Abstract
Plant growth and development is influenced by mutual interactions among plant hormones. The five classical plant hormones are auxins, cytokinins, gibberellins, abscisic acid and ethylene. They are small diffusible molecules that easily penetrate between cells. In addition, newer classes of plant hormones have been identified such as brassinosteroids, jasmonic acid, salicylic acid and various small proteins or peptides. These hormones also play important roles in the regulation of plant growth and development. This review begins with a brief summary of the current findings on plant hormones. Based on this knowledge, a conceptual model about interactions among plant hormones is built so as to link and develop an understanding of the diverse functions of different plant hormones as a whole in plants.
Key words: abscisic acid, auxin, brassinosteroids, cytokinins, ethylene, gibberellins, jasmonic acid, salicylic acid, plant peptide hormones
Introduction
Every stage of the plant's life cycle is regulated by plant hormones. In general, plant biological activity is manipulated by more than one hormone, thus the biological phenomenon often reflects the combined interplay of several different hormones. Meanwhile, unlike animals which can escape from harsh environments, plants can only survive through adjusting various biological activities when encountering biotic and abiotic stresses. During these situations, plant hormones also cooperate to modify biological responses for the formation and maintenance of plant stress tolerance.1 In this review we aim to develop a model that can be used as a foundation to investigate the interactions of plant hormones that allow plants to survive the onslaught of various environmental factors.
Hormones are compounds that work at low concentrations where they are able to signal and control the response, growth and development of living organisms via circulating through part or all of the organisms. The action of hormones involves processes of signal transduction. Signal transduction is a relay involving the conversion of intracellular or extracellular signals into cellular responses. Such signaling processes can be separated into three types according to transduction distances: (1) long distant (plant) or endocrine (animal) signaling where signal cells and target cells are at a long distance apart; (2) paracrine signaling where signal cells and target cells are adjacent cells; (3) autocrine signaling where signal cells and target cells are the same cells.2
The signaling process embodies synthesis of signal molecules (ligands), transport of signal molecules, binding of signal molecules with receptors, development of cellular responses and removal/degradation of signal molecules. A conformational change of receptor via ligand-binding plays a critical role in initiating the signal transduction process. Most receptors are present on the cell surface although some are localized in the cytosol. Furthermore, cell surface receptors can be classified as: receptors with intrinsic enzyme activities (e.g., receptor kinases), protein coupled receptors (e.g., G protein-coupled receptors) and ligandgated ion channel receptors.3 The cell surface receptors recognize and perceive stimuli (ligands) in the extracellular environment, thereafter they interact with the ligands and transduce the extra-cellular signal into the cells.
Usually when an extracellular signal molecule binds to its receptor, the initial cellular reaction involves activating the production of a second messenger or an intracellular signaling mediator, which consequently triggers a series of cellular responses. Substances acting as second messengers can vary in their chemical composition. They include lipids, sugar, ions, nucleotides or gases, such as Ca2+, cAMP, cGMP, cyclic ADP-ribose (cADPR), small GTPases (small or monomeric G proteins), 1,2-diacylglycerol (DAG), inositol-1,4,5-trisphosphate (IP3), NO, inositol phospholipids (phosphoinositides) and phosphatidic acid. Intracellular signaling mediators refer to several classes of intracellular signal proteins. They are mainly different kinds of enzymes including kinases, phosphatases, phospholipases, phosphodiesterases and so on.2,3 Notably, some of these intracellular signaling molecules are shared between different signaling pathways. Hence these molecules are most likely to serve as crosstalk points in the signaling network communication within the cell.
Auxin, cytokinin, gibberellin (GA), abscisic acid (ABA) and ethylene are well accepted as five classes of classic plant hormones. Nowadays, evidence has accumulated to extend this concept to include brassinosteroids, jasmonic acid and salicylic acid. Furthermore some biologically active peptides are also found to be key signaling players in many aspects of plant life, like systemin, polaris, phytosulfokines (PSKs), ENOD40, CLAVATA3 (CLV3), S-locus cysteine-rich proteins (SCPs) and plant natriuretic peptides (PNPs).4–6 Therefore it is likely that further newcomers may be added to this list in the future. One such example is strigolactones, which are essential for root colonization by arbuscular mycorrhizae but are also important hormonal compounds involved in inhibiting shoot branching.7 To understand signaling cross-talk among plant hormones, it is particularly important to elucidate the signaling mechanism of each plant hormone first. The main functions and signaling pathways of the major plant hormones are briefly reviewed as a prelude to developing a model to unravel the possible interactions among different hormones in terms of the dynamic maintenance of a normal plant.
