Abstract
The enzymes called lipoxygenases (LOXs) can dioxygenate unsaturated fatty acids, which leads to lipoperoxidation of biological membranes. This process causes synthesis of signaling molecules and also leads to changes in cellular metabolism. LOXs are known to be involved in apoptotic (programmed cell death) pathway, and biotic and abiotic stress responses in plants. Here, the members of LOX gene family in Arabidopsis and rice are identified. The Arabidopsis and rice genomes encode 6 and 14 LOX proteins, respectively, and interestingly, with more LOX genes in rice. The rice LOXs are validated based on protein alignment studies. This is the first report wherein LOXs are identified in rice which may allow better understanding the initiation, progression and effects of apoptosis, and responses to bitoic and abiotic stresses and signaling cascades in plants.
Key words: apoptosis, biotic and abiotic stresses, genomics, jasmonic acid, lipids
Lipoxygenases (linoleate:oxygen oxidoreductase, EC 1.13.11.-; LOXs) catalyze the conversion of polyunsaturated fatty acids (lipids) into conjugated hydroperoxides. This process is called hydroperoxidation of lipids. LOXs are monomeric, non-heme and non-sulfur, but iron-containing dioxygenases widely expressed in fungi, animal and plant cells, and are known to be absent in prokaryotes. However, a recent finding suggests the existence of LOX-related genomic sequences in bacteria but not in archaea.1 The inflammatory conditions in mammals like bronchial asthama, psoriasis and arthritis are a result of LOXs reactions.2 Further, several clinical conditions like HIV-1 infection,3 disease of kidneys due to the activation of 5-lipoxygenase,4,5 aging of the brain due to neuronal 5-lipoxygenase6 and atherosclerosis7 are mediated by LOXs. In plants, LOXs are involved in response to biotic and abiotic stresses.8 They are involved in germination9 and also in traumatin and jasmonic acid biochemical pathways.10,11 Studies on LOX in rice are conducted to develop novel strategies against insect pests12 in response to wounding and insect attack,13 and on rice bran extracts as functional foods and dietary supplements for control of inflammation and joint health.14 In Arabidopsis, LOXs are studied in response to natural and stress-induced senescence,15 transition to flowering,16 regulation of lateral root development and defense response.17
The arachidonic, linoleic and linolenic acids can act as substrates for different LOX isozymes. A hydroperoxy group is added at carbons 5, 12 or 15, when arachidonic acid is the substrate, and so the LOXs are designated as 5-, 12- or 15-lipoxygenases. Sequences are available in the database for plant lipoxygenases (EC:1.13.11.12), mammalian arachidonate 5-lipoxygenase (EC:1.13.11.34), mammalian arachidonate 12-lipoxygenase (EC:1.13.11.31) and mammalian erythroid cell-specific 15-lipoxygenase (EC:1.13.11.33). The prototype member for LOX family, LOX-1 of Glycine max L. (soybean) is a 15-lipoxygenase. The LOX isoforms of soybean (LOX-1, LOX-2, LOX-3a and LOX-3b) are the most characterized of plant LOXs.18 In addition, five vegetative LOXs (VLX-A, -B, -C, -D, -E) are detected in soybean leaves.19 The 3-dimensional structure of soybean LOX-1 has been determined.20,21 LOX-1 was shown to be made of two domains, the N-terminal domain-I which forms a β-barrel of 146 residues, and a C-terminal domain-II of bundle of helices of 693 residues21 (Fig. 1). The iron atom was shown to be at the centre of domain-II bound by four coordinating ligands, of which three are histidine residues.22
Figure 1.
