SUMMARY
We have identified human MBT domain-containing protein L3MBTL2 as an integral component of a protein complex that we termed Polycomb Repressive Complex 1 (PRC1)-like 4 (PRC1L4) given the co-presence of PcG proteins RING1, RING2 and PCGF6/MBLR. PRC1L4 also contained E2F6 and CBX3/ HPlγ known to function in transcriptional repression. PRCIL4-mediated repression necessitated L3MBTL2 that compacted chromatin in a histone modification-independent manner. Genome-wide location analyses identified several hundred genes simultaneously bound by L3MBTL2 and E2F6, preferentially around transcriptional start sites that exhibited little overlap with those targeted by other E2Fs or by L3MBTL1, another MBT-domain containing protein that interacts with RB1. L3MBTL2-specific RNAi resulted in increased expression of target genes that exhibited a significant reduction in H2A lysine 119 monoubiquitination. These findings highlight a PcG/MBT collaboration that attains repressive chromatin without entailing histone lysine methylation marks.
INTRODUCTION
Chromatin architectural states areimportant determinats for processes that require access to DNA (Campos and Reinberg, 2009). Placement, removal and specific recognition of histone lysine methylation marks are recognized as important chromatin regulatory events. Multiple protein families ‘reading’ (recognizing) histone methylation states have been discovered and their respective binding specificities have been mapped (Taverna et al., 2007). It is likely that the overall binding affinity of a chromatin reader is not solely determined by its interaction to a single methyl-mark but rather by a multivalent recognition of modifications (as discussed in (Ruthenburg et al., 2007) as well as by interactions with the surface of the nucleosome. However, in most instances methyl-binding is determined in experiments using synthetic peptides that correspond to the unstructured N- and C-terminal regions of histones. A more relevant approximation of chromatin are oligonucleosomes, but to date only a handful of studies used methyl marks installed on such complex substrates (for instance, see (Margueron et al., 2009; Simon et al., 2007; Trojer et al., 2007)). To date, there is still a significant gap between studies that focus mostly on binding properties through biochemistry and others that implicate chromatin-binding modules in the regulation of transcription through genetics. There are only a few instances in which histone methyl-binders were shown to directly modulate chromatin structure in a histone methylation dependent manner, one of which is the MBT family member L3MBTL (hereafter referred to as L3MBTL1) that compacts chromatin only in the presence of methyl marks, for instance monomethylated lysine 20 on histone H4 (H4K20me1) (Trojer et al., 2007; Trojer and Reinberg, 2008).
The human MBT protein family comprises at least 8 members with established functions in development and their dysfunction is implicated in disease (Bonasio et al., 2009). These proteins are characterized by two, three or four MBT domains arranged in tandem and recent structural studies indicate that all MBT domains within a protein form an interlocked superstructure (Eryilmaz et al., 2009; Grimm et al., 2007; Grimm et al., 2009; Guo et al., 2009; Li et al., 2007; Min et al., 2007; Santiveri et al., 2008; Sathyamurthy et al., 2003; Wang et al., 2003). To date, MBT proteins appear to accommodate a histone methyl-lysine in only one of their MBT domains. Binding occurs through caging of the methylated lysine by critical aromatic residues in the binding pocket with only a limited number of interactions with residues flanking the methylated lysine. MBT domain-containing proteins exhibit a strict preference in binding for mono- and di-methyl modification states (reviewed in (Bonasio et al., 2009; Taverna et al., 2007; Trojer and Reinberg, 2008).
Importantly, all MBT proteins studied to date have been implicated in transcriptional repression. Owing to genetic studies in Drosophila and later biochemical studies in mammalian models, we know that the mechanisms of MBT mediated repression intersect with those of Polycomb group (PcG) proteins. The molecular mechanisms of PcG protein mediated gene silencing include the catalysis of histone H3 lysine 27 (H3K27) methylation and histone H2A lysine 119 monoubiquitination (H2AK119ub1) by the Polycomb Repressive Complexes 2 (PRC2) and 1 (PRC1), respectively (for reviews see (Kerppola, 2009; Muller and Verrijzer, 2009). Of note, dependent upon their constituents, certain PRC complexes impact chromatin architecture by directly compacting chromatin in a histone modification independent manner; the PRC2 complex harbouring EZH1 in lieu of the EZH2 homologue (Margueron et al., 2008), the Drosophila PRC1 subunit Posterior Sexcomb (Psc) (Francis et al., 2004) and the mouse PRC1 subunit Ring1B (Eskeland et al., 2010). Six paralogs of Drosophila Psc exist in mammalian cells, none of which have been reported to exhibit chromatin compaction properties. There are multiple variations of PRC1 in mammals, all of which contain RING1/RING1A and RING2/RING1B as E3 ubiquitin ligases specific for H2AK119ub1 (Wang et al., 2004), but are distinguishable by their Psc homologs and additional PcG protein subunits (Kerppola, 2009). Although both BMI1/PCGF4 (Cao et al., 2005; Wei et al., 2006), MEL-18/PCGF2 and NSPC1 /PCGF1 (Wu et al., 2008) stimulate in vitro RING2 H2AK119 specific E3-ligase activity, we still do not fully appreciate the functional contribution of mammalian Psc homologs in PcG mediated gene silencing. Moreover, to date, we have only limited insight as to whether the six Psc homologs target distinct or overlapping gene subsets.
Here we report the identification of a human complex comprising the MBT-domain containing protein L3MBTL2 along with well-characterized PcG proteins. We describe the multiple activities exhibited by this complex that promote repressive chromatin architecture. We also found that the methyl-lysine binding capability of L3MBTL2 was not required for its transcriptional repressor function, yet the ability of the complex to compact chromatin and independently mediate H2AK119ub1 was important for maintaining repression of target genes.
RESULTS
Purification of a PRC1-like protein complex
Human L3MBTL2 contains an atypical C2/C2 type Zn finger in the N-terminal portion of the protein, followed by four MBT domains but, unlike L3MBTL1 and many other MBT proteins, it lacks the SPM dimerization domain at its C-terminus (Figure 1A). In order to identify L3MBTL2 associated polypeptides, we generated a 293F cell line that constitutively expresses full-length L3MBTL2 with a C-terminal FLAG-tag (L3MBTL2-F). Despite significant expression levels we detected L3MBTL2-F exclusively in the nucleus (Figure S1A in the Supplemental Data available with this article online). The purification scheme for isolating L3MBTL2-associated polypeptides from nuclear extracts (NE) is shown in Figure 1B. L3MBTL2-F was found in both the flow-through and the DE52-bound fractions (Figure S1B). Subsequent analysis of these fractions by Superose 6 gel filtration chromatography showed that L3MBTL2-F derived from the DE52 flow-through was a homogeneous complex of approximately 400 kDa (to be described elsewhere), while that derived from the DE52-bound material eluted from the column at various molecular weights (Figure S1C). These latter fractions were independently subjected to an anti-FLAG immunoaffinity resin and the bound proteins were eluted by competition with FLAG-peptide.
Figure 1. Purification of a PRC1-like complex containing L3MBTL2.

(A) Domain organization of human L3MBTL2. MBT domains are shown in red and the C2/C2 Zn finger in blue. For purification of L3MBTL2 associated polypeptides a 293F cell line that constitutively expresses a C-terminally FLAG-tagged version of L3MBTL2 was generated. (B) Schematic representation of the L3MBTL2-F purification strategy. (C) Purified L3MBTL2-F protein complex was resolved by gradient SDS-PAGE and visualized by silver staining. Polypeptides identified by tandem mass spectrometry are indicated on the right and molecular weight markers on the left. (D) Western blot analysis of flow through (FT), wash (W) and bound (B) fractions after the final anti-HPlγ immuno-affinity purification step. Nuclear extract (NE) is shown for comparison. Western blot was carried out with antibodies indicated on the right. (E) Histone E3-ubiquitin ligase assays were carried out in the absence or presence of the activating enzyme (E1), the conjugating enzyme (E2), purified PRC1L4 complex (L3MBTL2c), recombinant oligonucleosomes, ATP, and with recombinant ubiquitin. Reactions were resolved by SDS-PAGE and immunodetection of histone ubiquitination carried out with an H2AK119ub1 specific antibody. Ponceau Red Staining of histones was used as loading control (lower panel).
