conformations were compared, and their backbone coordinates were entered into the root mean square deviation (rmsd) calculator of the computer program, Visual Molecular Dynamics (VMD).10
Results
Ligand Binding Test
Pdb files of ligands created through the DockoMatic GUI using the integrated Treepack software were compared to structures of conotoxins deposited in the RCSB by two sets of experimental procedures. For experiment one, a model was created of a bound conotoxin in the ligand binding domain of the Ac-AChBPs. To do this, crystal structures of Ac-AChBP receptors with bound ImI or the PnIA analog PnIA[A10L:D14K] were selected from the RCSB. The conotoxin ligand was eliminated from the binding cavity of the Ac-AChBP followed by removal of water molecules from the receptor. Receptor cleaning eliminates the nonessential ligand and all water molecules. To test the ability of DockoMatic to run jobs through AutoDock and generate reliable results, the conotoxin peptide ligand that was extracted from the ligand bound receptor/crystal structure complex was redocked into the now vacant (i.e. cleaned) receptor. Table I includes the results of ligand redocking with respect to ligand binding orientation and binding energy as compared to the original crystal structure.
Table I.
DockoMatic redocking of X-ray crystal and NMR solution structures with associated estimated binding energy and RMSD.
| Receptor Ac-AChBP | Ligand | Estimated Binding Energy | RMSD |
|---|---|---|---|
| PDB codes | kcal*mol−1 | Angstrom (Å) | |
| 2BR8 | PnIA[A10L:D14K] | −15.44 | 1.0127 |
| 2BYP | ImI | −16.03 | 0.8779 |
| 2C9T | ImI | −13.87 | 1.2225 |
For native ImI, there were two different crystal structures in the RCSB that were used for this exercise (i.e. 2BYP and 2C9T). Redocking of the extracted ImI peptide to the 2BYP receptor provided a peptide ligand overlay between the crystal structure complex and the computationally determined binding complex with a rmsd of 0.8779 Å. Conotoxin peptide redocking to the Ac-AChBP receptor model 2C9T yielded an overlay of bound peptide ligand between experimentally determined structure and computationally calculated structure of 1.2225 Å. When the structure of the double mutant PnIA[A10L:D14K] was redocked into 2BR8, the rmsd was 1.0127 Å. A visual representation of the bound ligand demonstrating the structural orientation of each peptide overlaid in the ligand binding domain of the receptor is shown in Figure 2. This result is significant because it demonstrates that the peptide sequence files entered into DockoMatic, followed by submission to AutoDock, provide output that is within reasonable agreement to experimentally determined structure binding images (i.e. rmsd ≤ 1.2 Å). One goal of this work is to scale the process by several orders of magnitude relative to the number of ligands that can be simultaneously submitted to AutoDock for binding calculations (i.e. high throughput screening of potential drug candidates). In this control experiment, we have validated the successful integration of AutoDock into DockoMatic to yield good homology between expected binding and computationally predicted binding to a common receptor.4
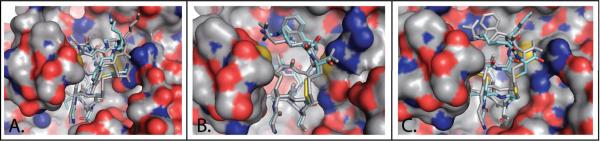
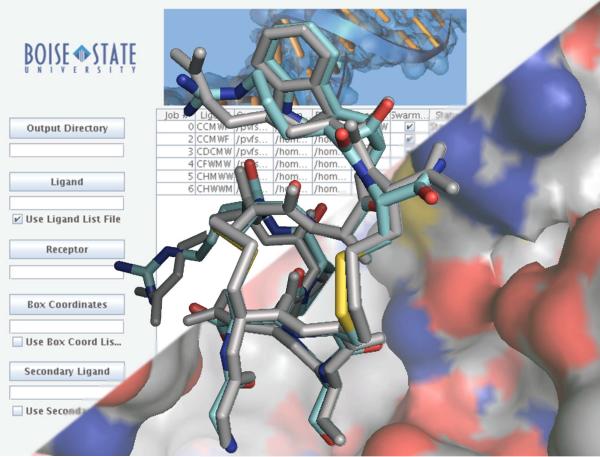
Figure 2.
Ac-AChBP structures with ligand redocked using DockoMatic. Original ligand (grey), and redocked ligand (blue). A) PnIA[A10L:D14K] rebound to 2BR8 with an RMSD of 1.0127 Å; B) ImI redocked to 2BYP, RMSD of 0.8779Å; C) ImI redocked to 2C9T, RMSD of 1.2225 Å.