Auxin
Auxin is an indispensable hormone for plant growth and development. It is synthesized from actively growing tissues such as shoot meristems, leaf primordia, young expanding leaves, developing seeds, fruits and pollens. A number of biological processes are regulated by auxin: cell division, cell expansion, ion fluxes, root initiation, phototropism, geotropism, apical dominance, ethylene production, fruit development, parthenocarpy, abscission and sex expression. One major cellular effect of auxin is to cause hyperpolarization of the plasma membrane. This is a necessary requirement for various auxin-triggered biological actions.1,8,9 Meanwhile, auxin also seems able to stimulate GA biosynthesis and suppress ethylene and ABA biosynthesis.10,11
Auxin has been one of the plant hormones attracting researchers to hunt for its perception mechanism(s). Several reports indicate that the levels of lysophospholipid and Ca2+ were increased after auxin addition.9 Thus these two components may be intracellular signals for auxin. Receptors for auxin-binding are separated into two types: soluble and those on the outer plasma membrane. Some soluble auxin-binding proteins appear to be directly linked with the plasma membrane ATPase as auxin binding can rapidly cause proton pumping,12 while others directly regulate gene expression and include TIR1, the auxin receptor. Auxin affects gene expression via interaction with the auxin responsive element (AuxRE) which contains the characteristic nucleotide sequence TGTCTC and is located in the promoter region of auxin-inducible genes. Transcription factors bind to AuxRE and are members of the auxin responsive factor (ARF) family which can be repressed by the Aux/IAA family of transcription factors. Aux/IAA is a target for ubiquitin degradation by TIR1 which is the auxin receptor and a member of the Skp1/Cullin/F-box (SCF) ubiquitin ligase (E3) family. The targeted Aux/IAA is degraded in the 26S proteasome after being attached to polyubiquitin chains and this process is accelerated by auxin application.8,9,13,14
The ubiquitin/proteasome system degrades targeted proteins. It plays a crucial role in maintaining proteomic plasticity in plants where it occupies nearly 6% of the proteome.15 In this system, the 76 amino acids protein ubiquitin acts as a covalent molecular tag, and is attached to a substrate by the catalytic activities of three enzymes named: E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating protein) and E3 (ubiquitin ligase), and this last group includes four different types of ligase enzyme (RING, HECT, U-Box and SFC). Ubiquitin chains are thus formed tagging the protein for degradation. The polyubiquitylated substrate is delivered to a proteasome via ubiquitinbinding proteins, and then is deubiquitylated and digested by various proteases and/or proteolytic complexes. The free ubiquitin molecules are then released and become available for another attachment. TIR1 and other auxin-signaling F-box proteins are the receptors for auxin which raises the question: are other members of the F-box protein family (>700 in Arabidopsis alone) functioning as plant hormone receptors? Many activities of plant hormones are reported to be closely linked with the ubiquitin/proteasome system.14–16
Cytokinin
Cytokinins have many functional activities throughout the life of plants. Cytokinins occur at highest concentrations in meristematic regions, roots, young leaves, developing fruits and seeds. Their main functions include regulation of cell division, induction of organ differentiation, control of stomatal movement, delay of chlorophyll breakdown and attenuation of leaf senescence.1,9 Two different kinds of cytokinin hormonal activities are present in plants. One is the local (paracrine/autocrine) activity that mainly regulates cell division; whereas the other is the long distance signaling (endocrine) that acts as a root to shoot signal to control the functional response of above ground organs.17,18
It is clear now that cytokinin is perceived by membrane located histidine kinase receptors. Signal transduction operates through a bicomponent system where cytokinins bind to the histidine kinases and then histidine phosphotransfer proteins transmit the signal to nuclear response regulators, leading to gene activation or repression.17,19 Moreover, cytokinins also influence gene expression in chloroplasts, revealing that its receptors may also be located in this subcellular site.9 Some research shows that auxin pretreatment may upregulate the expression of cytokinin receptors.20 Signaling manipulation of cytokinin shares some similarity with the auxin control mechanism(s). Cytokinin responsive genes are positively stimulated by transcription factors. These transcription factors are counteracted by a family of specific proteins which are also subjected to degradation by the ubiquitin recognizing proteasome system. Cytokinin promotes the degradation of these specific proteins.21,22 The removal of specific short-lived proteins may be essential feature of many hormone responsive processes. The unstable proteins possibly serve as transcriptional repressors.
Gibberellin
Gibberellins (GAs) stimulate plant elongation, promote flowering and release seed/tuber dormancy. It is believed that gibberellins are produced and function in the same cell acting as an autocrine signal.1 GAs are synthesized from geranylgeranyl diphosphate through several intermediate steps catalyzed by enzymes in plastids and the cytosol and bioactive GAs are deactivated by epoxidation or methylation processes.23 The regions of GA action are mainly located in the meristematic zone and transition zone.9,24,25
GA signaling also demonstrates the importance of protein degradation in plant hormone regulation. GA binds to a soluble, nuclear-localized receptor GA-INSENSITIVE DWARF1 (GID1) that in turn binds to members of a family of transcription repressors called DELLA after their signature Asp-Glu-Leu-Leu-Ala (DELLA) motif. This in turn activates the F-box proteins SLEEPY1 (SLY1)/GID2 culminating in ubiquitin mediated degradation of DELLA and transcription of GA responsive genes. In low GA levels, DELLA proteins sequester and inactivate PHYTOCHROME INTERACTING FACTORS (PIFs), which are basic helix-loop-helix transcription factors.14,15,26 The discovery that DELLA proteins sequester PIFs is particularly exciting as it provides a biochemical mechanism underpinning the involvement of GA in light regulated seedling development. Following GA perception, early responses of plants include the increase of intracellular Ca2+/calmodulin, decrease of intracellular pH and elevation of the second messenger cGMP.27
GA seems to cross-talk with several other hormones. In seeds, the GA-induced Ca2+/calmodulin signaling regulates the synthesis and secretion of hydrolase; while ABA blocks the expression of hydrolase.28 This may explain the antagonistic interaction between GA and ABA. GA shows overlap or downstream effects with auxin in numerous developmental reactions.24,29 One of the ultimate effects of GA is inducing cell elongation. The pre-requirement of cell elongation is cell wall plasticity and cytoplasmic protein synthesis.30 To achieve these results, the cooperation of other hormones such as auxin may be an underlying requirement. A recent study shows that auxin and DELLA proteins independently regulate the GA synthesis pathway and promote the accumulation of bioactive GAs.31 In addition, combined positive or negative interplays between GA and ethylene also exist, depending on internal and external conditions, tissue types and developmental stages.24 Transcript meta-analysis suggests that applying exogenous ethylene to plants represses expression of genes involved in GA metabolism while some ethylene synthesis genes are upregulated by GA treatment.24 Overall, GA functions in a complicated context with many interacting factors.