We previously identified CBX3 (hereafter referred to as HPlγ) as an L3MBTL1- and L3MBTL2- but not an L3MBTL3-associated polypeptide and showed that this interaction was exclusive for the γ-isoform of HP1 (Trojer et al., 2007). Thus, we subjected the FLAG affinity elution fractions to an anti-HPlγ immunoaffinity resin and eluted the bound proteins with glycine. This sequential affinity purification strategy led to the isolation of a number of polypeptides, relative to the case of the control purification using nuclear extracts from cells containing empty vector (Figure 1C). The HPlγ co-purified polypeptides were excised from the gel and their identity determined by tandem mass spectrometry (Figure 1C). In addition to HPlγ, we identified RING1, RING2 and PCGF6 (hereafter referred to as MBLR) that belong to the PcG family of proteins, the transcriptional repressor E2F6 and as expected, L3MBTL2. HSP70, PRMT5 and the human Enhancer of Rudimentary (EHR) were considered contaminants as they were found in multiple unrelated affinity-purifications (data not shown). Western blot analysis of the fractions from the final anti-HPlγ immunoaffinity purification step further established the identity of the purified polypeptides (Figure 1D; the sources of antibodies used are summarized in Table S1). RB1 specific antibodies were used as a negative control, and revealed that RB1 was present in the flow-through of the sequential affinity purification, as expected (Figure 1D). MBLR antibodies were generated and characterized is shown in Figure S1D-I. Using these MBLR antibodies (Figure 1D), we were able to detect this protein too, yet we failed to detect the histone lysine demethylase JARID1D/KDM5D previously found in a complex with MBLR (Lee et al., 2007). Thus, MBLR is likely a component of distinct protein complexes.
L3MBTL2 was previously identified as part of a large E2F6-complex containing the H3K9me2 specific HKMTs G9a/KMT1C and EuHMTase/GLP/KMT1D. The complex was found to silence genes in quiescent cells, in part by modifying chromatin structure (Ogawa et al., 2002). However, the presence of L3MBTL2 in this complex was never confirmed nor its biochemical properties determined. We detected some G9a in anti-L3MBTL2 co-immunoprecipitation (IP) experiments from extracts (Figure S1J). However, the purified L3MBTL2-F complex was devoid of detectable G9a, the expected MW of which is 200 kDa (Figure 1C).
RING1, RING2 and BMI1 are known to exhibit histone H2AK119 specific monoubiquitin-E3-ligase activity (Cao et al., 2005; Wang et al., 2004). We tested if the L3MBTL2-complex also contained E3-ligase activity using in vitro ubiquitin ligase assays with recombinant oligonucleosomes as substrate and an antibody that specifically recognizes the K119-ubiquitinated form of H2A (Figure 1E). Indeed, H2AK119ub1 was detected dependent on the presence of activating enzyme (E1), conjugating enzyme (E2), recombinant oligonucleosomes, ATP and the L3MBTL2-complex (Figure 1E, lane 6). Omission of the L3MBTL2-complex abrogated this activity (Figure 1E, lane 3). Thus, we termed the purified L3MBTL2-complex PRC1-like 4 (PRC1L4; see Discussion for details).
L3MBTL2 participates in PRC1L4 complex formation and directly interacts with H2AK119ub1 E3-ligases
We next tested if endogenous L3MBTL2 interacts with PRC1L4 complex components. To this end we generated L3MBTL2-specific antibodies directed against either the N- or C-termini of the protein (for characterization of antibodies see Figure S1K-R). The antibodies directed against the N-terminus were then used for IP experiments of endogenous L3MBTL2 using HeLa nuclear extracts. L3MBTL2 antibodies, but not an IgG control, precipitated L3MBTL2, RING2, HPlγ, and E2F6 (Figure 2A). The MBLR homologous protein BMI1 was not precipitated, bolstering the existence of multiple PRC1L complexes in mammalian cells. Moreover, antibodies specific for MBLR (Figure 2B), those for HPlγ, but not for HPlα (Figure 2C), and those for RING2 and for E2F6 (Figure 2D) precipitated L3MBTL2. To test if L3MBTL2 is associated with H2AK119ub1 E3-ligase activity we carried out IP experiments using FLAG-affinity purified L3MBL2-F complex as input material. BMI1, L3MBTL2 and HPlγ immunoprecipitates were subjected to E3-ligase assays using recombinant oligonucleosomes as substrate. In agreement with the results presented above, antibodies specific for HPlγ or L3MBTL2, but not for BMI1, precipitated H2AK119ub1 E3-ligase activity (Figure 2E).
Figure 2. Stable interaction of L3MBTL2-complex partners.
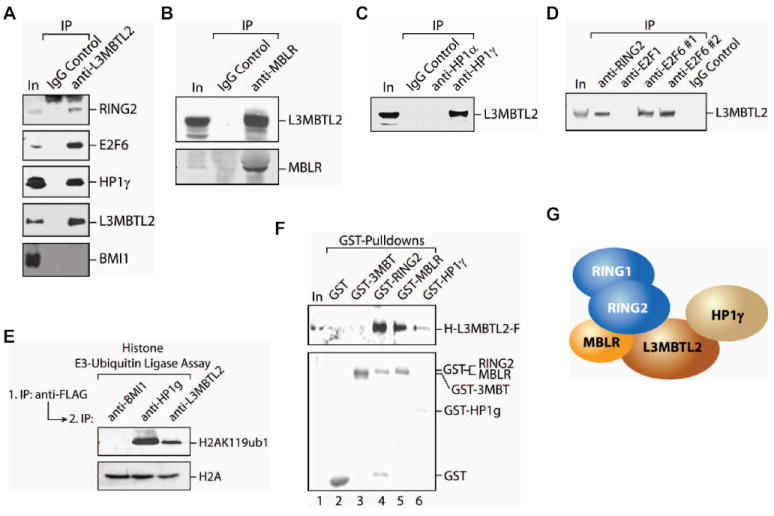
(A) Immunoprecipitation (IP) experiments from HeLa NE using anti-L3MBTL2 or an IgG control. Shown are input (50 μg of HeLa NE; lane 1) and immunoprecipitates from the IgG control (lane2) and anti-L3MBTL2 (lane 3). Western blots were carried out using the antibodies indicated on the right. (B). Western blot analyses using anti-L3MBTL2 and anti-MBLR specific antibodies, of IPs obtained from HeLa NE using anti-MBLR antibodies or an IgG control. (C) As in (B) but using anti-HPlα or anti-HPlγ specific antibodies or an IgG control for IPs. (D) As in (B) but using anti-E2F1, two different anti-E2F6 and anti-RING2 specific antibodies or an IgG control for IPs. (E) In vitro H2AK119 E3-ubiquitin ligase assays performed with IPs from FLAG affinity purified L3MBTL2-F complex using anti-BMI1, anti-HPlγ and anti-L3MBTL2 specific antibodies. Shown is a western blot from the E3-ligase reactions using anti-H2AK119ub1 and total H2A antibodies (as a substrate loading control). (F) GST pull-down experiments using GST, GST-3MBT, GST-RING2, GST-MBLR and GST-HPlγ recombinant proteins which were incubated with recombinant, full-length HIS-L3MBTL2-FLAG and precipitated with Glutathione Sepharose. Shown are western blots of input (lane 1) and precipitates (lanes 2-6) using anti-FLAG (upper panel) and anti-GST (lower panel) antibodies. (G) Schematic representation of the interactions between L3MBTL2 complex components based on the results shown in Figures 2F and S2.