The second experiment was to validate the Treepack driven utility for the automated creation of peptide analogs in DockoMatic. This was done by comparing conotoxin analog structures generated by DockoMatic with NMR solution structures deposited in the RCSB. DockoMatic requires a parent peptide file for analog creation; the ligands submitted were the NMR solution structure pdb files for PnIA and ImI, PDB codes 1PEN and 1IMI, respectively. Three analogs of ImI: ImI[R11E], ImI[R7L], and ImI[D5N], and one analog of PnIA: PnIA[A10L:D14K] were selected based on solution structure availability in the RCSB.
Two sets of experiments were conducted in parallel: 1) the solution structures were used as the ligands to bind to the Ac-AChBP receptor, and 2) the analog sequence was entered into DockoMatic, the ligand generated, and the ligand binding calculation automatically performed through AutoDock. The results were filtered using the result check feature in DockoMatic, followed by orientation and docking conformation comparisons evaluated by calculated rmsd and estimated binding energy. Peptide ImI[R11E] was bound to three receptor crystal structures of Ac-AChBP, 2BR8, 2BYP, and 2C9T, with estimated energy of −12.16, −10.67, and −10.62 kcal*mol−1 respectively (Table II). The overlay of the same analog created by DockoMatic resulted in a −11.13, −11.88, and −10.58 kcal*mol−1 binding energy to the three Ac-AChBPs showing a difference in energy of 1.03, 1.21, and 0.04 respectively. The rmsd of the three receptor overlays for peptide ImI[R11E] are 0.938, 0.564, and 0.811 Å, respectively.
Table II.
Comparative results listing the estimated binding energies. Column 1 lists the receptor pdb codes for each experiment. Column 2 lists the α-conotoxin mutations tested against column 1. Columns 3–4 list the results of the NMR solution structure and the DockoMatic created structure of the α-conotoxin mutations. Columns 5–6 compare the results of the experiment by standard deviation of the estimated binding energy and the rmsd of the backbone structures.
| Receptor Ac-AChBP | Structure* | NMR Solution Structure | DockoMatic Structure+ | Standard Deviation | RMSD |
|---|---|---|---|---|---|
| PDB code | Estimated Binding Energy (kcal*mol−1) | Estimated Binding Energy (kcal*mol−1) | Comparing Estimated Binding Energy | Angstrom (Å) | |
| 2BR8 | PnIA[A10L:D14K] | −15.44 | −14.60 | 0.594 | 0.4300 |
| 2BR8 | ImI[R11E] | −12.16 | −11.13 | 0.728 | 0.9380 |
| ImI[R7L] | −11.09 | −11.24 | 0.106 | 1.8156 | |
| ImI[D5N] | −13.80 | −13.13 | 0.474 | 1.1750 | |
| 2BYP | ImI[R11E] | −10.67 | −11.88 | 0.856 | 0.5635 |
| ImI[R7L] | −12.59 | −12.76 | 0.120 | 0.7594 | |
| ImI[D5N] | −14.88 | −12.84 | 1.442 | 1.4718 | |
| 2C9T | ImI[R11E] | −10.62 | −10.58 | 0.028 | 0.8113 |
| ImI[R7L] | −13.49 | −12.66 | 0.587 | 0.8489 | |
| ImI[D5N] | −15.54 | −12.08 | 2.447 | 1.1535 |
PDB codes solved structures: PnIA[A10L:D14K] 2BR8,ImI[R11E] 1E74, ImI[R7L] 1E75, and ImI[D5N] 1E76.
PDB codes for DockoMatic templates: PnIA 1PEN, ImI 1Im1
The result of the redocking experiment with ImI analog, ImI[R7L] as compared to the DockoMatic generated docking of the Treepack created ImI[R7L] with the three Ac-AChBP receptors produced energy differences of 0.15, 0.17, and 0.89 kcal*mol−1, and demonstrated ligand RMSDs of 1.816, 0.759, and 0.849 Å. The same experiment using peptide ImI[D5N] provided energy differences of 0.67, 2.04, and 3.46 kcal*mol−1 respectively, and rmsd differences of 1.175, 1.4718, and 1.1535 Å respectively. For final comparison, the extracted ligand analog of PnIA, PnIA[A10L:D14K] was redocked to the Ac-AChBP 2BR8 with a calculated binding energy of −15.44 kcal*mol−1. The same experiment was performed with the structure of native PnIA, 1PEN, yielding a binding energy of −14.6 kcal*mol−1, thus a difference of 0.84 and an RMSD between bound ligands of 0.43 Å.