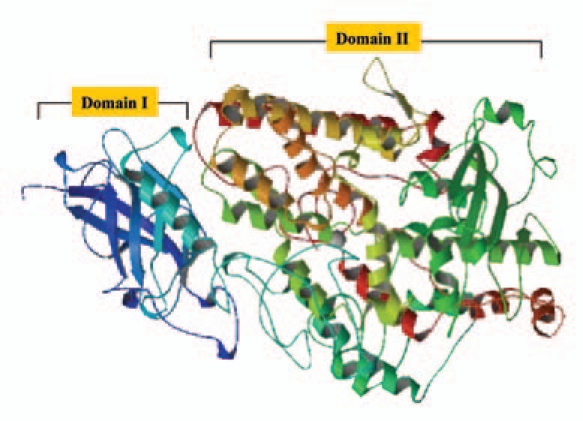
Three-dimensional structure of soybean lipoxygenase L-1. The domain I (N-terminal) and domain II (C-terminal) are indicated. The catalytic iron atom is embedded in domain II (PDB ID-1YGE).21
This article describes identification of LOX genes in Arabidopsis and rice. The Arabidopsis genome encodes for six LOX proteins23 (Table 1). The loci AT1G55020, AT3G45140, AT1G17420 and AT3G22400 are annotated as LOX1, LOX2, LOX3 and LOX5, respectively in the Arabidopsis genome database (www.arabidopsis.org) (Table 1). The loci AT1G67560 and AT1G72520 are annotated as “LOX family protein” and “LOX, putative,” respectively (Table 1). These two loci can be considered as LOX4 (AT1G67560) and LOX6 (AT1G72520) (Table 1). The LOX proteins are composed of ∼850–930 amino acids (aa) in Arabidopsis (Table 1).
Table 1.
Genes encoding lipoxygenases in Arabidopsis thaliana L.
| Locus | Annotation | Nomenclature | A* | B* | C* |
| AT1G55020 | lipoxygenase 1 (LOX1) | LOX1 | 859 | 98044.4 | 5.2049 |
| AT1G17420 | lipoxygenase 3 (LOX3) | LOX3 | 919 | 103725.1 | 8.0117 |
| AT1G67560 | lipoxygenase family protein | LOX4 | 917 | 104514.6 | 8.0035 |
| AT1G72520 | lipoxygenase, putative | LOX6 | 926 | 104813.1 | 7.5213 |
| AT3G22400 | lipoxygenase 5 (LOX5) | LOX5 | 886 | 101058.8 | 6.6033 |
| AT3G45140 | lipoxygenase 2 (LOX2) | LOX2 | 896 | 102044.7 | 5.3177 |
A, amino acids; B, molecular weight; C, isoelectric point.
Interestingly, the rice genome (rice.plantbiology.msu.edu) encodes for 14 LOX proteins as compared to six in Arabidopsis (Tables 1 and 2). Of these, majority of them are composed of ∼790–950 aa with the exception for loci, LOC_Os06g04420 (126 aa), LOC_Os02g19790 (297 aa) and LOC_Os12g37320 (359 aa) (Table 2). Four of the LOXs are annotated to contain a chloroplast precursor (Table 2). The Table 3 indicates homology percent identity/similarity for rice LOXs against Arabidopsis LOX proteins. The best probable hit with Arabidopsis protein database is taken in to consideration while preparing the Table 3. Interestingly, all the rice LOXs showed high level protein similarity with known LOXs from Arabidopsis indicating their novelty at the genomic level (Table 3). The protein alignment shows that LOX sequences are relatively well conserved in rice (Fig. 2).
Table 2.