To analyze the interactions between the complex components, we expressed and purified recombinant, affinity-tagged proteins from E.coli and Sf9 insect cells (Figure S2A, B). Recombinant, hexahistidine- (HIS) and FLAG-tagged, full-length L3MBTL2 (H-L3MBTL2-F, Figure S2A) directly interacted with RING2, MBLR and HPlγ in GST pull-down experiments (Figure 2F). Different GST pull-down experiments were performed to identify the direct physical contacts of RING1, RING2, MBLR and HPlγ (Figure S2C) and the results are summarized in a schematic (Figure 2G). RING1 did not interact with MBLR in our experiments. Importantly, L3MBTL2 appeared to be a key constituent of the PRC1L4 complex since it interacted with three of the six components (Figure 2F).
L3MBTL2 and E2F6 target the same genes for repression
Having established a physical interaction between E2F6 and L3MBTL2 and other associated polypeptides involved in transcription repression, we set out to determine the genomic targets of L3MBTL2, its close homolog L3MBTL1 and E2F6. ChIP assays in MCF7 breast adenocarcinoma cells were followed by hybridization to ENCODE arrays (representing about 1% of the human genome; a list of all arrays used for these ChIP-chip assays, including array platform and array identification number is provided in Table S2). Binding sites for L3MBTL2, L3MBTL1 and E2F6 were identified. Figure 3A shows L3MBTL2 and E2F6 enrichment peaks on chromosome 1, illustrating a high degree of correlation of the enrichment patterns reflecting a significant level of co-occupied genomic locations. L3MBTL2 was found preferentially within 2 kilobases (kbs) around the transcriptional start sites (TSSs) (Figure 3B). In contrast, L3MBTL1 also occupied gene regions more distal from the TSS to a high extent (Figure 3B).
Figure 3. L3MBTL2 and E2F6 binding sites correlate on a genome scalein various cell types.

(A) ENCODE ChIP-chip binding patterns of E2F6 (yellow) and L3MBTL2 (orange) for a region of chromosome 1. The Y-axis indicates fold enrichment of the ChIP sample. Transcribed regions are indicated as grey bars. Genomic coordinates are indicated on the bottom. (B) ChIP-chip campaign performed from MCF7 cells with an ENCODE array identified high confidence binding sites for L3MBTL2 and L3MBTL1 which were compared for their location relative to transcriptional start sites (TSS). The majority of L3MBTL2 binds within 2 kb of the TSS. In contrast, L3MBTL1 binding sites are more distributed and frequently found also in the body of genes. (C) A 1.5 kb promoter array was used to determine genome-wide binding of E2F6, L3MBTL2 and L3MBTL1 in MCF7 breast adenocarcinoma cells. Venn diagrams illustrate the overlap between E2F6 and L3MBTL2 (upper panel) or E2F6 and L3MBTL1 (lower panel) binding sites. (D) ChIP-seq was carried out to determine L3MBTL2 and E2F6 genomic binding sites in K562 erythroleukemia cells. A venn diagram illustrates that about 57% of all L3MBTL2 binding sites were co-occupied with E2F6. (E) Correlation of high confidence L3MBTL2, L3MBTL2/E2F6 and E2F6 genomic binding sites with the histone methylation marks H3K9me1, H4K20me1, H3K4me3, H3K9me3 and H3K27me3 as determined by genomewide ChIP-seq from K562 cells. The majority of L3MBTL2 target sites did not show any of these methylation marks (none).
Given the good correlation of L3MBTL2 binding sites with promoter regions we also performed a ChIP-chip analysis for L3MBTL2, E2F6 and L3MBTL1 using arrays containing 1.5 kb for each of the ~24,000 known human promoters. (Table S2). Of the top 1000 ranked promoters (Table S3), we found 53% overlap between L3MBTL2 and E2F6 target sites (Figure 3C, S3A), supporting the biochemical data that L3MBTL2 and E2F6 cooperate functionally in a single protein complex. In contrast, there was only a modest (~16%) overlap among the top 1000 target genes of E2F6 and L3MBTL1 (Figure 3C). Moreover, there was only a minor (<10%; below random) overlap of genes targeted by E2F1 and L3MBTL2 (Figure S3A, B), consistent with the idea that PRC1L4 is not present on active genes bound by E2F1. However, we noticed ~32% overlap of genes co-targeted by E2F4, a repressive E2F-type transcription factor, and L3MBTL2 (Figure S3A). Further inspection revealed a 24.5% overlap (which is in the range of random overlap) between L3MBTL2, E2F6 and E2F4 binding sites (Fig. S6C; Table S4). Gene ontology analysis of the L3MBTL2-E2F6 target genes showed that over 30% of all genes are of unknown molecular function in the nucleus. The second largest group (~17%) included target genes that encode proteins involved in nucleic acid binding. Closer examination revealed that ribosomal protein and histone encoding genes constituted more than 40% of this group (Figure S3D).
To validate these ChIP-chip results in an independent experiment, we performed manual ChIP assays in MCF7 cells using two different L3MBTL2 antibodies (see Figure S3E) and confirmed that L3MBTL2 and E2F6 bind to the high confidence binding sites identified by ChIP-chip, for instance to the promoter regions of RAD51C, UXT, RPA2 and CDC7 (Figure S3E). We also examined if L3MBTL2 and E2F6 target the same sites in other cell lines, specifically HeLa and 293F. Our ChIP assay analysis suggested that the majority of sites are also occupied in 293F cells including CDC7, CSTF3, MCM3, UXT, RPA2, RAD51C, RFC3 and HOXC5. Some genes were found to be occupied in all three cell lines, such as CDC7, while certain genes, for instance MCM3, were occupied by L3MBTL2 and E2F6 in MCF7 and 293F, but not in HeLa cells (Figure S3F, G). Well known PcG protein target genes like HOXC13 and MYT1 were not found among L3MBTL2 target genes, either by ChIP-chip or a manual ChIP assay approach (Table S3, Figure S3G). Re-examination of the CDC7 gene by manual ChIP analysis (with primer sets that hybridize to proximal and distal regions of the CDC7 TSS) showed that L3MBTL2 binding was only evident at the proximal promoter and not 2 kb upstream or downstream of the TSS (Figure S3H).
To corroborate our findings and to explore how L3MBTL2-E2F6 binding sites vary across different cell models, we carried out genome-wide ChIP sequencing for L3MBTL2 and E2F6 in K562 erythroleukemia cells and confirmed a substantial (57%) overlap of L3MBTL2 genomic binding sites with E2F6 (Figure 3D). The vast majority of L3MBTL2 binding sites were identified around TSSs (Figure S4A). Analyses of E2F6 and L3MBTL2 binding sites in MCF7 and K562 revealed a significant number of common targets (compare Tables S3 and S4), indicating that L3MBTL2 and E2F6 co-occupied a number of promoters, irrespective of the cell type investigated.