Abscisic acid
Abscisic acid (ABA) is a “stress hormone,” named for its roles in response to stress environments. One notable effect of ABA is causing stomatal closure and preventing water loss by transpiration.32 Besides the traits of stress response, ABA is also associated with normal physiological manipulations like storage of compounds, dehydration at later stages of embryogenesis, seed maturation, dormancy formation and abscission.9 ABA is a carotenoid derivative produced in chloroplasts and other plastids and its synthesis is increased under stress conditions such as drought, salinity and cold.1,33 ABA is also circulated throughout the plant as an inactive glucose ester that can be rapidly released into the active by β-glucosidases present in the apoplast and endoplasmic reticulum33 allowing a combination of long distance and paracrine/autocrine effects to occur.
Multiple candidates for ABA receptors have been reported but the core signaling pathway involves PYRABACTIN RESISTANCE (PYR)/REGULATORY COMPONENT OF ABA RECEPTOR (RCAR) family of ABA receptors. The PYR/RCAR receptors are found in the cytoplasm as an inactive dimer that dissociates upon ABA binding to inhibit PROTEIN PHOSPHATASE 2C (PP2C) activity and thereby allow SNF1-RELATED PROTEIN KINASE 2 (SnRK2) to activate various downstream effectors including ion channels and transcription factors. However other candidates including membrane bound proteins are still potential ABA receptors (reviewed in refs. 34–39); therefore ABA is capable of inducing different signaling effects. Phosphatidic acid and IP3 have been reported to be involved in the ABA signaling pathway where they probably act as second messengers. For instance in guard cells, IP3 activates Ca2+ channels within the endoplasmic reticulum and vacuoles, leading to the release of Ca2+ into the cytosol from internal stores.9 Along with the increase of Ca2+ level, Ca2+ inhibits the plasma membrane H+-ATPase to prevent K+ uptake and further drive K+, Cl−-efflux.40 As a result, guard cells are closed. Obviously, multiple signaling pathways are involved in ABA transduction, demonstrating that ABA functions in a network rather than in a chain. Signal molecules involved in ABA signaling include: increases in pH, phosphatidic acid, IP3, Ca2+, NO, cADPR, mitogen activated protein kinase (MAPK) cascades and so on.9
Interestingly, ABA biosynthesis is largely regulated by its endogenous levels via a positive feedback system.33,41 This kind of synthesis pattern can further stimulate ABA accumulation and represents a critical step of stress adaptation, since ABA plays an important role in stress responses by mediating considerable gene expression. ABA-responsive gene promoters often contain cis-acting elements. The cis-acting elements commonly have a G-box ACGT core motif. Most proteins binding to the DNA sequences with this ACGT core motif share a basic region that adjoins to a leucine zipper domain.34,42 However, these leucine zipper proteins are particular targets for ubiquitin mediated degradation involving RING E3 ligases.43 ABA can promote or protect proteins from ubiquitin mediated degradation.14,34
Ethylene
Ethylene is a gaseous hormone that activates fruit maturation, stimulates germination, accelerates senescence, inhibits elongation, increases horizontal growth, initiates adventitious roots and programs cell death. Ethylene is produced from 1-aminocyclopropane-1-carboxylic acid (ACC) and the production of this compound is rate-limited by the activity of ACC synthases, which are short-lived enzymes whose stability is increased by ethylene itself in a feed forward process.15 However, the production of ethylene is tissue-specific and depends upon the stage of plant development.1,44 Ethylene levels may also increase or decrease in response to abiotic and biotic stresses.9
Ethylene receptors like cytokinin receptors are members of the bicomponent regulatory system family. Ethylene binds to the receptors via a copper co-factor and ethylene binding induces a mitogen-activated protein kinase (MAPK) cascade.9,44 Ethylene receptors are localized in endoplasmic reticulum membranes. Since ethylene is a gas, such a binding site is in accord with the diffusion of ethylene in either aqueous or lipid environment. The ubiquitin 26S proteosome system plays a key part in ethylene responses. For instance the ETYLENE INSENSITIVE3 (EIN3) is constitutively synthesized and degraded by SCF E3 ligases co-assembled with F-box proteins in the absence of ethylene. The presence of ethylene inhibits EIN3 degradation and allows it to act as a transcription factor which means that a very rapid and robust response will occur when ethylene levels rise.15 In addition, microarray studies demonstrated that a high level of coordination in gene expression was present with ethylene, jasmonic acid and salicylic acid treatments. This implicates close linkages among these hormones.45
Brassinosteroid
Brassinosteroid is a plant steroid hormone. Brassinosteroids have been detected in pollens, leaves, shoots, flowers, stems and seeds, but not roots.1 They affect cell expansion and division, tissue differentiation, reproductive development and stress resistance.46 Brassinosteroids act in a paracrine/autocrine manner with limited signal transport between tissues and organs.47
Unlike animal steroids that are perceived by nuclear receptors, brassinosteroids are perceived by the leucine rich repeat receptor like kinases (LRR RLK), which are members of the family of membrane localized receptor kinases containing extracellular leucine rich repeat regions. Brassinosteroids bind to a subdomain of these repeats, thereby initiating intracellular signal transduction via activating a kinase cascade beginning with receptor autophosphorylation and culminating in altered gene expression.48 The sites of phosphorylation of the brassinosteroid receptor BRASSINOSTEROID INSENSITIVE1 (BRI1) and its downstream interacting partners are relatively well characterized.49,50 The homeostatic levels of brassinosteroids are manipulated by brassinosteroid dependent transcriptional controls whose genes are involved in brassinosteroid metabolism. Such controls lower brassinosteroid production by reducing the transcription of biosynthetic genes or increasing the transcription of catabolic genes, when brassinosteroid levels are too high.51