Genes encoding lipoxygenases in rice
| Chromosome | Locus Id | Putative function | A* | B* | C* |
| 2 | LOC_Os02g10120 | lipoxygenase, putative, expressed | 927 | 103585 | 6.0054 |
| 2 | LOC_Os02g19790 | lipoxygenase 4, putative | 297 | 33031.9 | 10.4799 |
| 3 | LOC_Os03g08220 | lipoxygenase protein, putative, expressed | 919 | 101959 | 7.4252 |
| 3 | LOC_Os03g49260 | lipoxygenase, putative, expressed | 868 | 97984.5 | 6.8832 |
| 3 | LOC_Os03g49380 | lipoxygenase, putative, expressed | 878 | 98697.5 | 7.3416 |
| 3 | LOC_Os03g52860 | lipoxygenase, putative, expressed | 871 | 97183.5 | 6.5956 |
| 4 | LOC_Os04g37430 | lipoxygenase protein, putative, expressed | 798 | 89304.6 | 10.5125 |
| 5 | LOC_Os05g23880 | lipoxygenase, putative, expressed | 848 | 95342.9 | 7.6352 |
| 6 | LOC_Os06g04420 | lipoxygenase 4, putative | 126 | 14054.7 | 6.3516 |
| 8 | LOC_Os08g39840 | lipoxygenase, chloroplast precursor, putative, expressed | 925 | 102819 | 6.2564 |
| 8 | LOC_Os08g39850 | lipoxygenase, chloroplast precursor, putative, expressed | 942 | 104494 | 7.0056 |
| 11 | LOC_Os11g36719 | lipoxygenase, putative, expressed | 869 | 98325.4 | 5.3574 |
| 12 | LOC_Os12g37260 | lipoxygenase 2.1, chloroplast precursor, putative, expressed | 923 | 104687 | 6.2242 |
| 12 | LOC_Os12g37320 | lipoxygenase 2.2, chloroplast precursor, putative, expressed | 359 | 40772.7 | 8.5633 |
A, amino acids; B, molecular weight; C, isoelectric point.
Table 3.
Percent homology of rice lipoxygenases against Arabidopsis
| Loci (Os) | Homolog (At) | Identity/similarity (%) | No. of aa compared |
| LOC_Os02g10120 | LOX2 | 60/76 | 534 |
| LOC_Os02g19790 | LOX5 | 54/65 | 159 |
| LOC_Os03g08220 | LOX3 | 66/79 | 892 |
| LOC_Os03g49260 | LOX5 | 56/73 | 860 |
| LOC_Os03g49380 | LOX5 | 60/75 | 861 |
| LOC_Os03g52860 | LOX1 | 56/72 | 877 |
| LOC_Os04g37430 | LOX3 | 61/75 | 631 |
| LOC_Os05g23880 | LOX5 | 49/66 | 810 |
| LOC_Os06g04420 | LOX5 | 49/62 | 114 |
| LOC_Os08g39840 | LOX2 | 49/67 | 915 |
| LOC_Os08g39850 | LOX2 | 53/70 | 808 |
| LOC_Os11g36719 | LOX5 | 52/67 | 837 |
| LOC_Os12g37260 | LOX2 | 53/67 | 608 |
| LOC_Os12g37320 | LOX2 | 48/60 | 160 |
Os, Oryza sativa L.; At, Arabidopsis thaliana L.; aa, amino acids.
Figure 2.
Protein alignment of rice LOXs and vegetative lipoxygenase, VLX-B,28 a soybean LOX (AA B67732). The 14 rice LOCs are indicated on left and sequence position on right. Gaps are included to improve alignment accuracy. Figure was generated using ClustalX program.
In plants, programmed cell death (PCD) has been linked to different stages of development and senescence, germination and response to cold and salt stresses.24,25 To conclude, this study indicates that rice genome encodes for more LOX proteins as compared to Arabidopsis. The LOX members are not been thoroughly investigated in rice. The more advanced knowledge on LOXs function might spread light on the significant role of LOXs in PCD, biotic and abiotic stress responses in rice.