The rmsd of all conotoxin peptide ligand structures ranged from 0.5635 to 1.8156 Å in binding comparisons between experimentally determined peptide structure and DockoMatic created ligand structure analogs. The difference in estimated binding energy from matching poses varied by less than 3.5 kcal*mol−1, the standard deviation ranged from 0.028 to 2.447 with the majority of the results falling below 1.00. These results demonstrate that the Treepack analog creation tool that has been incorporated into DockoMatic provides ligand structures that bind with similar orientation and affinity to RCSB structures determined experimentally. DockoMatic offers the ability to generate analog structures for peptides in a fraction of the time and expense of experimentally determined peptide ligand structures.
Evaluation of High Throughput Capability
Each ligand specified as input by the user in DockoMatic is automatically submitted to the cluster for processing as an AutoDock job. The Autodock job performs 100 ligand to receptor binding calculations and compiles the output into a single dlg file. For each completed AutoDock job, DockoMatic extracts in priority order the 100 receptor binding calculations into the pdb reference file. DockoMatic determines that an AutoDock job is complete when the dlg and the pdb reference file are created. Thus, for 1 ligand, 100 results are summarized and listed in a single dlg file, and the results are put into a user specified rank order in the pdb reference file (e.g. from lowest to highest binding energy). DockoMatic thus incurs a very small amount of computation time to setup up a job and submit it for processing, as well as a relatively small amount of computation (relative to the total job run-time) time to parse, process, and summarize each completed job. It is important to note that DockoMatic processes each job as it completes – i.e. it does not wait for all submitted jobs to complete before beginning the process of summarizing results.
There are a number of factors that must be considered in order to determine the high-throughput capacity of DockoMatic. For example, the maximum number of jobs that can be submitted as a set from DockoMatic is dependent on both the number of subdirectories a given file system can accommodate, and the amount of disk space that is available to store results. For most file systems, there is a set maximum number of subdirectories that can be created within each directory; for instance, the most common file systems used with a Linux kernel, ext3 and ext4, are limited to 32,000 and 64,000 subdirectories respectively. This file system limitation can become a factor because each AutoDock job in a set of jobs uses one subdirectory for output, and DockoMatic typically creates the output subdirectories for a set of jobs in a single output directory. The storage device used by the Andersen laboratory for storing output results employs Lustre, which is a parallel file system designed for use in a clustered environment. Lustre allows a maximum of 25 million subdirectories, although in practice this is not the predominant limitation. For instance, disk space becomes an issue for large numbers of jobs. Each DockoMatic job generates output files that take up an average of 115 MB of disk space. At 115 MB, 1 million jobs requires on the order of 115 Terabytes of disk space which easily exceeds the current raw capacity of 72 Terabytes of the Lustre file system.
There are also other factors that can be equally as important, such as the speed of the machine that is used to host DockoMatic, the amount of system memory available, the computational capacity of the cluster itself, etc. Nevertheless, while any assessment of capacity is dependent on the particular setup and environment, it is important to have a rough estimate of limition in order to assess the feasibility of a potential set of experiments using DockoMatic.
The DockoMatic GUI performs two primary computational functions for each AutoDock job. These are the following:
DockoMatic directs the flow of jobs by creating a folder for the assigned AutoDock output and then starts the AutoDock job.
DockoMatic monitors the output directory of each AutoDock job for the presence of a reference file that contains a summary of the job statistics.
Since the DockoMatic GUI is unaware of what the exact process is that creates the output file it is looking for, it is not actually necessary to initiate true AutoDock jobs in evaluating any throughput limitation of the DockoMatic GUI. So, to evaluate the high throughput capacity of the software, a series of mock jobs were created to populate the output folders with the required reference file, rather than running actual AutoDock experiments. The evaluation was performed in this manner due to time constraints, and the goal being to test how many jobs DockoMatic could adequately handle, regardless of job type. All aspects and functionality of DockoMatic were preserved. The experiment worked by the following series of steps: 1) a list of jobs was submitted to DockoMatic, 2) DockoMatic created all output folders, 3) DockoMatic populated the monitoring grid, listing each job and the output location, 4) each job (in this case a file copy) was submitted to swarm and queued to the cluster with its status changed to “Started”, 5) after the file was copied, DockoMatic recognized each job as being complete and the status was changed to “Done”. This process was timed for jobs ranging from 100 to 1,000,000 submissions (see table III). For mock job lists of 100 and 1000 it took DockoMatic less than one second to create directories and populate management grids. The process of distributing files and acknowledging job completion required 51 seconds and 8.7 minutes, respectively. To initiate 10,000 jobs required on the order of 8 seconds with the final recognition of all jobs completed just over an hour and a half later (1.57 hrs). The time to set up 100,000 jobs and submit them was on the order of 231 seconds with an estimated completion time on the order of 15 hours. To achieve one million jobs is estimated to be on the order of one week of computer cluster time. In summary, in its current configuration, DockoMatic can reasonably handle the submission of 10,000 to 100,000 jobs for binding calculation.