Jasmonic Acid
Jasmonic acid and its derivatives such as methyl jasmonate are cyclic fatty acid-derived regulators synthesized from linolenic acid.45 Jasmonic acid levels are the highest in areas of active division such as stem apex, young leaves, immature fruits and root tips.1 Jasmonic acid and its bioactive derivatives and precursors have various activities in plants. Firstly, jasmonic acid can inhibit germination in non-dormant seeds but stimulate germination of dormant seeds. Secondly, jasmonic acid also inhibits root-growth and tuber formation. Thirdly, insufficiency of jasmonic acid affects anther or ovule development and results in sterile flower organs. Fourthly, jasmonic acid may be associated with senescence.52
Many frontline plant defense genes are induced by jasmonic acid, which mediate numerous transcriptional responses to wounding, herbivory and pathogenesis.53 The signaling of jasmonic acid is via the ubiquitin system as the jasmonic acid receptor is CORONATINE INSENSITIVE1 (COI1) which is an F-box protein related to the auxin receptor TIR1. The other component of this system is the JA-ZIM domain (JAZ) repressor proteins which inhibit MYB2 directed transcription. JA binds COI1 and this assembles with JAZ to form an SCF complex that tags JAZ with ubiquitin for degradation via the 26S proeasome and so releases the transcription factors.14,15,54 Jasmonic acid is also a potent lipid regulator, playing a central role in regulating the biosynthesis of many secondary metabolites, so that plants can tolerate a wide variety of biotic and abiotic stresses.55
Salicylic Acid
Salicylic acid is a phenol hormone critical for signaling innate immunity in plants. It functions in flowering promotion, thermogenesis and disease resistance. Salicylic acid has been identified in leaves and reproductive structures.1
Like the immune system of vertebrates and insects, plants usually undergo apoptotic-like cell death in the sites surrounding pathogen entry, causing a hypersensitive response in the nearby region. During this period, plants continuously develop a broad range and long lasting defense to further similar infections, known as systemic acquired resistance.56 Salicylic acid is an endogenous signal for the activation of both local and systemic resistance and induces the production of “pathogenesis-related proteins” such as catalase, peroxidase and hydrolase.56,57 Microarray data demonstrates that salicylic acid treatments share a large number of coinduced and co-repressed genes with jasmonic acid treatments.58 Therefore, salicylic acid and jasmonic acid display cross regulation in gene expression, both being responsible for inducing the production of resistance related proteins.
The salicylic acid receptor may be a member of the large gene family, receptor like protein kinases (RLKs). Salicylic acid is able to induce the expression of numerous RLKs. The salicylic acid responsive RLKs often contain C-X8-C-X2-C motifs in the putative extracellular domain. These RLK genes also contain the TTGAC sequence in the promoter regions.59 Members of the RLK family bind to other ligands, such as brassinosteroids and several of the peptides discussed below.
Peptide Signaling Molecules
Peptide signaling molecules (or hormones) are a relatively new class of growth regulators in plants used across all the kingdoms that form a relatively ancient adaptation of paracrine/autocrine communication between cells. Several of the peptide signaling molecules have distinct roles in development where they regulate cell differentiation and organogenesis or modulating general growth in response to the environment. Other peptide signaling molecules seem to act as endogenous danger signals.6,60 Some relatively well known signaling peptides include: systemin, polaris, phytosulfokine (PSK), ENOD40, CLAVATA3 and the CLE family, S-locus cysteine rich proteins (SCP), PEP1, Rapid Alkalization Factor (RALF) and plant natriuretic peptide (PNP). Systemin is important in the induction of defense responses to wounding in Solanaceae species and is intimately associated with jasmonic acid signaling.61 Polaris and PSKs are required for proper root growth and vascular development. The expression of polaris is suppressed by ethylene and induced by auxin. PSK promotes cell proliferation and longevity and PSK-α appears to act in a cooperative manner with CLE41-44 to regulate vessel development. ENOD40 is associated with legume nodule formation. CLAVATA3 and other members of the CLE family regulate development of shoot and root apical meristems. SCP regulates pollen determination. PEP1 and RALF (and systemin) act as danger signals to stimulate the innate immune response in plants in response to pathogen attack. PNP plays a role in water and solute balance. Hence peptide hormones are involved in various biological processes, regulating growth, development, reproduction and defense responses of plants.4–6,60,62,63
Conserved domains of peptide hormones are found between animals and plants, indicating evolutionary parallels occurred in both kingdoms. However, receptors for peptide signaling molecules that have so far been identified are significantly different. In animals peptide hormones are generally perceived by tyrosine kinases and/or G protein-coupled receptors; whereas in plants peptide receptors are mainly serine/threonine kinases of the LRR RLK family which contain an extracellular domain with leucine-rich repeats, a transmembrane domain and a cytoplasmic kinase domain. These include the receptors of CLAVATA3, PSK and PEP1.6,64 Another interesting feature is that several of these receptors also contain a predicted guanylate cyclase domain embedded within their kinase domain,65 and in some instances this has been shown to have some catalytic activity.66 This indicates that multiple signaling systems may be controlled by the ligand binding the receptor.