Note
æLipoxygenase-mediated modification of insect elicitors has been recently proposed.26 The isolation and expression of the LOX gene indicated its vital role in regulating cell death related to flower senescence, and also in leaf response to phloem feeders in the tea plant.27
References
- 1.Ivanov I, Heydeck D, Hofheinz K, Roffeis J, O'Donnell VB, et al. Molecular enzymology of lipoxygenases. Arch Biochem Biophys. 2010;503:161–174. doi: 10.1016/j.abb.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 2.Kühn H, Borngräber S. Mammalian 15-lipoxygenases. Enzymatic properties and biological implications. Adv Exp Med Biol. 1999;447:5–28. [PubMed] [Google Scholar]
- 3.Maccarrone M, Bari M, Corasaniti MT, Nisticó R, Bagetta G, Finazzi-Agrò A. HIV-1 coat glycoprotein gp120 induces apoptosis in rat brain neocortex by deranging the arachidonate cascade in favor of prostanoids. J Neurochem. 2000;75:196–203. doi: 10.1046/j.1471-4159.2000.0750196.x. [DOI] [PubMed] [Google Scholar]
- 4.Maccarrone M, Taccone-Gallucci M, Meloni C, Cococcetta N, Di Villahermosa SM, Casciani CU, et al. Activation of 5-lipoxygenase and related cell membrane lipoperoxidation in hemodialysis patients. J Am Soc Nephrol. 1999;10:1991–1996. doi: 10.1681/ASN.V1091991. [DOI] [PubMed] [Google Scholar]
- 5.Montero A, Badr KF. 15-Lipoxygenase in glomerular inflammation. Exp Nephrol. 2000;8:14–19. doi: 10.1159/000020643. [DOI] [PubMed] [Google Scholar]
- 6.Manev H, Uz T, Sugaya K, Qu T. Putative role of neuronal 5-lipoxygenase in an aging brain. FASEB J. 2000;14:1464–1469. doi: 10.1096/fj.14.10.1464. [DOI] [PubMed] [Google Scholar]
- 7.Cathcart MK, Folcik VA. Lipoxygenases and atherosclerosis: protection versus pathogenesis. Free Radic Biol Med. 2000;28:1726–1734. doi: 10.1016/s0891-5849(00)00230-6. [DOI] [PubMed] [Google Scholar]
- 8.Feussner I, Wasternack C. The lipoxygenase pathway. Annu Rev Plant Biol. 2002;53:275–297. doi: 10.1146/annurev.arplant.53.100301.135248. [DOI] [PubMed] [Google Scholar]
- 9.Melan MA, Enriquez A, Peterman TK. The LOX1 gene of Arabidopsis is temporally and spatially regulated in germinating seedlings. Plant Physiol. 1994;105:385–393. doi: 10.1104/pp.105.1.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardner HW. 9-Hydroxy-traumatin, a new metabolite of the lipoxygenase pathway. Lipids. 1998;33:745–749. doi: 10.1007/s11745-998-0265-z. [DOI] [PubMed] [Google Scholar]
- 11.Gfeller A, Dubugnon L, Liechti R, Farmer EE. Jasmonate biochemical pathway. Sci Signal. 2010;3:3. doi: 10.1126/scisignal.3109cm3. [DOI] [PubMed] [Google Scholar]
- 12.Zhou G, Qi J, Ren N, Cheng J, Erb M, Mao B, et al. Silencing OsHI-LOX makes rice more susceptible to chewing herbivores, but enhances resistance to a phloem feeder. Plant J. 2009;60:638–648. doi: 10.1111/j.1365-313X.2009.03988.x. [DOI] [PubMed] [Google Scholar]
- 13.Wang R, Shen W, Liu L, Jiang L, Liu Y, Su N, et al. A novel lipoxygenase gene from developing rice seeds confers dual position specificity and responds to wounding and insect attack. Plant Mol Biol. 2008;66:401–414. doi: 10.1007/s11103-007-9278-0. [DOI] [PubMed] [Google Scholar]
- 14.Roschek B, Jr, Fink RC, Li D, McMichael M, Tower CM, et al. Pro-inflammatory enzymes, cyclooxygenase 1, cyclooxygenase 2 and 5-lipooxygenase, inhibited by stabilized rice bran extracts. J Med Food. 2009;12:615–623. doi: 10.1089/jmf.2008.0133. [DOI] [PubMed] [Google Scholar]