L3MBTL2 genome binding sites do not correlate with H3K9me1 and H4K20me1
Next, we confirmed that components of the PRC1L4 complex were also found on L3MBTL2-E2F6 target genes. We detected RING2 and HPlγ in addition to L3MBTL and E2F6 on several target genes (Figure S4B). RING2 was also bound to the HOXC13 promoter but in this case independently of PRC1L4 since we could not detect L3MBTL2, E2F6 and HPlγ. None of the PRC1L4 components was present on the GAPDH promoter (Figure S4B). We were curious to see if the L3MBTL2-E2F6 binding sites are decorated with particular histone modifications given that the purified complex exhibited H2AK119-ubiquitin ligase activity. To this end, we used an antibody that specifically recognizes the H2AK119ub1 (Cao et al., 2005; Wang et al., 2004). As expected, we found H2AK119ub1 on L3MBTL2-E2F6 target genes such as RPA2 and CDC7, and on well characterized PcG target genes such as HOXC13, but not on GAPDH (Figure S4B). Given that the fourth MBT-domain of L3MBTL2 was shown previously to bind directly to H3K9me1 and H4K20me1 in vitro, we explored if L3MBTL2 binding correlated with these modifications. Surprisingly, ChIP-seq revealed that L3MBTL2 binding sites were completely devoid of the presence of H3K9me1 and H4K20me1 on a genome scale (Figures 3E and S4E, F).
PRC1 binding sites are often correlated with the presence of H3K27me3 that is catalyzed by PRC2, thereby functionally linking these two PcG complexes. Interestingly, we did not find H3K27me2 or -me3 on all inspected L3MBTL2-E2F6 target genes (data not shown). A genome-wide comparison of L3MBTL2-E2F6 binding sites with H3K27me3 confirmed that there is only a poor correlation (Figure 3E), indicating that PRC1L4 recruitment and function is independent of H3K27me3. Given that the PRC1L4 component HPlγ recognizes H3K9me3, we also compared PRC1L4 occupancy with H3K9me3, but failed to detect any correlation (Figure 3E). Interestingly, a genome-wide comparison of L3MBTL2 binding sites with H3K4me3 did reveal a certain degree of correlation (Figure 3A and S4C). However, the vast majority of L3MBTL2 binding sites lacked H3K4, H3K9 and H3K27 trimethylation (Figure 4B and S4D-F).
Figure 4. L3MBTL2 functions in transcriptional repression.

(A) (Upper panel) A schematic of the stably integrated luciferase reporter used to study the effect of L3MBTL2 on transcription is shown. The luciferase transgene is under the control of a promoter containing GAL4 DNA recognition sequences. Changes in luciferase activity were monitored upon induction of GAL4-L3MBTL2 with doxycycline (+DOX; see schematic). (Lower, left panel) Western blot analysis of whole cell extracts prepared from cells containing either empty vector (GAL4) or a vector expressing GAL4-L3MBTL2 using the antibodies indicated on the right. Molecular weight markers are indicated on the left. (Lower, right panel) Bar graph of luciferase activity scored as a function of doxycycline treatment of cells expressing GAL4 or GAL4-L3MBTL2, as indicated. (B) Examples of MCF7 1.5 kb promoter array ChIP-chip and K562 ChIP-seq signals illustrate a similar binding pattern for E2F6 and L3MBTL2 in both cell types. Shown are the CDC7 and UXT genes with ChIP-chip data shown on the top row and ChIP-seq data on the bottom. The plots show unprocessed enrichment ratios for L3MBTL2 (red) and E2F6 (blue) for all probes within a genomic region (ChIP versus whole genomic DNA) and peak tag numbers (ChIP-seq). (C) E2F6 protein levels were reduced by various amounts of siRNA (left panel). Transcript levels for E2F6, CDC7 and UXT in 293F cells treated with 5 nM of either Control (Ctrl) or E2F6 siRNA were determined by qPCR (right panel). GAPDH transcript levels were used to normalize the data across samples. Data are represented as the average of three independent experiments ±SD. (D) L3MBTL2 protein levels were effectively reduced by two distinct siRNAs. GAPDH siRNA served as control (left panel). Transcript levels of L3MBTL2, CDC7 and UXT upon treatment with 5 nM of Control, GAPDH or L3MBTL2 siRNAs were determined by qPCR. B2M transcript levels were used to normalize the data across samples. Data are represented as the average of three independent experiments ±SD.
L3MBTL2 functions as a transcriptional repressor and affects PRC1L4 chromatin residency
To gain insight into L3MBTL2 transcriptional functions, we took advantage of a stably integrated luciferase reporter system successfully used previously (Vaquero et al., 2004). This cell line constitutively expresses the tetracycline repressor protein (TetR) and luciferase gene expression is under the control of a promoter containing GAL4 DNA recognition sequences (Figure 4A, top panel). We stably transfected these cells with constructs that express either the GAL4 DNA binding domain (GAL4) or GAL4 fused to L3MBTL2 (GAL4-L3MBTL2), under the control of a Tet-operator sequence. GAL4-L3MBTL2 was expressed upon addition of doxycycline (DOX) as evidenced by western analysis of cell extracts using either anti-GAL4 or anti-L3MBTL2 antibodies (Figure 4A, bottom left panel). The presence of GAL4-L3MBTL2 also correlated with a significant reduction in luciferase gene expression (Figure 4A, bottom right panel), suggesting that L3MBTL2 functions as a transcriptional repressor.
Given that E2F6 was previously implicated in transcriptional repression (Giangrande et al., 2004; Oberley et al., 2003; Ogawa et al., 2002; Pohlers et al., 2005; Storre et al., 2005; Xu et al., 2007), we examined its transcriptional impact in proliferating MCF7 cells that were treated with control or E2F6 siRNAs. Effective down-regulation of E2F6 was confirmed at the protein and transcript levels (Figure 4B, left and right panels, respectively). CDC7 and UXT gene promoters were directly bound by E2F6 and L3MBTL2 as determined by ChIP-chip (Figure 4C). Upon E2F6 knockdown all these genes showed increased expression, strongly suggesting that E2F6 was functionally repressive (Figure 4B). A similar analysis was performed in the case of L3MBTL2. Effective down-regulation of L3MBTL2 was confirmed at the protein and transcript levels (Figure 4D, left and right panels, respectively). Similar to the case of E2F6 knockdown, CDC7 and UXT showed increased expression upon L3MBTL2 knockdown, confirming that L3MBTL2 and E2F6 were functionally repressive (Figure 4D, right panel).
Next, we generated 293F cell lines that constitutively express L3MBTL2 specific short hairpin RNAs (shRNAs). Two of five tested shRNAs (#4 and #5) significantly reduced the levels of L3MBTL2 transcript and protein (Figures 5A and B, respectively). L3MBTL2-E2F6 target gene expression was increased in cells containing L3MBTL2 shRNAs compared to those expressing control shRNAs (Figure 5C), while the expression of E2F6 was not affected (Figure 5C). We also observed an increase in CDC7 and UXT protein levels upon L3MBTL2 knockdown, while HPlγ levels were unchanged (Figure 5D). We detected a significant up-regulation of RPA2 transcript levels in the context of stable L3MBTL2 knockdown (Figure 5C) although this was not reflected by a detectable difference in global RPA2 protein levels (Figure 5B). We conclude that L3MBTL2 functions as a transcriptional co-repressor on E2F6-L3MBTL2 occupied genes.
Figure 5. L3MBTL2 is required to promote H2AK119ub1 on PRC1L4 target genes.

(A) 293F cells with stably integrated, constitutively expressing Control (Ctrl) or L3MBTL2 shRNA constructs were analyzed for L3MBTL2 transcript levels by PCR. Five different L3MBTL2 shRNA constructs were used of which #4 and #5 showed substantial reduction of L3MBTL2 transcript. (B) As in (A) but L3MBTL2 protein levels were analyzed by western blot. GAPDH levels served as loading control. RPA2 protein levels were only marginally increased upon significant decrease in L3MBTL2 levels. (C) 293F ells expressing Control or L3MBTL2 shRNAs (#4 and #5) were analyzed for the transcript levels of L3MBTL2, E2F6, CDC7, UXT, and RPA2 by qPCR. B2M transcript levels were used to normalize the data across samples. Data are represented as the mean of three independent experiments ±SD. (D) 293F cells expressing Control (Ctrl) or L3MBTL2 shRNAs were analyzed for the protein levels of L3MBTL2, UXT and CDC7. β-ACTIN and HPlγ served as loading controls. (E) ChIP assays from 293F cells with stably integrated, constitutively expressing Control (Ctrl) or L3MBTL2 shRNA (#4) to determine occupancy of PRC1L4 complex components on various L3MBTL2 target genes (indicated at the bottom of each graph). L3MBTL2 (left), H2AK119ub1 (middle) and E2F6 (right) specific antibodies were used for ChIP. Relative enrichments were determined by qPCR and represented as specifically precipitated material above IgG control and normalized to Input DNA. Shown is a representative of three independent experiments.