Peptide signaling molecules are often generated from the cleavage of their precursor polypeptides. Proteolysis is the common cleavage manner by various peptidases located within the cell and also extracellular. A polypeptide can be processed into multiple peptides, each with independent functions. The pep-tides can also undergo a series of post-translational modification such as hydroxyprolination, tyrosine sulphation or glycosylation before they can function properly. Such complicated maturation processes are usually completed in the endoplasmic reticulum or Golgi apparatus.6,67,68
Model of Hormone Interaction
Plants need to maintain homeostasis of water, ion and other nutrients for survival. Various themes can be elucidated from the above discussion where plants have developed common signaling processes. One such process involves the ubiquitin 26 proteasome as a means to remove abnormal and useless proteins or, in the case of hormone signaling, proteins that are no longer required.14,15 Another common theme is that many of the plant hormone receptors identified to date are intracellular, which suggests that plants also have developed transport systems to not only to allow the hormone to move over long distances (where it does) but also to enter the destination cells. Auxin transport proteins are relatively well characterized and recently ATP binding cassette (ABC) transporters for ABA have been identified.35,39 Also another feature to emerge is that antagonist and competitive interactions are also able to reduce the dominance of one hormone. An example is the interplay between ABA and other hormones such as GA and auxin32 which is discussed in more detail below. Thus there is the possibility of several hormone signals acting on one response pathway simultaneously. The final output of a physiological activity can be viewed as a combined result of the effects of several different hormones that are raised in response to various biological stimuli. This results in signaling interaction networks which include positive and/or negative feedback loops rather than simple linear signaling pathways. The above overview was used to develop a hypothetical conceptual model (Fig. 1) about the signaling network of plant hormones. This overview has been developed employing the traditional Chinese thoughts regarding mutual restraint and mutual promotion within the natural world. The model is intended to build up a framework or guideline for research thinking and to assist in the development of testable hypotheses.
Figure 1.
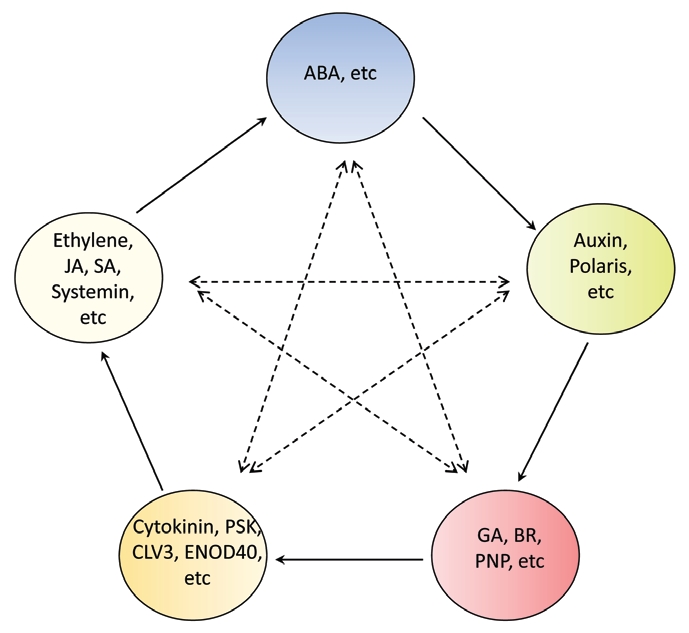
A conceptual model of potential interactions amongst plant hormones. The hormones within the same circle have complementary or similar actions. The solid arrows point to a new circle containing hormones whose action has been promoted or substantially prepared by the hormones in the previous circle. The dashed lines link the hormones that have competitive or antagonistic interactions in plants.
Since hormonal influences mainly affect regulation of gene expression and/or the interaction among signaling pathways, it is therefore essential to adopt two approaches to unravel the action of plant hormones. Firstly, a particular developmental process of plants is generally controlled by some key players in a temporal and spatial manner. Thus a single hormone influencing the quantitative and qualitative changes of gene expression becomes a crucial concern in a hormone study. Microarray studies are now identifying coordinated expression of genes by one or more hormones in time and spatially separated studies. This data can be used to develop critical points that need to be investigated to confirm hormone interaction and we will use an example to demonstrate how the model predicts such interactions. Secondly, in a systemic view, any biological phenomenon of plants is due to the mixed responses of different signaling factors operating via a complex and flexible network. Discovering the points of cross talk among various signaling pathways is a central feature to dissecting the integration of hormones. There appears to be mutual restraint and mutual promotion within plant hormones signaling networks and where this is described, it has influenced the development of the model. Hence during the dissection of a plant hormone signaling pathway, conflicting and complementary effects from other plant hormones should be monitored simultaneously (this is indicated by the dashed intersecting arrows in Fig. 1) to obtain a view of hormonal interaction in the plant as a whole.
How effective is this model in practice? We use a recent publication69,70 looking at whole plant responses in a spatial and temporal manner to test the model. The model predicts that the ethylene group will provide a fundamental state for the plant to respond to ABA and that ABA in turn does this for the auxin group. The model also predicts that the actions of the ethylene and ABA groups can be competed with or counteracted by GA for instance although the auxin group will compete/antagonize with the ethylene group and the cytokinin group with ABA. Skirycz et al.69 used microarray and metalabolomic analysis to probe leaf growth and development under mild growth limiting osmotic stress conditions. They found growing leaves with dividing and expanding cells were regulated by ethylene and GAs but not ABA, whereas mature leaves were regulated by ABA under these conditions.69,70 That is contrary to the current dogma, ABA did not affect the leaves undergoing division although it contributed to the response in the expanding cells and was the main player in mature leaves. These exciting results demonstrate that different hormonal responses are employed at different developmental stages to adapt young leaves to stress. Furthermore, the model independently developed in this review (Fig. 1) also predicts that these hormones could be candidates involved in the plant's adaptations to this environmental situation. Naturally, the model will need to be examined and modified further in practice.