- 15.Seltmann MA, Stingl NE, Lautenschlaeger JK, Krischke M, Mueller MJ, Berger S. Differential impact of lipoxygenase 2 and jasmonates on natural and stress-induced senescence in Arabidopsis. Plant Physiol. 2010;152:1940–1950. doi: 10.1104/pp.110.153114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bañuelos GR, Argumedo R, Patel K, Ng V, Zhou F, Vellanoweth RL. The developmental transition to flowering in Arabidopsis is associated with an increase in leaf chloroplastic lipoxygenase activity. Plant Sci. 2008;174:366–373. doi: 10.1016/j.plantsci.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vellosillo T, Martínez M, López MA, Vicente J, Cascón T, Dolan L, et al. Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. Plant Cell. 2007;19:831–846. doi: 10.1105/tpc.106.046052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Axelrod B, Cheesbrough TM, Laakso S. Lipoxygenase from soybeans. Methods Enzymol. 1981;71:441–451. [Google Scholar]
- 19.Bunker TW, Koetje DS, Stephenson LC, Creelman RA, Mullet JE, Grimes HD. Sink limitation induces the expression of multiple soybean vegetative lipoxygenase mRNAs while the endogenous jasmone acid level remains low. Plant Cell. 1995;7:1319–1331. doi: 10.1105/tpc.7.8.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyington JC, Gaffney BJ, Amzel LM. The three-dimensional structure of an arachidonic acid 15-lipoxygenase. Science. 1993;260:1482–1486. doi: 10.1126/science.8502991. [DOI] [PubMed] [Google Scholar]
- 21.Minor W, Steczko J, Stec B, Otwinowski Z, Bolin JT, Walter R, et al. Crystal structure of soybean lipoxygenase L-1 at 1.4 Å resolution. Biochemistry. 1996;35:10687–10701. doi: 10.1021/bi960576u. [DOI] [PubMed] [Google Scholar]
- 22.Minor W, Steczko J, Bolin JT, Otwinowski Z, Axelrod B. Crystallographic determination of the active site iron and its ligands in soybean lipoxygenase L-1. Biochemistry. 1993;32:6320–6323. doi: 10.1021/bi00076a003. [DOI] [PubMed] [Google Scholar]
- 23.Bannenberg G, Martínez M, Hamberg M, Castresana C. Diversity of the enzymatic activity in the lipoxygenase gene family of Arabidopsis thaliana. Lipids. 2009;44:85–95. doi: 10.1007/s11745-008-3245-7. [DOI] [PubMed] [Google Scholar]
- 24.Morel JB, Dangl JL. The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 1997;4:671–683. doi: 10.1038/sj.cdd.4400309. [DOI] [PubMed] [Google Scholar]
- 25.Maccarrone M, Melino G, Finazzi-Agrò A. Lipoxygenases and their involvement in programmed cell death. Cell Death Differ. 2001;8:776–784. doi: 10.1038/sj.cdd.4400908. [DOI] [PubMed] [Google Scholar]
- 26.Vandoorn A, Baldwin IT, Bonaventure G. Lipoxygenase-mediated modification of insect elicitors: Generating chemical diversity on the leaf wound surface. Plant Signal Behav. 2010;5:1674–1676. doi: 10.4161/psb.5.12.14036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu S, Han B. Differential expression pattern of an acidic 9/13-lipoxygenase in flower opening and senescence and in leaf response to phloem feeders in the tea plant. BMC Plant Biol. 2010;10:228. doi: 10.1186/1471-2229-10-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Youn B, Sellhorn GE, Mirchel RJ, Gaffney BJ, Grimes HD, Kang C. Crystal structures of vegetative soybean lipoxygenase VLX-B and VLX-D, and comparisons with seed isoforms LOX-1 and LOX-3. Proteins. 2006;65:1008–1020. doi: 10.1002/prot.21182. [DOI] [PMC free article] [PubMed] [Google Scholar]