Table III.
Evaluation of high throughput capability of DockoMatic. Mock experiment results showing the number of jobs in each trial, the length of time to submit jobs, and the job completion time based on output file preparation.
| Number of Jobs | Initiation time (sec)* | Completion time** |
|---|---|---|
| 100 | < 1 | 51 sec |
| 1000 | < 1 | 8.7 min |
| 10,000 | 8 | 1.57 hrs |
| 100,000 | 231 | ~15 hrsξ |
| 1,000,000 | ~1 weekξ |
Time required to create job submission directories and populate grid boxes.
Time required to copy dlg and pdb reference files into output directories.
Estimated time.
Conclusion
We have demonstrated that DockoMatic has been upgraded to include the Treepack program for automated peptide analog structure creation and subsequent AutoDock job submission. To validate these two functions, three ImI analogs and one PnIA analog were created and tested against solved peptide structures obtained from the RCSB. The result of these experiments was a successful structure creation and binding comparison with differences in root mean square deviation from mean structure and estimated binding energy variation to within 1.8 Å and 3.5 kcal*mol−1 respectively. DockoMatic as a standalone utility consists of an intuitive GUI: 1) to create linear peptide ligands; 2) to produce analogs based on structure templates; 3) to perform high throughput submission to AutoDock; and 4) to facilitate result analysis during post processing. AutoDock has been successfully integrated into DockoMatic for the routine submission of 10,000 to 100,000 jobs with more possible based on computer cluster access. The expanded functionality of DockoMatic to perform in silico site directed mutagenesis using the Treepack utility offers the opportunity for chemists and biologists to apply the extraordinary tools developed by computer scientists toward predictive science. DockoMatic is freeware that calls upon other freeware software (TreePack and AutoDock) for high throughput ligand creation and receptor binding calculation.
Figure 3.
Acknowledgments
Research funded by the Defense Threat Reduction Agency under contract number W81XWH-07-1-000, DNA Safeguard. This publication was made possible by NIH Grant #P20 RR0116454, the Idaho IDeA Network of Biomedical Research Excellence, Research Corporation Cottrell College Scholars program, and Mountain States Tumor Medical Research Institute. The authors wish to thank Greg Hampikian, Julie Oxford, and Barb Jibben for editorial comments and conversations that have strengthened the presentation of content described in this manuscript.
References
- 1.Sousa SF, Fernandes PA, Ramos MJ. Proteins. 2006;65(1):15. doi: 10.1002/prot.21082. [DOI] [PubMed] [Google Scholar]
- 2.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. Journal of computational chemistry. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Journal of computational chemistry. 1998;19(14):1639. [Google Scholar]
- 4.Bullock C, Jacob R, McDougal O, Hampikian G, Andersen T. BMC Research Notes. 2010;3(1):289. doi: 10.1186/1756-0500-3-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dutertre S, Lewis RJ. Biochemical pharmacology. 2006;72(6):661–670. doi: 10.1016/j.bcp.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 6.Millard EL, Daly NL, Craik DJ. European Journal of Biochemistry. 2004;271(12):2320–2326. doi: 10.1111/j.1432-1033.2004.04148.x. [DOI] [PubMed] [Google Scholar]
- 7.Dutertre S, Lewis RJ. European Journal of Biochemistry. 2004;271(12):2327–2334. doi: 10.1111/j.1432-1033.2004.04147.x. [DOI] [PubMed] [Google Scholar]
- 8.Groebe DR, Dumm JM, Levitan ES, Abramson SN. Molecular pharmacology. 1995;48(1):105–111. [PubMed] [Google Scholar]
- 9.McIntosh JM, Azam L, Staheli S, Dowell C, Lindstrom JM, Kuryatov A, Garrett JE, Marks MJ, Whiteaker P. Molecular pharmacology. 2004;65(4):944. doi: 10.1124/mol.65.4.944. [DOI] [PubMed] [Google Scholar]
- 10.Humphrey W, Dalke A, Schulten K. Journal of Molecular Graphics. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 11.Xu J. In: Research in Computational Molecular Biology. Miyano S, Mesirov J, Kasif S, Istrail S, Pevzner P, Waterman M, editors. Springer Berlin; Heidelberg: 2005. pp. 423–439. [Google Scholar]
- 12.Xu J, Berger B. J ACM. 2006;53(4):533–557. [Google Scholar]
- 13.Sali A, Blundell T. Journal of Molecular Biology. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]