Conclusion
The last decade has revealed a lot about plant hormone signaling pathways. Receptors for several hormones have been identified at the genetic level and confirmed biochemically and in some cases structural information has been gained. The common theme of protein degradation generally involving the ubiquitin 26S proteasome system has been clearly established. Microarray experiments are revealing coordinated interactions among plant hormones. The functional activity and signaling mechanism of plant hormones is a very complicated system where the entire biological response is the combined result of different hormone actions. We have used the available data to develop a conceptual model to describe how hormones interact. This model can be used to develop testable hypotheses and further insights into the mechanisms of maintenance of plant homeostasis by continuing to explore coordinated regulation by different hormones.
Acknowledgments
This work was supported by the Australian Research Council's Discovery project funding scheme (DP0878194). Y.H.W. was supported by an Australian Postgraduate Award.
Abbreviations
- ABA
abscisic acid
- ACC
1-aminocyclopropane-1-carboxylic acid
- cADPR
cyclic ADP-ribose
- ARF
auxin responsive factor
- AuxRE
auxin responsive element
- CLV3
CLAVATA3
- DAG
1,2-diacylglycerol
- GA
gibberellin
- IP3
inositol 1,4,5-trisphosphate
- LRR RLK
leucine rich repeat receptor like kinase
- MAPK
mitogen activated protein kinase
- PNP
plant natriuretic peptide
- PSK
phytosulfokine
- RALF
rapid alkalization factor
- RLK
receptor like kinase
- SCF
Skp1/Cullin/F-box
- SCP
S-locus cysteine-rich protein
References
- 1.Arteca RN. Plant growth substances: principles and applications. New York: Chapman and Hall; 1996. [Google Scholar]
- 2.Norman AW, Litwack G. Hormones. New York: Academic Press; 1997. [Google Scholar]
- 3.Esbenshade TA, Duzic E. Current Protocols in Pharmacology. John Wiley & Sons, Inc; 2005. [Google Scholar]
- 4.Bahyrycz A, Konopinska D. Plant signalling pep-tides: Some recent developments. J Peptide Sci. 2007;13:787–797. doi: 10.1002/psc.915. [DOI] [PubMed] [Google Scholar]
- 5.Stacey G, Koh S, Granger C, Becker JM. Peptide transport in plants. Trends Plant Sci. 2002;7:257–263. doi: 10.1016/s1360-1385(02)02249-5. [DOI] [PubMed] [Google Scholar]
- 6.Wheeler JI, Irving HR. Evolutionary advantages of secreted peptide signalling molecules in plants. Funct Plant Biol. 2010;37:382–394. [Google Scholar]
- 7.Xie X, Yoneyama K, Yoneyama K. The strigolactone story. Ann Rev Phytopath. 2010;48:93–117. doi: 10.1146/annurev-phyto-073009-114453. [DOI] [PubMed] [Google Scholar]
- 8.Kepinski S, Leyser O. Ubiquitination and auxin signaling: a degrading story. Plant Cell. 2002:81–95. doi: 10.1105/tpc.010447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulaeva ON, Prokoptseva OS. Recent advances in the study of mechanisms of action of phytohormones. Biochemistry-Moscow. 2004;69:233–247. doi: 10.1023/b:biry.0000022053.73461.cd. [DOI] [PubMed] [Google Scholar]
- 10.Koshioka M, Nishijima T, Yamazaki H, Liu Y, Nonaka M, Mander LN. Analysis of gibberellins in growing fruits of Lycopersicon esculentum after pollination or treatment with 4-chlorophenoxyacetic acid. J Hort Sci. 1994;69:171–179. [Google Scholar]
- 11.Nitsch LMC, Oplaat C, Feron R, Ma Q, Wolters-Arts M, Hedden P, et al. Abscisic acid levels in tomato ovaries are regulated by LeNCED1 and SICYP707A1. Planta. 2009;229:1335–1346. doi: 10.1007/s00425-009-0913-7. [DOI] [PubMed] [Google Scholar]
- 12.Kim GT, Tsukaya H. Regulation of the biosynthesis of plant hormones by cytochrome P450s. J Plant Res. 2002;115:169–177. doi: 10.1007/s102650200022. [DOI] [PubMed] [Google Scholar]
- 13.Paciorek T, Friml J. Auxin signaling. J Cell Sci. 2006;119:1199–1202. doi: 10.1242/jcs.02910. [DOI] [PubMed] [Google Scholar]
- 14.Santner A, Estelle M. Recent advances and emerging trends in plant hormone signalling. Nature. 2009;459:1071–1078. doi: 10.1038/nature08122. [DOI] [PubMed] [Google Scholar]
- 15.Viestra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nature Rev Mol Cell Biol. 2009;10:385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- 16.Dreher K, Callis J. Ubiquitin, hormones and biotic stress in plants. Ann Bot. 2007;99:787–822. doi: 10.1093/aob/mcl255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heyl A, Werner T, Schmulling T. Cytokinin metabolism and signal transduction. In: Hedden P, Thomas SG, editors. Plant Hormone Signaling. Oxford, UK: Blackwell Publishing Ltd; 2006. pp. 93–124. [Google Scholar]
- 18.Kudo T, Kiba T, Sakakibara H. Metabolism and long-distance translocation of cytokinins. J Integ Plant Biol. 2010;52:53–60. doi: 10.1111/j.1744-7909.2010.00898.x. [DOI] [PubMed] [Google Scholar]