H2AK119ub1 is dependent on the presence of L3MBTL2
To study how L3MBTL2 affects chromatin residency of the PRC1L4 complex, we performed ChIP analysis in 293F cells comparing chromatin binding of PRC1L4 components in cells that are wild type versus those expressing L3MBTL2 shRNA. On several inspected target genes, L3MBTL2 occupancy was significantly reduced upon shRNA mediated L3MBTL2 knockdown (Figure 5E, left panel). Most importantly, H2AK119ub1 levels decreased accordingly on these promoters, while those of E2F6 were virtually unchanged (Figure 5E, middle and right panels, respectively). This suggested that L3MBTL2 is important in promoting H2AK119ub1. Furthermore, while E2F6 was still able to bind to these target genes, its ability to repress transcription was impaired upon loss of L3MBTL2, suggesting that one mechanism of E2F6-mediated repression is through the recruitment of the H2AK119ub1 machinery in an L3MBTL2-dependent manner.
L3MBTL2 binds chromatin independently of histone modifications
Collectively, our results suggest that L3MBTL2 acts as a repressor of transcription in association with E2F6. Given the presence of its MBT domains, L3MBTL2 function might be dependent on histone methyl-lysine binding. A recent study demonstrated that the four MBT domains of L3MBTL2 bound to mono- and di-methylated histone lysine residues. A co-crystal structure revealed that the fourth MBT domain accommodated the H4K20me1 histone peptide (Guo et al., 2009).
We focused on the chromatin binding properties of full-length L3MBTL2 protein using GST pull-down experiments with various candidate histone species, initially in vitro. GST-L3MBTL2 precipitated native and recombinant histone octamers (Figure 6A) and did not require the N-terminal histone tails of H3 and H4 for this interaction (Figure 6B). This was surprising given that all the methylated lysines shown to bind L3MBTL2 are located on the N-terminal regions of histones H3 and H4 (Guo et al., 2009). Thus, we performed streptavidin pull-down experiments with biotinylated peptides that correspond to the N-terminal regions of H3 and H4. Both peptides, although unmethylated, interacted with L3MBTL2 while the H4-specific histone lysine methyltransferase PR-SET7 only bound to the H4 peptide, as expected. The three MBT domains of L3MBTL1 did not bind any peptide (Figure S5), consistent with previous reports demonstrating that L3MBTL1 binding to H4 requires mono-methylation (Trojer et al., 2007).
Figure 6. L3MBTL2 compacts nucleosomal arrays devoid of histone modifications.
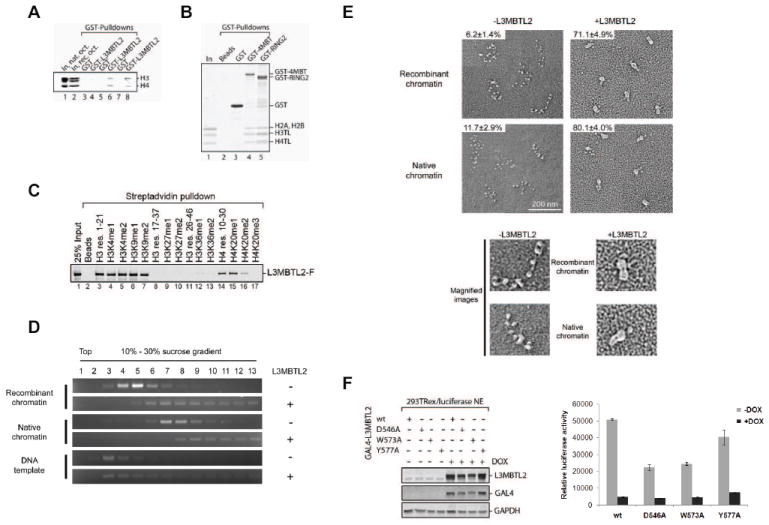
(A) GST pull-down experiment in which recombinant GST (lanes 3, 5 and 7) or GST-L3MBTL2 (lanes 4, 6 and 8) were incubated in the absence of histones (lanes 3 and 4) or with native (lanes 5 and 6) or recombinant (lanes 7 and 8) histone octamers and precipitated with Glutathione Sepharose. Shown are inputs (lanes 1-2) and precipitates (lanes 3-8) resolved by SDS-PAGE with proteins visualized by CBB-staining. Notably, histone octamers separate as histone H2A-H2B dimers and H3-H4 tetramers in low salt solutions. Only H3 and H4 were precipitated in the presence of GST-L3MBTL2. (B) GST pull-down experiment as described in (A) but using Beads, GST, GST-4MBT (comprising the four MBT domains of L3MBTL2) and GST-RING2 with recombinant H3 and H4 tail-less (TL) histone octamers. (C) Biotinylated peptides comprising the N-terminal amino acid residues of histones H3 (res. 1-21, 17-37 and 26-46) and H4 (res. 10-30) with the methylated lysines indicated on top were incubated with full-length, FLAG-tagged L3MBTL2. Streptavidin agarose was used to precipitate peptides and peptide-bound L3MBTL2. Shown is a western blot of the input (In) and precipitates (lanes 2-17) using anti-FLAG(M2) antibodies. (D) Sucrose gradient sedimentation of recombinant, full length L3MBTL2 in the absence or presence of recombinant and native chromatin. Fractions of the sucrose gradients were analyzed for the presence of chromatin by agarose gel electrophoresis and subsequent staining with ethidium bromide. (E) Electron micrographs visualizing recombinant and native oligonucleosomal arrays in the absence or presence of L3MBTL2. The percentage of compacted molecules is indicated on the top left of each image. The lower panel shows a magnification of a single oligonucleosomal array in an extended (-L3MBTL2) or compacted (+L3MBTL2) conformation. (F) Expression constructs encoding GAL4 fusion proteins containing L3MBTL2, either wild type or with single amino acid substitutions in the fourth MBT domain (D546A, W573A, Y577A), were transfected in 293 T-ReX cell lines harbouring a luciferase transgene. The effect on luciferase activity was measured upon induction of GAL4-L3MBTL2 expression with DOX. Whole cell extracts were prepared and analyzed for the induction of GAL4-L3MBTL2 protein by western blot (left panel). Luciferase activity was significantly reduced by GAL4-L3MBTL2, irrespective of an intact methyl-binding pocket of the fourth MBT domain (right panel).
That L3MBTL2 bound to unmethylated histones and in a manner independent of the presence of the histone H3 and H4 tails was unexpected. We therefore performed a comprehensive comparison of methylated and unmethylated peptides with respect to their ability to bind full-length L3MBTL2. L3MBTL2 bound to peptides corresponding to the first 21 amino acids of histone H3 regardless of H3K4 or H3K9 mono- and di-methylation (Figure 6C, lanes 3-7). Similarly, histone H4 peptides (residues 10-30) were bound by L3MBTL2 in the absence or presence of H4K20me1 and -me2 (Figure 6C, lanes 14-16). Only tri-methylation of H4K20 prevented L3MBTL2-binding (Figure 6C, lanes 14-17). Our data suggest a sequence specific and not a charge density specific binding event given that peptides corresponding to H3 residues 17-37 did not interact with L3MBTL2, irrespective of the presence or absence of H3K27 or H3K36 methylation (Figure 6C, lanes 8-13). We conclude that there is no significant preference for mono- and di-methylated H3K4, H3K9 or H4K20 peptides, compared to their unmethylated counterparts.