- 19.Perelli S, Moubayidin L, Sabatini S. The molecular basis of cytokinin function. Curr Opin Plant Biol. 2010;13:21–26. doi: 10.1016/j.pbi.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 20.Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci USA. 2009;106:16529–16534. doi: 10.1073/pnas.0908122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girod PA, Fu H, Zryd JP, Vierstra RD. Multiubiquitin chain binding subunit MCB1 (RPN10) of the 26S proteasome is essential for developmental progression in Physcomitrella patens. Plant Cell. 1999;11:1457–1471. doi: 10.1105/tpc.11.8.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smalle J, Kurepa J, Yang P, Babiychuk E, Kushnir S, Durski A, et al. Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell. 2002;14:17–32. doi: 10.1105/tpc.010381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yammaguchi S. Gibberellin metabolism and its regulation. Ann Rev Plant Biol. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- 24.Dugardeyn J, Vandenbussche F, Van Der Straeten D. To grow or not to grow: What can we learn on ethylene-gibberellin cross-talk by in silico gene expression analysis? J Exp Bot. 2008;59:1–16. doi: 10.1093/jxb/erm349. [DOI] [PubMed] [Google Scholar]
- 25.Thomas SG, Hedden P. Gibberellin metabolism and signal transduction. In: Hedden P, Thomas SG, editors. Plant Hormone Signaling. Oxford, UK: Blackwell Publishing Ltd; 2006. pp. 147–184. [Google Scholar]
- 26.Schwechheimer C, Willige BC. Shedding light on gibberellic acid signalling. Curr Opin Plant Biol. 2009;12:57–62. doi: 10.1016/j.pbi.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 27.Richards DE, King KE, Ait-ali T, Harberd NP. How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signaling. Ann Rev Plant Physiol Plant Mol Biol. 2001;52:67–88. doi: 10.1146/annurev.arplant.52.1.67. [DOI] [PubMed] [Google Scholar]
- 28.Sun TP, Gubler F. Molecular mechanism of gibberellin signaling in plants. Ann Rev Plant Biol. 2004;55:197–223. doi: 10.1146/annurev.arplant.55.031903.141753. [DOI] [PubMed] [Google Scholar]
- 29.Weiss D, Ori N. Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 2007;144:1240–1246. doi: 10.1104/pp.107.100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoddart JL. Gibberellin receptors. In: Chadwick CM, Garrod DR, editors. Hormones, Receptors and Cellular Interactions in Plants. New York: Cambridge University Press; 1986. pp. 91–114. [Google Scholar]
- 31.O'Neill DP, Davidson SE, Clarke VC, Yamauchi Y, Yamaguchi S, Kamiya Y, et al. Regulation of the gibberellin pathway by auxin and DELLA proteins. Planta. 2010;232:1141–1149. doi: 10.1007/s00425-010-1248-0. [DOI] [PubMed] [Google Scholar]
- 32.Grill E, Himmelbach A. ABA signal transduction. Curr Opin Plant Biol. 1998;1:412–418. doi: 10.1016/s1369-5266(98)80265-3. [DOI] [PubMed] [Google Scholar]
- 33.Wasilewska A, Vlad F, Sirichandra C, Redko Y, Jammes F, Valon C, et al. An update on abscisic acid signaling in plants and more…. Mol Plant. 2008;1:198–217. doi: 10.1093/mp/ssm022. [DOI] [PubMed] [Google Scholar]
- 34.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: Emergence of a core signaling network. Ann Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 35.Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 2010;24:1695–1708. doi: 10.1101/gad.1953910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim TH, Böhmer M, Hu H, Nishimura N, Schroeder JI. Guard cell signal transduction network: advances in understanding abscisic acid, CO2 and Ca2+ signaling. Ann Rev Plant Biol. 2010;61:561–591. doi: 10.1146/annurev-arplant-042809-112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klinger JP, Batelli G, Zhu JK. ABA receptors: The START of a new paradigm in phytohormone signalling. J Exp Bot. 2010;61:3199–3210. doi: 10.1093/jxb/erq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCourt P, Creelman R. The ABA receptors: we report you decide. Curr Opin Plant Biol. 2008;11:474–478. doi: 10.1016/j.pbi.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 39.Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, et al. Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 2010;51:1821–1839. doi: 10.1093/pcp/pcq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas TL, Chung HJ, Nunberg AN. ABA signaling in plant development and growth. In: Aducci P, editor. Signal Transduction in Plants. Berlin: Birkhauser Verlag; 1997. pp. 23–44. [Google Scholar]
- 41.Jia WS, Xing Y, Lu CM, Zhang JH. Signal transduction from water stress perception to ABA accumulation. Acta Botanica Sinica. 2002;44:1135–1141. [Google Scholar]
- 42.Leung J, Giraudat J. Abscisic acid signal transduction. Ann Rev Plant Physiol Plant Mol Biol. 1998;49:199–222. doi: 10.1146/annurev.arplant.49.1.199. [DOI] [PubMed] [Google Scholar]
- 43.Zhang YY, Xie Q. Ubiquitination in abscisic acid-related pathway. J Integr Plant Biol. 2007;49:87–93. [Google Scholar]
- 44.Guo H, Ecker JR. The ethylene signaling pathway: new insights. Curr Opin Plant Biol. 2004;7:40–49. doi: 10.1016/j.pbi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 45.Chen YF, Etheridge N, Schaller GE. Ethylene signal transduction. Ann Bot. 2005;95:901–915. doi: 10.1093/aob/mci100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Belkhadir Y, Chory J. Brassinosteroid signaling: a paradigm for steroid hormone signaling from the cell surface. Science. 2006;314:1410. doi: 10.1126/science.1134040. [DOI] [PubMed] [Google Scholar]
- 47.Szekeres M, Bishop GJ. Integration of brassinosteroid biosynthesis and signaling. In: Hedden P, Thomas SG, editors. Plant Hormone Signaling. Oxford, UK: Blackwell Publishing Ltd; 2006. pp. 67–92. [Google Scholar]
- 48.Karlova R, de Vries SC. Advances in understanding brassinosteroid signaling. Science's STKE. 2006:36. doi: 10.1126/stke.3542006pe36. [DOI] [PubMed] [Google Scholar]
- 49.Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J, et al. Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell. 2005;17:1685–1703. doi: 10.1105/tpc.105.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, et al. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15:220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 51.Szekeres M. Brassinosteroid and systemin: two hormones perceived by the same receptor. Trends Plant Sci. 2003;8:102–104. doi: 10.1016/S1360-1385(03)00010-4. [DOI] [PubMed] [Google Scholar]
- 52.Wasternack C. Oxylipins: biosynthesis, signal transduction and action. In: Thomas SG, Hedden P, editors. Plant Hormone Signaling. Oxford, UK: Blackwell Publishing Ltd; 2006. pp. 185–228. [Google Scholar]
- 53.Liechti R, Gfeller A, Farmer EE. Jasmonate signaling pathway. Science's STKE. 2006:2. doi: 10.1126/stke.3222006cm2. [DOI] [PubMed] [Google Scholar]
- 54.Farmer EE. Plant biology: Jasmonate perception machines. Nature. 2007;448:659–660. doi: 10.1038/448659a. [DOI] [PubMed] [Google Scholar]
- 55.Chen H, Jones AD, Howe GA. Constitutive activation of the jasmonate signaling pathway enhances the production of secondary metabolites in tomato. FEBS Lett. 2006;580:2540–2546. doi: 10.1016/j.febslet.2006.03.070. [DOI] [PubMed] [Google Scholar]
- 56.Kumar D, Klessig DF. High-affinity salicylic acid-binding protein 2 is required for plant innate immunity and has salicylic acid-stimulated lipase activity. Proc Natl Acad Sci USA. 2003;100:16101–16106. doi: 10.1073/pnas.0307162100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klessig DF, Durner J, Noad R, Navarre DA, Wendehenne D, Kumar D, et al. Nitric oxide and salicylic acid signaling in plant defense. Proc Natl Acad Sci USA. 2000;97:8849–8855. doi: 10.1073/pnas.97.16.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcion C, Metraux JP. Salicylic acid. In: Thomas SG, Hedden P, editors. Plant Hormone Signaling. Oxford, UK: Blackwell Publishing Ltd; 2006. pp. 229–256. [Google Scholar]
- 59.Ohtake Y, Takahashi T, Komeda Y. Salicylic acid induces the expression of a number of receptor-like kinase genes in Arabidopsis thaliana. Plant Cell Physiol. 2000;41:1038–1044. doi: 10.1093/pcp/pcd028. [DOI] [PubMed] [Google Scholar]
- 60.Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Ann Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 61.Schilmiller AL, Howe GA. Systemic signaling in the wound response. Curr Opin Plant Biol. 2005;8:369–377. doi: 10.1016/j.pbi.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 62.Chilley PM, Casson SA, Tarkowski P, Hawkin N, Wang KL, Hussey PJ, et al. The POLARIS peptide of Arabidopsis regulates auxin transport and root growth via effects on ethylene signaling. Plant Cell. 2006;18:3058–3072. doi: 10.1105/tpc.106.040790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wisniewska J, Trejgell A, Tretyn A. Plant signalling pep-tides. Acta Physiologiae Plantarum. 2003;25:105–122. [Google Scholar]
- 64.Franssen HJ, bisseling T. Peptide signaling in plants. Proc Natl Acad Sci USA. 2001;98:12855–12856. doi: 10.1073/pnas.231490798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kwezi L, Meier S, Mungur L, Ruzvidzo O, Irving H, Gehring C. The Arabidopsis thaliana brassinosteroid receptor (AtBRI1) contains a domain that functions as a guanylyl cyclase in vitro. PLoS one. 2007;2:449. doi: 10.1371/journal.pone.0000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qi Z, Verma R, Gehring C, Yamaguchi Y, Zhao Y, Ryan CA, et al. Ca2+ signaling by plant Arabidopsis thaliana Pep peptides depends on AtPepR1, a receptor with guanylyl cyclase activity and cGMP-activated Ca2+ channels. Proc Natl Acad Sci USA. 2010;107:21193–21198. doi: 10.1073/pnas.1000191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lindsey K. Plant peptide hormones: the long and the short of it. Curr Biol. 2001;11:741–743. doi: 10.1016/s0960-9822(01)00435-3. [DOI] [PubMed] [Google Scholar]
- 68.Matsubayashi Y, Sakagami Y. Peptide hormones in plants. Ann Rev Plant Biol. 2006;57:649–674. doi: 10.1146/annurev.arplant.56.032604.144204. [DOI] [PubMed] [Google Scholar]
- 69.Skirycz A, De Bodt S, Obata T, De Clercq I, Claeeys H, De Rycke R, et al. Development stage specificty and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol. 2010;152:226–244. doi: 10.1104/pp.109.148965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Verelst W, Skirycz A, Inzé D. Abscisic acid, ethylene and gibberellic acid act at different development stages to instuct the adapatation of young leaves to stress. Plant Sig Behav. 2010;5:473–475. doi: 10.4161/psb.5.4.11421. [DOI] [PMC free article] [PubMed] [Google Scholar]