We next reconstituted chromatin with recombinant and native histones and fractionated them in the presence or absence of full length L3MBTL2 by sucrose gradient sedimentation (as described in (Sims et al., 2006). The L3MBTL2-chromatin complex was shifted towards the bottom of the gradient, relative to chromatin alone, suggesting a chromatin conformational change upon L3MBTL2-binding (Figure 6D). L3MBTL2 binding to chromatin was observed with recombinant and native chromatin, suggesting that posttranslational modifications on histones did neither facilitate nor preclude L3MBTL2 binding. To further explore the effects of L3MBTL2 binding to recombinant and native chromatin we examined chromatin in the absence or presence of L3MBTL2 by electron microscopy. Electron micrographs illustrated that chromatin was compacted upon the addition L3MBTL2, regardless of the presence of posttranslational modifications on chromatin (Figure 6E).
Since L3MBTL2 binding to and compaction of chromatin occurred in the absence of methyl-lysines, we next tested for L3MBTL2-mediated repression as a function of the absence of any of its three amino acid residues shown to be essential for histone methyl-lysine binding in structural studies. We generated GAL4-L3MBTL2 constructs encoding an alanine substitution mutant at either residue D546, W573 or Y577 and compared cells stably transfected with these candidates relative to the GAL4-wild type L3MBTL2 case in the luciferase reporter system. GAL4-LMBTL2, either wild type or mutant versions were detected in extracts after cells were treated for 48 hours with DOX (Figure 6F, left panel). Similar to the wild type case, the mutant versions of GAL4-L3MBTL2 led to strong repression of luciferase expression (Figure 6F, right panel). This result suggested that caging of the methylated histone-lysines does not contribute to the steady state transcriptionally repressive effect of L3MBTL2 when tethered to a promoter. To rule out that the mutant L3MBTL2 candidates might exhibit a defect during the early onset of L3MBTL2-mediated gene silencing in this system, we scored for luciferase expression as a function of time (3, 8, 12 and 24 hours) post doxycycline treatment. No such defect was detected as luciferase repression progressed during the first 12 hours and reached a steady state level at 24 hours post-induction in the case of wild type as well as mutant L3MBTL2 proteins (Figure S6).
DISCUSSION
Our findings here demonstrated that the MBT domain containing protein L3MBTL2 interacted with a number of PcG proteins previously identified in PRC1-like (PRC1L) complexes. Interestingly, these PRC1L complexes purified from human cells contain overlapping but not identical subunit compositions (Table 1). RING1 and RING2 are invariant components found in all PRC1L complexes and are required to mediate H2AK119ub1 (de Napoles et al., 2004). We suggest that a major determinant for the classification of human PRC1L complexes be the presence of only one of six RING domain containing human homologs of Drosophila Psc, termed BMI1/PCGF4, MEL18/PCGF2, MBLR/PCGF6, PCGF1, PCGF3 and PCGF5. The L3MBTL2-complex purified here contained several PRC1 subunits including RING1, RING2 and the Psc homolog MBLR, and exhibited H2AK119ub1 E3-ligase activity. We therefore termed this complex PRC1L4 (Table 1). Complexes containing BMI1/PCGF4, MEL18/PCGF2 and PCGF1 have been reported earlier (Table 1) and we predict that at least two more PRC1L complexes will be discovered, being characterized by the presence of PCGF3 and PCGF5, respectively (Table 1). This claim is supported by a recent proteomics study focused on identifying RING2 interaction partners in which all six PCGF homologous proteins were recovered (Sanchez et al., 2007).
Table 1.
Polycomb Repressive Complex 1 (PRC1) variations in human cells
| Human PRC1-like complex variations | Complex composition
|
Reference | ||
|---|---|---|---|---|
| Human homologs of Drosophila Posterior sex comb (Psc) | Human homologs of Drosophila Ring (Invariant subunits) | Other (variable) subunits | ||
| PRC1 | BMI1/PCGF4 | RING1/RING2 | PHC1, PHC2, PHC3, SCMH1, HPC1, HPC2 | Levine et al, 2002 |
|
| ||||
| PRC1L1 | BMI1/PCGF4 | RING1/RING2 | HPH2 | Wang et. al., 2004 |
|
| ||||
| PRC1L2/BCOR complex | NSPC1/PCGF1 | RING1/RING2 | BCOR, FBXL10, RYBP, SKP1 | Gearhart et al., 2006 |
|
| ||||
| PRC1L3/melPRC1 | MEL-18/PCGF2 | RING1/RING2 | HPH2, CBX8 | Elderkin et al., 2007 |
|
| ||||
| PRC1L4 | MBLR/PCGF6 | RING1/RING2 | L3MBTL2, HP1γ, E2F6 | this study |
|
| ||||
| PRC1L5 | PCGF3 | RING1/RING2 | ? | |
|
| ||||
| PRC1L6 | PCGF5 | RING1/RING2 | ? | |
The direct physical interaction between L3MBTL2 and multiple PcG proteins suggests a tight functional cooperation, and given that RING1, RING2 and E2F6 null mice showed similar developmental defects (del Mar Lorente et al., 2000; Storre et al., 2002; Voncken et al., 2003), we anticipate that mice null for L3MBTL2 will also exhibit abnormalities during embryogenesis. Previously, a larger protein assembly containing L3MBTL2 was shown to repress genes in quiescent cells (Ogawa et al., 2002). Importantly, we have shown here that L3MBTL2 and PRC1L4 also play a gene regulatory role in actively dividing cells which is consistent with earlier studies that implicated E2F6 in gene regulatory events at particular cell cycle stages (Giangrande et al., 2004). Prior to our study, E2F6 was shown to interact with a number of different PcG proteins (Attwooll et al., 2005; Deshpande et al., 2007; Ogawa et al., 2002), and we speculate that such interactions might arise as a function of and/or be specific to cell cycle stages and/or differentiation states of the cell.
In this study and a previous one (Trojer et al., 2007), we investigated two MBT-domain containing proteins L3MBTL2, and -L1, as to their functional import in gene regulation. Our results showed negligible genomic co-occupancy of L3MBTL1 with L3MBTL2 (Figure 3C, S3A), suggesting that the two disparate MBT-domain containing proteins function independently and in a non-redundant manner. H2AK119ub1 was present on all L3MBTL2 target genes tested. Its catalysis by PRCIL4 likely bears directly on L3MBTL2-mediated repression given that L3MBTL2 down-regulation not only resulted in up-regulated gene expression, but also in loss of H2AK119ub1 (Figure 5E). Since L3MBTL2 did not stimulate RING2 E3-ligase activity in vitro (data not shown), we conclude that L3MBTL2 is an important factor in recruiting RING1 and RING2 on PRC1L4 genomic binding regions.
In earlier reports, the removal of H2AK119ub1 correlated with an increase in productive transcript but did not change overall RNA polymerase II levels on target promoters, suggesting that H2AK119ub1 affects the establishment of a mature elongation complex (Stock et al., 2007; Zhou et al., 2008). Consistently, studies established that H2AK119ub1 suppresses ongoing transcription proximal to double strand breaks (DSB) in an ATM-dependent manner. DSB repair leads to rapid transcriptional de-repression and coincides with H2AK119ub1 loss (Shanbhag et al., 2010). However, the molecular mechanism of H2AK119ub1-mediated transcriptional repression remains unknown such that it is still possible to consider that H2AK119ub1 might directly obstruct elongation factor dependent chromatin disassembly or prevent transcription initiation (Nakagawa et al., 2008). Interestingly, we have found a correlation of PRC1L4 occupancy with H3K4me3 (Figure 3E, S4C-F). This finding supports H2AK119ub1 functioning as a roadblock in elongation on the H3K4me3-marked subset of PRC1L4 target genes, given that H3K4me3 is placed co-transcriptionally and not prior to transcription initiation (Pavri et al., 2006) and that H3K4me3 marked genes might lack productive transcripts but have an already initiated and probably stalled RNA polymerase II transcription complex in their 5’-region. A recent study suggested that PRC1L complexes repress transcription of Hox genes independently of H2AK119ub1 since both a Ring1b (the mouse homolog of human RING2) wild type and E3-ligase deficient mutant could rescue Hox gene cluster chromatin decondensation in mouse embryonic stem cells (Eskeland et al., 2010). It is likely that loss of RING2 would destabilize the entire PRC1L complex, and thus up-regulate gene expression. More mechanistic studies are required to examine the direct effect of H2AK119ub1 on chromatin structure.
Surprisingly, we did not find any correlation of L3MBTL2 binding sites and the occurrence of repressive histone methylation marks in our genome-wide ChIP-seq analyses. The lack of H3K27me3 on PRC1L4 target genes clearly indicated that the recruitment and repressive function of PRC1L4 is independent of the H3K27me3. Moreover, the lack of H3K9me3 on PRC1L4 target genes suggested that PRC1L4 recruitment is not dependent of HP1γ-mediated binding to H3K9me3. These findings are also consistent with our earlier data from Ntera2 cells in that E2F6 binding sites did not correlate with the presence of H3K9me3 or H3K27me3 (Xu et al., 2007).
Previously, the four MBT domains of L3MBTL2 were shown to bind to H4K20me1 and H3K9me1 histone peptides by isothermal calorimetry (ITC) and a crystallographic analysis determined multiple aromatic residues in the fourth MBT domain to be critical for histone methyl-lysine binding (Guo et al., 2009). Surprisingly, we did not find any correlation of H4K20me1 and H3K9me1 with L3MBTL2 binding sites on a genome scale (Figure 3E), suggesting that these two marks do not play a major role in L3MBTL2 recruitment to chromatin. Recombinant, full-length L3MBTL2 bound to unmodified and mono- and di-methylated histone peptides in a similar manner (Figure 6C). Also peculiar is the lack of binding to H3K27 methylated peptides (Figure 6C), while such binding was previously detected by ITC. This could be explained by the weak binding affinity (KD =40-60 μM) towards H3K27me1 and -me2. Alternatively, the peptide length and the position of the methylated lysine within the peptide sequence might contribute to the observed differences in binding. Regardless, we found that L3MBTL2 could bind equally effectively to recombinant and native histones (Figure 6A), and its histone binding was not dependent on the N-terminal H3 and H4 sequences (Figure 6B). We concluded that in the context of full-length L3MBTL2 histone recognition did not require histone lysine methylation. Formally, it is still possible that L3MBTL2 has a binding preference for methyl marks on the linker histone, for instance H1.4K26 methylation.
While L3MBTL1 required H1.4K26 methylation for chromatin compaction, L3MBTL2 interacted with chromatin devoid of histone H1 or histone modifications (Figure 6D). Electron microscopy allowed us to visualize chromatin compaction resultant to L3MBTL2 binding (Figure 6E). Thus, L3MBTL2 likely functions like the Psc subunit of PRC1 (Francis et al., 2004), that mediates chromatin compaction in a histone modification-independent manner.
EXPERIMENTAL PROCEDURES
Biochemical purification of L3MBTL2-F and associated polypeptides
Nuclear extracts (~350 mg) were prepared from 30 l of culture (1.2×10ˆ6 cells/ml) following the Dignam protocol (Dignam et al., 1983). For a detailed description of the biochemical purification see Supplemental Data.
Antibodies, immunoprecipitations (IPs)
For a description of the antibodies used in this study see Supplemental Data.
In vitro ubiquitin ligase assays
Reactions were carried out as previously described (Wang et al., 2004). Briefly, reconstituted, recombinant oligonucleosomes (approx. 5 μg) were incubated with protein fractions or recombinant proteins for 1 h at 37°C in a 40 μl reaction in ubiquitin ligase buffer (50 mM Tris-HCl, pH 7.9, 5 mM MgCl2, 2 mM NaF, 0.6 mM DTT, 2 mM ATP) with 0.1 μg ubiquitin activating enzyme E1 (Boston Biochemicals), 0.6 μg ubiquitin conjugating enzyme GST-UbcH5C (Boston Biochemicals) and 1 μg FLAG-ubiquitin (Sigma). Reactions were mixed with Laemmli sample buffer, resolved by 15% Tris/glycine SDS-PAGE and subsequent western blotting.
Sucrose gradients and EM
These experiments were carried out as described previously (Trojer et al., 2007).
Determination of transcript levels
Total RNA was isolated from cells using TRIzol® Plus (Invitrogen). RNA concentration was determined by a NanoDrop spectrophotometer and 1.5 μg used for the generation of cDNA using the Superscript III kit and Random Primers (Invitrogen). PCRs were carried out using a Mx3000P qPCR instrument (Stratagene) with transcript specific primer sets and amplicon specific UPL probes (Roche). Primer sequences are available upon request. Relative changes in transcript levels were calculated using the ΔΔCt method.
Chromatin Immunoprecipitation (ChIP), ChIP-chip and ChIP-seq analysis
ChIP assays were performed as previously described (Trojer et al., 2007). ChIP samples were prepared from HEK293, HeLa, MCF7, 293F/shControl and 293F/shL3MBTL2. Precipitated DNA was amplified using an Mx3000P qPCR instrument (Stratagene) with genomic location specific primer sets and amplicon specific UPL probes (Roche). ChIP primer sequences are available upon request. Relative enrichment of specifically bound material above an IgG control background and normalized to Input DNA was quantified using the ΔΔCt method. For a detailed description of ChIP-chip and ChIP-seq experiments see Supplemental Data. The E2F6, L3MBTL1, and L3MBTL2 ChIP-chip and the ChIP-seq data are available at the GEO repository of the NCBI under provisional accession numbers GSE28161 and GSE28162, respectively. The H3K9me3 ChIP-seq data (GSM607493) and the input used for scoring the ChIP-seq datasets (GSM467649) have been previously submitted to GEO.
Supplementary Material
HIGHLIGHTS.
L3MBTL2 is a component of a PcG protein complex with H2AK119ub1 E3-ligase activity
L3MBTL2 is preferentially bound to TSSs and functions as transcriptional repressor
L3MBTL2 and E2F6 cooperate on a genome scale to regulate transcription
L3MBTL2 compacts chromatin independent of histone lysine methylation marks
Acknowledgments
We are grateful to Dr. Lynne Vales for critical reading of our manuscript and for helpful discussion. We thank Drs. Y. Zhang and S. Novak for providing reagents. We thank the members of the Farnham lab for helpful discussion, especially Kimberly Blahnik for helping with the bioinformatics work required for the data analysis. We also thank Dr. Pierre Chambon for reagents and reading of the manuscript. This work is supported by grants from the National Institutes of Health (GM-64844 to D. R. and CA45250 and 1U54HG004558 to P. F.), the National Cancer Institute (Support Grant P30 CA08748 to H. E.-B. and P. Tempst) and the Howard Hughes Medical Institute to D. R.
We would like to bid our last farewell to Dr. Regine Losson, a great person and excellent scientist who untimely passed away after courageously battling cancer.
Footnotes
SUPPLEMENTAL DATA Data include Full Experimental Procedures, 6 Figures and Figure Legends and 5 Supplemental Tables and can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Attwooll C, Oddi S, Cartwright P, Prosperini E, Agger K, Steensgaard P, Wagener C, Sardet C, Moroni MC, Helin K. A novel repressive E2F6 complex containing the polycomb group protein, EPC1, that interacts with EZH2 in a proliferation-specific manner. J Biol Chem. 2005;280:1199–1208. doi: 10.1074/jbc.M412509200. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Lecona E, Reinberg D. MBT domain proteins in development and disease. Semin Cell Dev Biol. 2009 doi: 10.1016/j.semcdb.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- del Mar Lorente M, Marcos-Gutierrez C, Perez C, Schoorlemmer J, Ramirez A, Magin T, Vidal M. Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development. 2000;127:5093–5100. doi: 10.1242/dev.127.23.5093. [DOI] [PubMed] [Google Scholar]
- Deshpande AM, Akunowicz JD, Reveles XT, Patel BB, Saria EA, Gorlick RG, Naylor SL, Leach RJ, Hansen MF. PHC3, a component of the hPRC-H complex, associates with E2F6 during G0 and is lost in osteosarcoma tumors. Oncogene. 2007;26:1714–1722. doi: 10.1038/sj.onc.1209988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eryilmaz J, Pan P, Amaya MF, Allali-Hassani A, Dong A, Adams-Cioaba MA, Mackenzie F, Vedadi M, Min J. Structural studies of a four-MBT repeat protein MBTD1. PLoS ONE. 2009;4:e7274. doi: 10.1371/journal.pone.0007274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, Bickmore WA. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- Giangrande PH, Zhu W, Schlisio S, Sun X, Mori S, Gaubatz S, Nevins JR. A role for E2F6 in distinguishing G1/S- and G2/M-specific transcription. Genes Dev. 2004;18:2941–2951. doi: 10.1101/gad.1239304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, de Ayala Alonso AG, Rybin V, Steuerwald U, Ly-Hartig N, Fischle W, Muller J, Muller CW. Structural and functional analyses of methyl-lysine binding by the malignant brain tumour repeat protein Sex comb on midleg. EMBO Rep. 2007;8:1031–1037. doi: 10.1038/sj.embor.7401085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Matos R, Ly-Hartig N, Steuerwald U, Lindner D, Rybin V, Muller J, Muller CW. Molecular recognition of histone lysine methylation by the Polycomb group repressor dSfmbt. Embo J. 2009;28:1965–1977. doi: 10.1038/emboj.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Nady N, Qi C, Allali-Hassani A, Zhu H, Pan P, Adams-Cioaba MA, Amaya MF, Dong A, Vedadi M, et al. Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2. Nucleic Acids Res. 2009;37:2204–2210. doi: 10.1093/nar/gkp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola TK. Polycomb group complexes--many combinations, many functions. Trends Cell Biol. 2009;19:692–704. doi: 10.1016/j.tcb.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Norman J, Shilatifard A, Shiekhattar R. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell. 2007;128:877–887. doi: 10.1016/j.cell.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Li H, Fischle W, Wang W, Duncan EM, Liang L, Murakami-Ishibe S, Allis CD, Patel DJ. Structural Basis for Lower Lysine Methylation State-Specific Readout by MBT Repeats of L3MBTL1 and an Engineered PHD Finger. Mol Cell. 2007;28:677–691. doi: 10.1016/j.molcel.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Allali-Hassani A, Nady N, Qi C, Ouyang H, Liu Y, Mackenzie F, Vedadi M, Arrowsmith CH. L3MBTL1 recognition of mono- and dimethylated histones. Nat Struct Mol Biol. 2007 doi: 10.1038/nsmb1340. [DOI] [PubMed] [Google Scholar]
- Muller J, Verrijzer P. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr Opin Genet Dev. 2009;19:150–158. doi: 10.1016/j.gde.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kajitani T, Togo S, Masuko N, Ohdan H, Hishikawa Y, Koji T, Matsuyama T, Ikura T, Muramatsu M, Ito T. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 2008;22:37–49. doi: 10.1101/gad.1609708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberley MJ, Inman DR, Farnham PJ. E2F6 negatively regulates BRCA1 in human cancer cells without methylation of histone H3 on lysine 9. J Biol Chem. 2003;278:42466–42476. doi: 10.1074/jbc.M307733200. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Pohlers M, Truss M, Frede U, Scholz A, Strehle M, Kuban RJ, Hoffmann B, Morkel M, Birchmeier C, Hagemeier C. A role for E2F6 in the restriction of male-germcell-specific gene expression. Curr Biol. 2005;15:1051–1057. doi: 10.1016/j.cub.2005.04.060. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C, Sanchez I, Demmers JA, Rodriguez P, Strouboulis J, Vidal M. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- Santiveri CM, Lechtenberg BC, Allen MD, Sathyamurthy A, Jaulent AM, Freund SM, Bycroft M. The malignant brain tumor repeats of human SCML2 bind to peptides containing monomethylated lysine. J Mol Biol. 2008;382:1107–1112. doi: 10.1016/j.jmb.2008.07.081. [DOI] [PubMed] [Google Scholar]
- Sathyamurthy A, Allen MD, Murzin AG, Bycroft M. Crystal structure of the malignant brain tumor (MBT) repeats in Sex Comb on Midleg-like 2 (SCML2) J Biol Chem. 2003;278:46968–46973. doi: 10.1074/jbc.M306469200. [DOI] [PubMed] [Google Scholar]
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Trojer P, Li G, Reinberg D. Methods to identify and functionally analyze factors that specifically recognize histone lysine methylation. Methods. 2006;40:331–338. doi: 10.1016/j.ymeth.2006.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Storre J, Elsasser HP, Fuchs M, Ullmann D, Livingston DM, Gaubatz S. Homeotic transformations of the axial skeleton that accompany a targeted deletion of E2f6. EMBO Rep. 2002;3:695–700. doi: 10.1093/embo-reports/kvf141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storre J, Schafer A, Reichert N, Barbero JL, Hauser S, Eilers M, Gaubatz S. Silencing of the meiotic genes SMC1beta and STAG3 in somatic cells by E2F6. J Biol Chem. 2005;280:41380–41386. doi: 10.1074/jbc.M506797200. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P, Li G, Sims RJ, 3rd, Vaquero A, Kalakonda N, Boccuni P, Lee D, Erdjument-Bromage H, Tempst P, Nimer SD, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- Trojer P, Reinberg D. Beyond histone methyl-lysine binding: how malignant brain tumor (MBT) protein L3MBTL1 impacts chromatin structure. Cell Cycle. 2008;7:578–585. doi: 10.4161/cc.7.5.5544. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Voncken JW, Roelen BA, Roefs M, de Vries S, Verhoeven E, Marino S, Deschamps J, van Lohuizen M. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci U S A. 2003;100:2468–2473. doi: 10.1073/pnas.0434312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Wang WK, Tereshko V, Boccuni P, MacGrogan D, Nimer SD, Patel DJ. Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets. Structure (Camb) 2003;11:775–789. doi: 10.1016/s0969-2126(03)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Zhai L, Xu J, Wang H. Role of Bmi1 in H2A ubiquitylation and Hox gene silencing. J Biol Chem. 2006;281:22537–22544. doi: 10.1074/jbc.M600826200. [DOI] [PubMed] [Google Scholar]
- Wu X, Gong Y, Yue J, Qiang B, Yuan J, Peng X. Cooperation between EZH2, NSPc1-mediated histone H2A ubiquitination and Dnmt1 in HOX gene silencing. Nucleic Acids Res. 2008;36:3590–3599. doi: 10.1093/nar/gkn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Bieda M, Jin VX, Rabinovich A, Oberley MJ, Green R, Farnham PJ. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 2007;17:1550–1561. doi: 10.1101/gr.6783507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell. 2008;29:69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


